 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
With unprecedented development in technology, epigenetics is recognized as a substantial and flexible regulatory pathway for phenotyping. Cytosine methylation and its subsequent oxidization have attracted significant attention due to their direct impact on gene regulation, in association with methyl-CpG-binding domain proteins (MBDs) and transcription related factors. In this study we record the dynamics of DNA demethylation using the recombinant MBD3-GFP protein in living cells under hypoxia and Decitabine treatment using Fluorescence Correlation Spectroscopy (FCS) by monitoring the diffusion dynamics of MBD3. Our study shows a DNA-replication-independent decrease of 5-methylcytosine (5mC)/5-hydroxymethylcytosine (5hmC) under hypoxia vs. a dependent decrease under Decitabine treatment. Further, we define a significantly faster diffusion of MBD3 in the nucleus as a precursory event for active demethylation rather than the Decitabine induced passive demethylation. By monitoring the diffusion of bound and unbound MBD3 in the nucleus we were able to identify and characterize hypoxia-sensitive cells from insensitive/tolerant cells, as well as the respective contribution to active demethylation in a time-dependent manner. Last, we quantitatively describe the concurrent decreasing trend in all of the three oxidized products of 5mC, which points to the potential involvement of ten-eleven-translocation proteins (TETs) in hypoxia induced active demethylation. Overall, for the first time we correlate the dynamic process of DNA demethylation with the biophysical properties of the corresponding DNA binding proteins in live single cells by single molecule spectroscopy.
Introduction
Epigenetic regulation and aberration play pivotal roles in a considerable number of physiological and pathological processes.Citation1 Of the previously elucidated epigenetic mechanisms, DNA modification is one of the most comprehensively investigated, due to its direct effect in bridging the relationship between genotype and phenotype.Citation2 Cytosine derivatives, especially those located in the CpG islands (CGIs) of promoters, exert the main switching function to turn on/off the downstream transcription.Citation3 The 5th carbon position of cytosine can be enzymatically methylated by DNA methyltransferases (DNMTs) with S-adenosyl methionine (SAM) as the methyl-donor.Citation4-Citation6 Once methylated, the originally active chromatin site will become compacted and inaccessible to the transcription initiation complex. However, the epigenetically silenced CGIs are subject to reactivation by demethylation.
In mammalian cells, the methylated cytosine (5mC) is further oxidized to hydroxy- (5hmC), formyl- (5fC), and carboxyl- (5caC) derivatives by ten-eleven-translocation proteins (TETs), and the final 5fC/5caC could be excised and replenished by thymine-DNA glycosylase (TDG).Citation7-Citation9 This unidirectional cytosine conversion chain should be under rigorous control to maintain normal embryogenesis, development and differentiation in the right order (). In most cases, our knowledge of epigenetic regulation primarily comes from ensemble biochemical approaches, either through quantification of specific makers or genomic mapping by DNA-sequencing technologies.Citation10-Citation13 Studies elucidating the sequence of events governing the intermediate epigenetic process are sparse. Advanced fluorescence microscopy and spectroscopy have the ability to provide exquisite real-time account of events in the nucleus of live single cells down to single molecule resolution.Citation14-Citation19 In this effort, we use the recombinant green fluorescent protein tagged methyl-CpG-binding domain protein 3 (MBD3-GFP) and fluorescence correlation spectroscopy (FCS) to quantitatively describe the biophysical relationship between the diffusion dynamics of MBD3 and DNA demethylation patterns in live single cells.
Figure 1. Cytosine derivatives and demethylation patterns. (A) The entire cytosine conversion cycle comprising of 5mC/5hmC/5fC/5caC, and final base excision repair (BER) are serially catalyzed by DNMTs, TETs and TDG enzymes. Although other dehydroxymethylation pathways have been proposed (deaminating 5hmC to 5hmU and then through BER by TDG/SMUG), TETs mediated oxidation is the only biochemically confirmed in mammalian cells by in vivo experiments. (B) Passive demethylation (induced by Decitabine) and active demethylation (induced by hypoxia) with the involvement of MBD3. The incorporation of Decitabine molecule depends on DNA replication and the incorporation of this analog accelerates the passive dilution of cytosine modifications, since its 5th nitrogen is not an accessible DNMTs substrate. On the other hand, the local physical and chemical environment (e.g., pH, ion concentration, ROS molecule etc.) or protein-DNA conformation, under hypoxic stress, would experience considerable changes to weaken the binding affinity between MBD3 and DNA, or even cause MBD3 detachment. The decrease in steric hindrance between MBD3 and the binding site could facilitate the involvement of TETs in further demethylation, which is DNA replication independent.
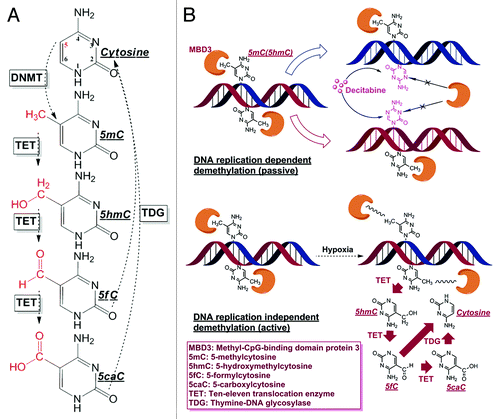
Global DNA hypomethylation is one representative epigenetic anomaly in cancer, and the consequent genome instability acts as a key contributor to tumor progression and metastasis.Citation20-Citation24 Yet, still unclear is the triggering mechanism of cancerous demethylation, which has been a stumbling block to an in-depth elucidation of the malignant behavior and better medical management. Interestingly, loss of 5hmC has also been recognized as an epigenetic hallmark of some cancers.Citation25-Citation27 As the direct product of 5mC oxidization by the TETs, 5hmC, constitutes only approximately 1% of 5mC and is in the midst of the demethylation pathway,Citation28,Citation29 it has been postulated that it is highly possible that cancer hypomethylation could be accompanied by hypohydroxymethylation or even further cytosine oxidization by the TETs. Theoretically, DNA demethylation can be categorized as a passive or active process (). For passive demethylation, the reduction of 5mC is achieved through DNA replication dependent dilution, such as hemimethylation induced by Decitabine, which is an incorporable cytosine analog but cannot be modified at its 5th nitrogen.Citation30-Citation32 For active demethylation to proceed, the pre-existing 5mC would be excessively erased by the TETs catalyzed conversion, probably until the completion of base excision repair (BER) after the formation of 5fC/5caC. Methods available to date cannot easily and simultaneously quantify 5hmC/5fC/5caC in cancer cells, hence detailed investigation of the demethylation process has not been extensive.Citation33 In addition to developing single molecule tools for exploring the dynamics of MBD3, we also have developed ELISA-based methods to quantify the various epigenetic markers at a limit-of-detection (LOD) not possible using the current commercialized kits (Fig. S3).
A hypoxic microenvironment is typical of most solid cancers.Citation34-Citation36 To some extent, low oxygen tension is critical for cancer stem cell maintenance and therefore responsible for fatal tumor progression and reoccurrence.Citation37,Citation38 A past study has shown that culturing of mouse and human fibroblasts under hypoxia can improve the reprogramming efficiency of the induced pluripotent stem cell (iPS).Citation39 It has also been reported that stem cells possess low level of global methylation compared with terminally differentiated cells.Citation40,Citation41 Experiments have shown that hypoxia can evoke a demethylation response in some cancer cells/tissues.Citation42-Citation44 In other words, hypoxia is highly related to hypomethylation and may necessarily be active in maintaining low levels of DNA modifications required for stemness or tumorigenesis.
MBD3 has been recognized as one of the few known proteins capable of binding to the genomic sites of 5hmC and has a negative correlation to the recruitment of RNA polymerase-II.Citation45 However, latest research utilizing quantitative mass-spectrometry-based proteomics did not support such specific interaction between MBD3 and 5hmC, but observed a more specific binding between the recombinant MBD3 protein and 5mC in vitro.Citation46 While the exact in vivo distribution and function of MBD3 are still elusive and require further studies, it is still rational to hypothesize that the spatial positioning of MBD3 would prevent the oxidation of either 5mC or 5hmC due to a possible steric hindrance. For active demethylation to progress, the MBD3 “cap” has to be removed so that the methylated site can be exposed, while the already bound MBD3 will not experience any detachment during passive demethylation. Biophysically, MBD3 proteins can be construed to have distinct diffusion properties in terms of mobility based on whether it is found in the bound or unbound state. Thus, precise determination of the diffusion properties of MBD3 would facilitate the differentiation of active from passive demethylation, and extend the diagnosis window to report on the initiation of a condition or abnormality by monitoring MBD3 diffusion even before an active demethylation event.
However, limited tools exist to capture in real-time the mobility of the targeted cellular or specific nuclear components. Due to the significant progress in microscopy and spectroscopy, single molecule tools have been shown to detect epigenetic modifications in single cells.Citation47 We will advance these methods to quantitatively describe the movement of selectively labeled molecules in live single cell nucleus. The confocal alignment of FCS allows the recording of FCS data in a volume < 1 femtoliter at laser power as low as 1 μW within minutes at physiological conditions not accessible by other methods.Citation48-Citation50 GFP fused to targets of interest in cells has become the norm to examine intracellular interactions of the target with other components.Citation51,Citation52 To obtain quantitative single molecule diffusion measurements by FCS monitoring, low levels of fluorescent targets in live cells is a prerequisite (optimal measurement range is from 100 pM to 200 nM) because a high concentration of fluorescing molecules in the detection volume will not produce useful diffusion information due to the averaging out of fluorescence fluctuations.Citation53 Our study will be the first to show the diffusion dynamics of DNA demethylation associated with MBD3 at single molecule precision. Further, compared with other MBDs such as MeCP2 and MBD1 etc., it has been reported that MBD3 possesses the lowest binding affinity with cytosine derivatives, and hence a likely candidate to be easily detached from the DNA binding site.Citation54 The single molecule resolution of FCS and the sensitivity of MBD3 make it an ideal candidate for monitoring the diffusion dynamics and exploring its association with demethylation events. With low transfection of the pGFP-MBD3 plasmid DNA we were able to obtain 100–150 nM (average 120 nM) of expression of functional pGFP-MBD3 and characterize its diffusion within the confocal volume using appropriate diffusion models.Citation14,Citation15
In our study, active or passive demethylation was induced in Hela cells by the respective hypoxia or Decitabine treatment and the demethylation products were biochemically confirmed by a modified ELISA immunoassay approach. Further, live cell FCS experiments were conducted to correlate the hypoxia induced active demethylation with the labile diffusion states of MBD3 protein. The methodology presented in this report provides a unique strategy to unravel the transitional biological events and to expand our understanding of epigenetic processes through live single cell studies.
Results
Experimentally induced hypomethylation and hypohydroxymethylation
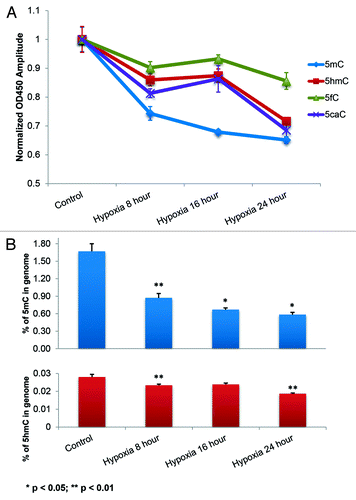
Optimized colorimetric ELISA was developed to quantify 5mC and 5hmC after 3 μM Decitabine treatment or hypoxia induced by 1 mM sodium hydroxysulfite (). Time points of 1 d and 8 h, were respectively selected to extract DNA from cells exposed to these two conditions. Theoretically, Decitabine demands a time equivalent of at least one cell cycle (16 h for Hela cellsCitation55) to take effect since DNA replication is required for this analog incorporation, while exposure to a hypoxic microenvironment has the ability to rapidly decrease the 5mC and 5hmC contents independent of cell proliferation. The two treatments had a similar effect on the reduction of 5hmC, although treatment with Decitabine resulted in a greater loss of 5mC within the selected durations.
Figure 2. Reductions of 5mC and 5hmC induced under different conditions. (A) Improved ELISA-based immunoassay quantified the genomic contents of 5mC and 5hmC. Significant decrease in both the cytosine derivatives can be noted within the Decitabine and Hypoxia treated groups. Each data column was averaged from triplicate measurements and the experiments were replicated another two times (data not shown). (B) Immunofluorescence staining of 5mC and 5hmC further confirmed the demethylation effect in 4% paraformaldehyde fixed cells. ImageJ 3D plugin was used to visualize the signal intensity in arbitrary units based on the 8-bit intensity scale (0–255). Image scale bar: 10 μm.
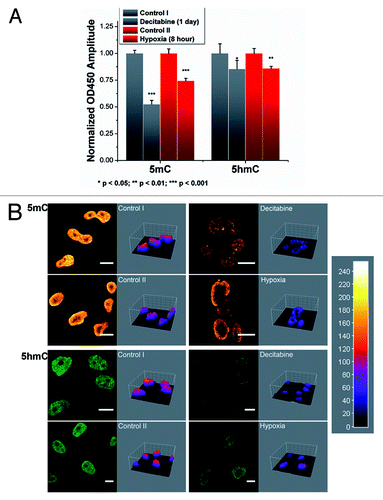
Immunofluorescence staining of 5mC and 5hmC were used to further confirm the observation from ELISA (). To ensure comparable fluorescence intensity, the power from pulsed diode laser was precisely controlled via a Sepia PDL 808 driver whose resolution can reach sub-μW level. Collected images were further visualized by ImageJ 3D plugin. Representative images show drastic loss of 5mC and significant decrease of 5hmC under both treatments. Within control groups, the levels of 5mC and 5hmC were consistent with respect to the time lapse (control I for 1 d and control II for 8 h). While under hypoxic stress, the cellular content of 5mC and 5hmC showed considerable variability in terms of intensity profile, especially for 5mC (Fig. S4B).
Diffusion kinetics of MBD3-GFP and its role in active and passive demethylation
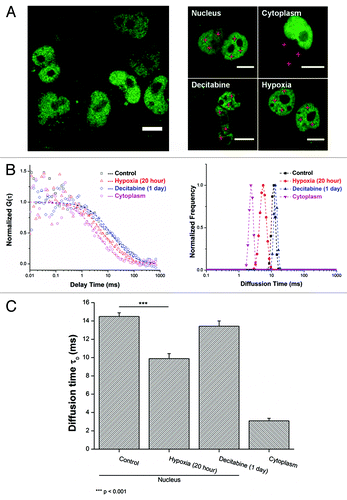
Since FCS observation has restricted concentration requirement (maximum of around 400 nM), optimization for the amount of plasmid DNA was necessary to achieve successful transfection efficiency. For the 12-well culture plate as low as 100 ng of DNA per well provided a satisfactory expression (). The basic diffusion characteristics of MBD3 in the nucleus and in the cytoplasm were then obtained. Distinct diffusion times: 14.49 ± 0.40 ms (slower diffusion of mobility) in the nucleus and 3.10 ± 0.27 ms (relatively faster diffusion of mobility) in the cytoplasm () were noted. The significantly slower diffusion (characterized by the longer diffusion time) in the nucleus, as expected, can be attributed to the specific binding characteristics between MBD3 and the genomic substrate, implying that the transfected MBD3-GFP was functional. Interestingly the raw autocorrelation function (ACF) could be well fitted by a single-component 3D diffusion model after 24 h of transfection (, left panel) or an anomalous diffusion model with “α”-value greater than 0.8 (Fig. S2C). The unique and pure diffusion component in the nucleus suggests that this group of MBD3 was not redundant and could bind to DNA at a steady-state without complex interactions, which complements the final FCS interpretation; otherwise, the unbound protein and crowded environment might necessitate a multi-component or anomalous diffusion fitting using more complex mathematical models, although in the initial 16 h of transfection two components that characterize MBD3 diffusion as free and bound (Fig. S2B) were observed. Next, the diffusion dynamics was monitored under Decitabine and hypoxia treatment. Consistent with the hypothesis, the diffusion time of MBD3 was the same (13.43 ± 0.57 ms) after treatment with Decitabine since it had no effect on the already bound MBD3. However, in hypoxic cells, where ongoing active demethylation requires the detachment of MBD3, a remarkably faster diffusion time of 9.89 ± 0.54 ms was noted. MEMFCS fitting further confirmed the single-component diffusion fitting differences (, right panel).
Figure 3. MBD3 tends to detach from DNA substrate and diffuse more freely under active demethylation, but retains its binding affinity with DNA under passive demethylation as concluded from FCS measurement and mathematical fitting. (A) Transfection of a low amount of pGFP-MBD3 recombinant protein and representative FCS data collection at different points (red crosses) from different cellular compartments and treatment conditions. For FCS, data from each cell was measured 2–3 times and averaged. (B) Distinguishable autocorrelation curves fitted by single-component diffusion model (left panel) indicated free MBD3 diffusion in the cytoplasm but restricted diffusion in the nucleus, and faster intranuclear diffusion after hypoxia treatment rather than Decitabine treatment, all of which were simultaneously analyzed by MEMFCS algorithm fitting (right panel). (C) Summarized statistics of diffusion time. Raw data were collected from more than 15 cells for each set. Mean value is presented along with the SEM. Statistical analysis was conducted with the Student t–test by Origin8.5 software package.
Together, the set of designed experiments enabled us to suggest a MBD3-based DNA demethylation surveillance platform in living cells. By comparing the dynamic properties under different scenarios, we were able to verify the attached and detached state of MBD3 in relation to cytosine derivatives. FCS can thus prove to be a highly sensitive tool and instrumental in the acquisition of highly resolved information to distinguish active from passive demethylation.
Time-course change of MBD3 diffusion time under active demethylation
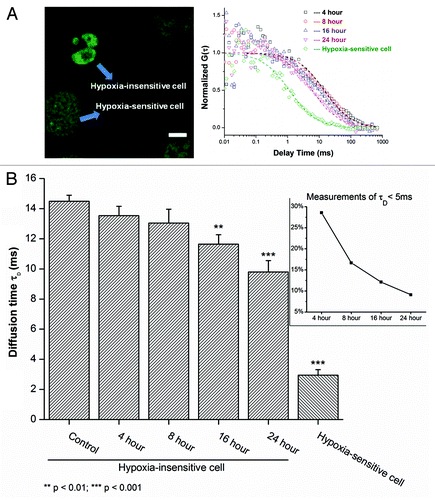
Following the assessment of active demethylation by monitoring the diffusion of MBD3 by FCS, further detailed experiments were performed to clarify this observation. In the subsequent experiments, FCS data were separately obtained from cells under hypoxia at the 4th, 8th, 16th, and 24th hour. Surprisingly, in the first 4 h a heterogeneous response in the diffusion time of MBD3 was observed. Two distinct diffusing species were observed: MBD3 in hypoxia-sensitive and in hypoxia-insensitive cells (). The diffusion time of MBD3 in hypoxia-insensitive cells was noted to be ~14 ms, while a number of nuclear measurements displayed a much faster diffusion time of 2.95 ± 0.36 ms, which is nearly the same as the diffusion of MBD3 in the cytoplasm, noting that the nucleoplasm shares a similar hydrodynamic viscosity with cytoplasm.Citation56 From this, we deduce that MBD3 thoroughly dissociated from the DNA binding sites in hypoxia-sensitive cells. Visualization of these hypoxia-sensitive cells revealed a cellular morphology akin to cells undergoing apoptosis or necrosis. Propidium iodide (PI) uptake experiments after 4 h of hypoxia (Fig. S4) confirmed that about one-third of cells were stained by PI and these cells exhibited a morphology characteristic of cells where the MBD3 diffusion was observed as the free-form (i.e., unbound). Taken together, we deduce that the active detachment of MBD3 is representative of an early demethylation event that could result in the death of hypoxia-sensitive (demethylation-sensitive) cells due to acute hypoxia within the first 4–8 h. During the later period, the measurements of freely diffusing MBD3 as well as the cells with abnormal morphology significantly decreased from approximately 30% to 10% ( inset), implying the eventual elimination of severely damaged cells. However, the dynamics of MBD3 diffusion in hypoxia resistant cells tended to increase after approximately 16 h of hypoxia and the diffusion time gradually decreased to 9.80 ± 0.75 ms (implying faster diffusion), consistent with our previous findings ( and ). Although during the 24 h observation window of hypoxia, MBD3 in hypoxia-insensitive cells did not seem to completely detach from the DNA binding site compared with the hypoxia-sensitive cells but exhibited a gradual increase in average diffusion rate. This is indicative of a subsequent weakening of the binding affinity and an initiation of the detaching process, which may facilitate further enzymatic catalysis.
Figure 4. Time-course response of MBD3 under hypoxic conditions in sensitive and insensitive cells. (A) The mobility of MBD3 protein could facilitate recognizing a heterogeneous cellular response to hypoxia (left panel) with distinct change in diffusion patterns (right panel). (B) In the first 8 h of hypoxia a considerable number of hypoxia-sensitive cells experiencing significant demethylation were noted as reflected by the detachment of MBD3 from the DNA (τD < 5 ms). After 8 to16 h, the proportion of sensitive cells and the corresponding τD counts decreased from about 30% to 10% (inset panel). The gradually accelerated mobility of MBD3 in hypoxia-insensitive cells was found to be dominant.
From our observations, it can be inferred that active demethylation under hypoxia gives rise to different diffusion conditions based on the cell susceptibility. The hypoxia-sensitive cells can dominantly account for an early ensemble loss of 5mC/5hmC and the insensitive cells could contribute to the long-term hypomethylation. This conclusion hinted us to further ask the question: whether active demethylation terminated at 5hmC. Thus, it was worthwhile to assess the amount of 5fC and 5caC in hypoxic environment to postulate the potential TET-mediated active demethylation.
Altering patterns of 5hmC, 5fC, and 5caC under active demethylation share a nearly identical trend
To study this, the three cytosine derivatives (5hmC, 5fC, and 5caC), along with 5mC, were quantified by a newly developed ELISA approach. A decrease in 5mC and 5hmC with hypoxia, consistent with the prediction of MBD3 diffusion under active demethylation, was noted. The demethylation peaked within the first 4 h of hypoxia and may be attributed to the massive demethylation of hypoxia-sensitive cells. But this trend did not last long due to the excessive DNA damage and cell death reflected by the following slower loss. Moreover, 5fC and 5caC showed a concurrent reduction pattern with 5hmC (). The stacked curves indirectly hinted that active demethylation further oxidized 5hmC to 5fC and 5caC, possibly by an abnormally high activity of the TETs. Even absolute quantification of 5hmC is difficult, the biotin-streptavidin coupled signal amplification strategy is capable of detecting as low as ~0.02% of genomic 5hmC in Hela cells and is consistent with the previously reported data (),Citation12 which indirectly validates our method. The 8–16 h plateau stage depicting a concurrent change in trend in the levels of 5hmC/5fC/5caC implied a transition of the contribution to the overall demethylation from sensitive cells to insensitive cells (fraction of hypo- hydroxy/formyl/carboxyl methylated DNA from sensitive cells decreased due to the rapid cell death), consistent with the MBD3 FCS experiment. Notably, the asynchronous activity between TETs and TDG should also contribute to the lagged response in terms of loss of 5hmC/5fC/5caC comparing to 5mC in hypoxia insensitive cells.
Figure 5. Concurrent decrease in 5fC and 5caC with 5hmC due to hypoxia induced active demethylation. (A) Stacked quantitative curves of 5mC, 5hmC, 5fC and 5caC; the simultaneous decrease in 5hmC, 5fC and 5caC is indicative of the TETs mediated active demethylation pathway (). (B) Absolute quantities of 5mC and 5hmC calculated from standard curve (Fig. S2B) indicated two main reduction periods within 24 h: in the first 8 h and from 16 to 24 h. The plateau period from 8 to 16 h, in terms of 5hmC, 5fC and 5caC changes, represents the gradual transition of the contribution of ensemble demethylation from sensitive to insensitive cells. Two-sample t-test was conducted to compare each neighboring time point.
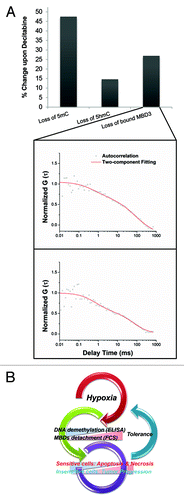
Putative validation of MBD3 detachment as the indicator of demethylation
To validate the credibility of FCS, Hela cells were treated with Decitabine first and then transfected with the redundant MBD3-GFP (1 µg) to validate the relationship between the loss of hydroxyl/methylation and bound MBD3. A significant positive correlation was noted (). As an important component of NuRD/Mi-2 chromatin-remodeling complex,Citation57 the exact nature and genomic binding preferences of MBD3 is still unclear, although several components of NuRD/Mi-2 could bind to chromatin directly. In most biophysical analysis using recombinant MBD3, including ours, the in vivo binding preferences (probably through the help of MBD2 or other proteins) to 5mC should not be overlooked. Since the diffusion statistics reflected by FCS is based on multiple measurements, the overall shift in the average diffusion rate could partially be attributed to the dissociation, providing an opportunity to study the dynamics of methylation and demethylation process in real-time in live cells, not possible with conventional methods.
Figure 6. (A) The putative validation of the main MBD3-GFP in vivo binding target by FCS. Upon Decitabine treatment, redundantly expressed MBD3-GFP showed two diffusing components and the loss of bound-component was positively correlated with the loss of 5mC and 5hmC. (B) Overview of the influence of hypoxia on DNA methylation and fate of tumor cell.
Discussion
Although global DNA hypomethylation has been characterized as one of the typical epigenetic events that result in an abnormal condition, such as cancer, increased emphasis was still placed on the hypermethylation of the promoters of specific tumor suppressor genes.Citation22,Citation58 As the presence of 5hmC and its role in DNA demethylation is being clarified, it has been generally found that the dynamic process of DNA methylation/demethylation interchange plays a pivotal role in stem cell renewal, differentiation and carcinogenesis. This highlights the importance of understanding the DNA demethylation process necessitating live single cell studies.
Tumor proliferation is often accompanied by a rapid increase in the number of cells, demanding an increase in oxygen and other nutrients supplied by the blood. Hypoxia acts as a common tumor microenvironment and gives rise to cells possessing stem cell-like properties. In fact, low oxygen concentration has been found to correlate with more aggressive tumor behavior often resulting in poorer prognosis at least via the upregulation of hypoxia-inducible factors (HIFs) and other related oncogenic proteins through DNA demethylation or epigenetic regulation.Citation59 Current technologies, such as immunocytochemistry (ICC), liquid chromatography-mass spectrometry (LC-MS) and DNA-sequencing strategies can provide either a distribution or quantitative information of specific epigenetic modification, but none is capable of elucidating the dynamic transition process or recording the ongoing epigenetic aberrations.Citation60 Our work clearly shows that cells cultured under hypoxic conditions has distinct demethylation () mechanism (termed as “active”) compared with the Decitabine treated cells (termed as “passive” demethylation).
Taking advantage of single molecule fluorescence techniques, for the first time we lay the groundwork to quantitatively describe the DNA demethylation events characterized by the altered diffusion of GFP labeled MBD3 protein in real-time. Our study is also the first to report on the spatiotemporal landscape of related nuclear proteins relevant to epigenetics in single living cells. Unlike other MBD proteins which have nM level binding affinity even with hemi-methylated CpG, the binding affinity of MBD3 is at the μM level and hence is more sensitive to environmental factors that trigger demethylation via its binding characteristics from the DNA binding site. However, the uncertainty is in the lack of information of the MBD3-DNA binding site in vivo. For the first time, we have shown that FCS, because of its exquisite sensitivity has the ability to distinguish between active and passive demethylation based on the diffusion time of MBD3. Under hypoxic condition, active demethylation is initially characterized by an intense detachment of MBD3 from DNA binding sites in hypoxia-sensitive cells, leading to acute cell apoptosis and necrosis due to extensive loss of DNA methylationCitation61,Citation62 within the first 4–8 h. In hypoxia-insensitive cells the demethylation process is more relaxed as inferred from the gradual increase in mobility (diffusion) of MBD3. Admittedly, under high hypoxic stress conditions, active demethylation and the subsequently triggered death-pathway are among a myriad of factors that contribute to cell death, in addition to playing a critical role in tumor progression (). Previous studies have shown that depletion of MBD3 can cause engineered mice to die early during embryogenesis, further signifying the role of MBD3 in balancing DNA methylation.Citation63 To maintain cell function, both 5mC and 5hmC needs to be regulated at tolerable levels, partially accomplished by the interaction with MBD3Citation45,Citation64 at the relevant DNA binding sites. Future work will focus on obtaining a systematic map of the distribution of MBD proteins by high-throughput sequencing to determine the effect of MBD proteins (e.g., MBD1, MBD2, and MBD3 etc.) on the susceptibility of DNA demethylation.
If we assume 5mC as the major starting point of active demethylation, the first oxidized product as understood from the literature is 5hmC. Further removal of 5hmC could occur by two possible pathways: either through the direct deamination by AID or APOBEC enzyme to 5hmU (5-hydroxymethyluracil) followed by TDG abasic repair; or through the TETs mediated oxidation pathways (). Empirically, our DNA quantification studies support the latter pathway due to the concurrent reduction of 5hmC with 5fC and 5caC (). However the specific TET protein or uncharacterized enzymes that might be involved in active demethylation under hypoxia remain unclear. TETs belong to the group of Fe2+- and α-ketoglutarate-dependent dioxygenases. TET3 plays an important role in early embryogenesis and paternal genome demethylation; TET2 mutations and dysfunction has been found to be closely related with hematopoietic malignancies; TET1 has been shown to act mostly on germline DNA imprint erasure to give rise to 5hmC.Citation65,Citation66 Identifying the major candidates that catalyze the oxidization of 5hmC to other downstream products (such as 5fC, 5caC, etc.) is critical to furthering our understanding of the sequence of hypomethylation events during malignant cell transformation. Previously, most of the cancerous hypomethylation were found in short and long interspersed nuclear elements (SINEs and LINEs) and other repeat genomic regions, which were long-thought to be the non-coding junk DNA.Citation20,Citation67-Citation71 With the release of the Encyclopedia of DNA Elements (ENCODE) data, nearly 80% of the human genome were found to be useful and could be assigned specific biochemical functions necessitating careful interrogation of the epigenetic state at these regions.Citation72 Considering the fact that the hypomethylated site varies among different cancers, an emerging thought is to study DNA demethylation in a locus-specific manner upon mutagenic stimulation so that targeted therapeutic strategies can be developed depending upon the type and state of the disease.
Materials and Methods
Cell culture and plasmid transfection
Hela cells were routinely cultured in DMEM/F-12 medium (Gibco, 11330) with 10% fetal bovine serum (Atlantic Biologicals, S11050) at 37 °C and a humidified atmosphere containing 5% CO2, 100 IU/ml penicillin and 100 μg/ml streptomycin (Gibco, 15140). pGFP-MBD3 plasmid was a gift from Dr Adrian Bird (Edinburgh, UK).Citation73 The plasmid DNA transfection was conducted by Lipofectamine®LTX&Plus Reagent kit (Invitrogen, 15338-100) according to manufacturer’s instruction with minor modifications. Before transfection, cells were seeded onto sterilized No.1 coverslips (VWR International) and placed in 12-well plate with 2 ml culture medium. After reaching 70–80% confluence the media was removed and replaced with serum-free DMEM/F-12 medium for another 24 h. Then 100 ng plasmid DNA, 2.5 μl LTX reagent and 0.1 μl Plus reagent were mixed into 200 μl Opti-MEM® Reduced-Serum Medium (Gibco, 31985-062) for each well (1 μg plasmid DNA per well was used for redundant expression). Transfection lasted for 24 h before further treatment.
Hypoxia induction and decitabine treatment
For efficient and acute hypoxia induction, 1 mM sodium dithionite (Na2S2O4, Sigma, 157953) was used to chemically consume the dissolved oxygen in the culture medium.Citation74-Citation76 It is reported that Na2S2O4 can lower pO2 to 10 Torr compared with 30 Torr pO2 induced by 100% N2.Citation77 After adding Na2S2O4, the pH of the DMEM/F-12 medium was adjusted to 7.4 and maintained by HEPES buffer considering the acidification potential of Na2S2O4. To sustain long-term hypoxic conditions, the media containing Na2S2O4 was changed every 8 h. 5-Aza-2'-deoxycytidine (Decitabine, Sigma, A3656) was dissolved in DMSO as 2 mM stock solution until use. For treatment, the stock solution was diluted (to 3 μM concentration) using fresh culture medium.
Biotinylation of antibody
Secondary antibodies (Goat anti-mouse IgG for 5mC, Mouse anti-Rabbit IgG for 5hmC, 5fC and 5caC, Pierce) for ELISA in PBS were labeled with 50-fold molar excess of succinimidyl-6-[biotinamido]-6-hexanamidohexanoate (NHS-LC-LC-biotin, Pierce, 21343) dissolved in DMSO. The reaction was performed at room temperature for 1 h and the biotinylated antibody was purified by dialysis and size exclusion chromatography on Sephadex G-15 gel column (gel volume: 10 ml), concentrated and stored at 4 °C until use.
Conjugation streptavidin with horseradish peroxidase (HRP)
Streptavidin (Prozyme, SA10) and HRP dissolved in PBS were coupled with succinimidyl 6-(3[2-pyridyldithio] propionamido) hexanoate (LC-SPDP, Pierce, 21651) and succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxy-(6-amidocaproate) (LC-SMCC, Pierce, 22362) in the ratio 1:50 respectively at room temperature for 1 h. The LC-SPDP was treated with dithiothreitol (DTT, final 10 mM) for activation at room temperature for 30 min. Chemically modified proteins were purified by Sephadex G-15 gel filtration equilibrated in 5 mM EDTA and then immediately mixed and reacted at room temperature for 2 h. This mixture was stored at 4 °C until use.
DNA extraction and ELISA quantification
The standard DNA used was: 5-Methylcytosine and 5-Hyodroxymethylcytosine DNA Standard set (Zymo Research, D5405); 5-Carboxylcytosine DNA Standard Kit (Active Motif, 55014). DNA from Hela cells was extracted and purified with DNeasy Blood and Tissue Kit (Qiagen, 69504), according to manufacturer’s instruction. Purified DNA (50 ng for 5mC, 200 ng for 5hmC, 5fC and 5caC in 100 μl buffer) was added to the polystyrene microwell plate followed by mixing with 100 μl of DNA coating solution (Pierce, 17250) at room temperature for 2 h with gentle agitation. The substrate was then washed with 300 μl of DI water in the microwell. The process was repeated three times. To block residual surfaces, the microwell plate was treated with 0.5% (w/v) casein dissolved in PBS (200 μl) at 37 °C for 1 h. The washing and reaction steps will be adopted in all subsequent steps. For primary antibodies, we used: mouse anti-5-methylcytosine (Eurogentec, BI-MECY-0100), rabbit anti-5-hydroxymethylcytosine (Active Motif, 39791), rabbit anti-5-formylcytosine (Active Motif, 61223), rabbit anti-5-carboxylcytosine (Active Motif, 61225). An amount of 0.5 μg/ml of primary antibody diluted in PBS containing 0.5% casein (100 μl) was added and then 1 μg/ml of biotinylated secondary antibody (100 μl) was reacted with the primary antibody. To form avidin-biotin complex, 0.5 μg/ml of streptavidin-HRP conjugate (100 μl) was finally added into the microwell. For signal generation, colorimetric substrate mixture (50 mM sodium acetate buffer, pH 5.1: 1% (w/v) TMB: 3% (v/v) H2O2 = 1000:10:1 as volume ratio) (200 μl) was reacted and maintained for 15 min at room temperature followed by the application of 50 μl of 2 M sulfuric acid to stop the reaction. The optical intensity was measured using the VersamaxTM absorbance microplate reader (Molecular Device).
Immunofluorescence
Immunostaining of 5hmC was done per the methods in the literature with suitable modification.Citation78 Generally, cells after different treatments were fixed with 4% paraformaldehyde for 15 min at 4 °C and washed 3 times with 1 × PBS buffer (Gibco, 10010). Cell membrane was permeabilized by 0.4% Triton X-100 (Sigma, T8787) for 30 min and washed with PBS for 15 min. Then cells were treated with 1 M HCl for 30 min at 37 °C and washed three times. Cells were blocked by PBS containing 5% goat serum and 0.3% Triton X-100 for 1 h. Primary antibody was 1:1000 diluted in PBS containing 1% BSA and 0.3% Triton X-100 and incubated with cells overnight at 4 °C. 1:1000 diluted Alexa Fluor®488 F(ab')2 fragment of Goat anti-Rabbit IgG (for 5hmC, Invitrogen, A-11070) or Alexa Fluor®546 F(ab')2 fragment of Goat anti-Mouse IgG (for 5mC, Invitrogen, A-11018) was applied for 1 h and the cells were again washed 3 times prior to imaging.
Confocal fluorescence imaging instrumentation
Fluorescence confocal imaging and FCS experiments were performed using a scanning confocal time-resolved microscopy (Microtime 200 from Picoquant GmbH); 465 nm picosecond pulse laser was employed to excite the MBD3-GFP and Alexa488 fluorophore. The laser beam was delivered to the sample through an apochromatic 60×, 1.2 N.A. water immersion objective and the emitted fluorescence was collected using the same objective from which the excitation beam was separated by a dual band dichroic (z467/638rpc, Chroma). A 50 um pinhole was used to reject the off-focus photons from the excitation volume, and the signal beam was subsequently filtered by a specific emission filter (500–540 nm, Chroma) before being detected by a single photon avalanche photodiodes (SPAD) (SPCM-AQR, PerkinElmer Inc.), as shown in Fig. S1. Fluorescence signal was measured using the time-correlated single photon counting (TCSPC) module in the time-tagged time-resolved (TTTR) mode (Time Harp200, PicoQuant GmbH). Raw fluorescence images and correlation data were first analyzed and exported by the SymPhoTime software package (PcoQuant GmbH).
Data analysis for FCS
Autocorrelation function from fluorescence fluctuation measurements describes the similarity between observations as a function of the time lag (τ) between events.Citation53 For fluorescent molecules diffusing through an extremely small detection volume, the autocorrelation function (normalized by average fluorescence < F >) can be obtained from:
where δF(t) = F(t) – [ F(t) ] . When the molecules diffuse freely in a 3D space, EquationEquation 1(1)
(1) can be written as:
where , w0 and z0 are lateral and axial radii of the detection volume, respectively. τD is diffusion time, which reflects the average time the fluorescent molecules reside in the confocal volume and can be expressed as
, where D is diffusion coefficient. For live cell FCS data analysis, two mathematical models were considered:
This anomalous diffusion model could accurately describe the mobility of molecules which are in extremely crowded and complex environment. In EquationEquation 3(3)
(3) , α represents the degree of anomalous behavior (a value of 0.5–0.8 can indicate an anomalous diffusionCitation79,Citation80). When the molecules can quickly switch or co-exist between free and bound states, the two-component fitting model need be used.
Where y is the fraction of bound molecules. Before data collection, fluorophore with known D (e.g., Rhodamine 123 in Fig. S1B)Citation81 was used for calibration. For spherical particles, D can be obtained from the Stokes-Einstein equation as:
where k is the Boltzmann’s constant, η is the viscosity and R the molecular radius or hydrodynamic diameter which is inversely proportional to D. A 3D Gaussian approximation was used and the effective volume was estimated by: . Thus a blue laser gives rise to a confocal volume of 0.6 fl. Additionally, raw autocorrelation curves were filtered by Fluorescence Lifetime Correlation Spectroscopy (FLCS) function to remove the background noise and after-pulsing artifacts.Citation82
Maximum entropy method analysis of FCS data (MEMFSC) is a multi-component fitting model, which simultaneously minimizes χ2 and maximizes entropy S without a given priori. Thus the MEMFCS output represents a continuous distribution of τD instead of the preset discrete values. Hence G(τ) can be rewritten as
S is defined as
where . MEMFCS is particularly attractive because of its ability to effectively differentiate the diffusion of different components in a heterogeneous biological system.Citation83
Propidium iodide uptake experiment
An amount of 1 mg/ml PI solution (Sigma, P4864) was dissolved in Hank’s Balanced Salt Solution (Gibco, 14025) to 0.1 mg/ml. After 4 h of hypoxia, cells were washed with sterilized PBS and then incubated with PI solution for half an hour. PI was immediately removed and the cells were rinsed three times before imaging.
Additional material
Download Zip (976.9 KB)Acknowledgments
We thank Dr Adrian Bird for providing the MBD3-GFP plasmid, and Dr Amy C Lossie and Dr Chiao-Ling Lo for plasmid maintenance and technical assistance. Support from the Purdue Center for Cancer Research through the innovative grant and the W.M. Keck Foundation is appreciated. Partial student support for BC by the National Institutes of Health, National Cancer Institute R25CA128770 Cancer Prevention Internship Program administered by the Oncological Sciences Center and the Discovery Learning Research Center at Purdue University is appreciated. This publication was made possible in part, with support from the Indiana Clinical and Translational Sciences Institute funded, in part by Grant Number (TR000006) from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/epigenetics/article/25958
References
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell 2007; 128:635 - 8; http://dx.doi.org/10.1016/j.cell.2007.02.006; PMID: 17320500
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012; 13:484 - 92; http://dx.doi.org/10.1038/nrg3230; PMID: 22641018
- Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem 2012; 81:97 - 117; http://dx.doi.org/10.1146/annurev-biochem-052610-091920; PMID: 22404632
- Ross SA. Diet and DNA methylation interactions in cancer prevention. Ann N Y Acad Sci 2003; 983:197 - 207; http://dx.doi.org/10.1111/j.1749-6632.2003.tb05974.x; PMID: 12724224
- McCabe DC, Caudill MA. DNA methylation, genomic silencing, and links to nutrition and cancer. Nutr Rev 2005; 63:183 - 95; http://dx.doi.org/10.1111/j.1753-4887.2005.tb00136.x; PMID: 16028562
- Salbaum JM, Kappen C. Genetic and epigenomic footprints of folate. Prog Mol Biol Transl Sci 2012; 108:129 - 58; http://dx.doi.org/10.1016/B978-0-12-398397-8.00006-X; PMID: 22656376
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324:930 - 5; http://dx.doi.org/10.1126/science.1170116; PMID: 19372391
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010; 466:1129 - 33; http://dx.doi.org/10.1038/nature09303; PMID: 20639862
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011; 333:1303 - 7; http://dx.doi.org/10.1126/science.1210944; PMID: 21817016
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 2011; 473:394 - 7; http://dx.doi.org/10.1038/nature10102; PMID: 21552279
- Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res 2011; 39:5015 - 24; http://dx.doi.org/10.1093/nar/gkr120; PMID: 21378125
- Li W, Liu M.. Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids 2011; 2011:870726
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol 2011; 29:68 - 72; http://dx.doi.org/10.1038/nbt.1732; PMID: 21151123
- Chen J, Irudayaraj J. Quantitative investigation of compartmentalized dynamics of ErbB2 targeting gold nanorods in live cells by single molecule spectroscopy. ACS Nano 2009; 3:4071 - 9; http://dx.doi.org/10.1021/nn900743v; PMID: 19891423
- Chen J, Irudayaraj J. Fluorescence lifetime cross correlation spectroscopy resolves EGFR and antagonist interaction in live cells. Anal Chem 2010; 82:6415 - 21; http://dx.doi.org/10.1021/ac101236t; PMID: 20586411
- Siegel AP, Hays NM, Day RN. Unraveling transcription factor interactions with heterochromatin protein 1 using fluorescence lifetime imaging microscopy and fluorescence correlation spectroscopy. J Biomed Opt 2013; 18:25002; http://dx.doi.org/10.1117/1.JBO.18.2.025002; PMID: 23392382
- Dong C, Irudayaraj J. Hydrodynamic size-dependent cellular uptake of aqueous QDs probed by fluorescence correlation spectroscopy. J Phys Chem B 2012; 116:12125 - 32; http://dx.doi.org/10.1021/jp305563p; PMID: 22950363
- Schwille P. Fluorescence correlation spectroscopy and its potential for intracellular applications. Cell Biochem Biophys 2001; 34:383 - 408; http://dx.doi.org/10.1385/CBB:34:3:383; PMID: 11898862
- Weidtkamp-Peters S, Weisshart K, Schmiedeberg L, Hemmerich P. Fluorescence correlation spectroscopy to assess the mobility of nuclear proteins. Methods Mol Biol 2009; 464:321 - 41; http://dx.doi.org/10.1007/978-1-60327-461-6_18; PMID: 18951193
- Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene 2002; 21:5400 - 13; http://dx.doi.org/10.1038/sj.onc.1205651; PMID: 12154403
- Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics 2009; 1:239 - 59; http://dx.doi.org/10.2217/epi.09.33; PMID: 20495664
- Ehrlich M, Lacey M. DNA hypomethylation and hemimethylation in cancer. Adv Exp Med Biol 2013; 754:31 - 56; http://dx.doi.org/10.1007/978-1-4419-9967-2_2; PMID: 22956495
- Esteller M. Epigenetics in cancer. N Engl J Med 2008; 358:1148 - 59; http://dx.doi.org/10.1056/NEJMra072067; PMID: 18337604
- Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 2003; 300:455; http://dx.doi.org/10.1126/science.1083557; PMID: 12702868
- Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 2012; 150:1135 - 46; http://dx.doi.org/10.1016/j.cell.2012.07.033; PMID: 22980977
- Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2011; 2:627 - 37; PMID: 21896958
- Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 2013; 32:663 - 9; http://dx.doi.org/10.1038/onc.2012.67; PMID: 22391558
- Münzel M, Globisch D, Carell T. 5-Hydroxymethylcytosine, the sixth base of the genome. Angew Chem Int Ed Engl 2011; 50:6460 - 8; http://dx.doi.org/10.1002/anie.201101547; PMID: 21688365
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet 2012; 13:7 - 13; PMID: 22083101
- Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, Klimek V, Slack J, de Castro C, Ravandi F, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer 2006; 106:1794 - 803; http://dx.doi.org/10.1002/cncr.21792; PMID: 16532500
- Kantarjian HM, O’Brien S, Cortes J, Giles FJ, Faderl S, Issa JP, Garcia-Manero G, Rios MB, Shan J, Andreeff M, et al. Results of decitabine (5-aza-2’deoxycytidine) therapy in 130 patients with chronic myelogenous leukemia. Cancer 2003; 98:522 - 8; http://dx.doi.org/10.1002/cncr.11543; PMID: 12879469
- Pandiyan K, You JS, Yang X, Dai C, Zhou XJ, Baylin SB, Jones PA, Liang G. Functional DNA demethylation is accompanied by chromatin accessibility. Nucleic Acids Res 2013; 41:3973 - 85; http://dx.doi.org/10.1093/nar/gkt077; PMID: 23408854
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009; 324:929 - 30; http://dx.doi.org/10.1126/science.1169786; PMID: 19372393
- Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev 2007; 26:225 - 39; http://dx.doi.org/10.1007/s10555-007-9055-1; PMID: 17440684
- Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer 2011; 11:393 - 410; http://dx.doi.org/10.1038/nrc3064; PMID: 21606941
- Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer 2004; 4:437 - 47; http://dx.doi.org/10.1038/nrc1367; PMID: 15170446
- Thomas S, Harding MA, Smith SC, Overdevest JB, Nitz MD, Frierson HF, Tomlins SA, Kristiansen G, Theodorescu D. CD24 is an effector of HIF-1-driven primary tumor growth and metastasis. Cancer Res 2012; 72:5600 - 12; http://dx.doi.org/10.1158/0008-5472.CAN-11-3666; PMID: 22926560
- Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell 2007; 129:465 - 72; http://dx.doi.org/10.1016/j.cell.2007.04.019; PMID: 17482542
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell 2009; 5:237 - 41; http://dx.doi.org/10.1016/j.stem.2009.08.001; PMID: 19716359
- Li J, Harris RA, Cheung SW, Coarfa C, Jeong M, Goodell MA, White LD, Patel A, Kang SH, Shaw C, et al. Genomic hypomethylation in the human germline associates with selective structural mutability in the human genome. PLoS Genet 2012; 8:e1002692; http://dx.doi.org/10.1371/journal.pgen.1002692; PMID: 22615578
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet 2013; 14:204 - 20; http://dx.doi.org/10.1038/nrg3354; PMID: 23400093
- Shahrzad S, Bertrand K, Minhas K, Coomber BL. Induction of DNA hypomethylation by tumor hypoxia. Epigenetics 2007; 2:119 - 25; http://dx.doi.org/10.4161/epi.2.2.4613; PMID: 17965619
- Liu Q, Liu L, Zhao Y, Zhang J, Wang D, Chen J, He Y, Wu J, Zhang Z, Liu Z. Hypoxia induces genomic DNA demethylation through the activation of HIF-1α and transcriptional upregulation of MAT2A in hepatoma cells. Mol Cancer Ther 2011; 10:1113 - 23; http://dx.doi.org/10.1158/1535-7163.MCT-10-1010; PMID: 21460102
- Pal A, Srivastava T, Sharma MK, Mehndiratta M, Das P, Sinha S, Chattopadhyay P. Aberrant methylation and associated transcriptional mobilization of Alu elements contributes to genomic instability in hypoxia. J Cell Mol Med 2010; 14:2646 - 54; http://dx.doi.org/10.1111/j.1582-4934.2009.00792.x; PMID: 19508390
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 2011; 147:1498 - 510; http://dx.doi.org/10.1016/j.cell.2011.11.054; PMID: 22196727
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Münzel M, Wagner M, Müller M, Khan F, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 2013; 152:1146 - 59; http://dx.doi.org/10.1016/j.cell.2013.02.004; PMID: 23434322
- Chen J, Miller A, Kirchmaier AL, Irudayaraj JM. Single-molecule tools elucidate H2A.Z nucleosome composition. J Cell Sci 2012; 125:2954 - 64; http://dx.doi.org/10.1242/jcs.101592; PMID: 22393239
- Kim SA, Sanabria H, Digman MA, Gratton E, Schwille P, Zipfel WR, Waxham MN. Quantifying translational mobility in neurons: comparison between current optical techniques. J Neurosci 2010; 30:16409 - 16; http://dx.doi.org/10.1523/JNEUROSCI.3063-10.2010; PMID: 21147979
- Sako Y, Yanagida T. Single-molecule visualization in cell biology. Nat Rev Mol Cell Biol 2003; Suppl SS1 - 5; PMID: 14587519
- Kim SA, Heinze KG, Schwille P. Fluorescence correlation spectroscopy in living cells. Nat Methods 2007; 4:963 - 73; http://dx.doi.org/10.1038/nmeth1104; PMID: 17971781
- Prendergast FG, Mann KG. Chemical and physical properties of aequorin and the green fluorescent protein isolated from Aequorea forskålea. Biochemistry 1978; 17:3448 - 53; http://dx.doi.org/10.1021/bi00610a004; PMID: 28749
- Tsien RY. The green fluorescent protein. Annu Rev Biochem 1998; 67:509 - 44; http://dx.doi.org/10.1146/annurev.biochem.67.1.509; PMID: 9759496
- Lakowicz JR. Principles of fluorescence spectroscopy. New York: Springer, 2006.
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 2012; 40:4841 - 9; http://dx.doi.org/10.1093/nar/gks155; PMID: 22362737
- Kumei Y, Nakajima T, Sato A, Kamata N, Enomoto S. Reduction of G1 phase duration and enhancement of c-myc gene expression in HeLa cells at hypergravity. J Cell Sci 1989; 93:221 - 6; PMID: 2693468
- Liang L, Wang X, Xing D, Chen T, Chen WR. Noninvasive determination of cell nucleoplasmic viscosity by fluorescence correlation spectroscopy. J Biomed Opt 2009; 14:024013; http://dx.doi.org/10.1117/1.3088141; PMID: 19405743
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev 1999; 13:1924 - 35; http://dx.doi.org/10.1101/gad.13.15.1924; PMID: 10444591
- Roossink F, de Jong S, Wisman GB, van der Zee AG, Schuuring E. DNA hypermethylation biomarkers to predict response to cisplatin treatment, radiotherapy or chemoradiation: the present state of art. Cell Oncol (Dordr) 2012; 35:231 - 41; http://dx.doi.org/10.1007/s13402-012-0091-7; PMID: 22836879
- Watson JA, Watson CJ, McCann A, Baugh J. Epigenetics, the epicenter of the hypoxic response. Epigenetics 2010; 5:293 - 6; http://dx.doi.org/10.4161/epi.5.4.11684; PMID: 20418669
- Song CX, Yi C, He C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat Biotechnol 2012; 30:1107 - 16; http://dx.doi.org/10.1038/nbt.2398; PMID: 23138310
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet 2001; 27:31 - 9; http://dx.doi.org/10.1038/83730; PMID: 11137995
- Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, Biniszkiewicz D, Bates B, Lee PP, Kuhn R, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci 2001; 21:788 - 97; PMID: 11157065
- Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev 2001; 15:710 - 23; http://dx.doi.org/10.1101/gad.194101; PMID: 11274056
- Reese KJ, Lin S, Verona RI, Schultz RM, Bartolomei MS. Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3. PLoS Genet 2007; 3:e137; http://dx.doi.org/10.1371/journal.pgen.0030137; PMID: 17708683
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 2011; 25:2436 - 52; http://dx.doi.org/10.1101/gad.179184.111; PMID: 22156206
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 2013; 339:448 - 52; http://dx.doi.org/10.1126/science.1229277; PMID: 23223451
- Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005; 33:6823 - 36; http://dx.doi.org/10.1093/nar/gki987; PMID: 16326863
- Cadieux B, Ching TT, VandenBerg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res 2006; 66:8469 - 76; http://dx.doi.org/10.1158/0008-5472.CAN-06-1547; PMID: 16951158
- Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, Moreno V, Esteller M, Capellà G, Ribas M, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res 2006; 66:8462 - 9468; http://dx.doi.org/10.1158/0008-5472.CAN-06-0293; PMID: 16951157
- Estécio MRH, Gharibyan V, Shen LL, Ibrahim AEK, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One 2007; 2:e399; http://dx.doi.org/10.1371/journal.pone.0000399; PMID: 17476321
- Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. . Biochim Biophys Acta 2007; 1775:138 - 62
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57 - 74; http://dx.doi.org/10.1038/nature11247; PMID: 22955616
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol 1998; 18:6538 - 47; PMID: 9774669
- Tao Z, Goodisman J, Souid AK. Oxygen measurement via phosphorescence: reaction of sodium dithionite with dissolved oxygen. J Phys Chem A 2008; 112:1511 - 8; http://dx.doi.org/10.1021/jp710176z; PMID: 18229902
- Mancarella S, Wang Y, Deng X, Landesberg G, Scalia R, Panettieri RA, Mallilankaraman K, Tang XD, Madesh M, Gill DL. Hypoxia-induced acidosis uncouples the STIM-Orai calcium signaling complex. J Biol Chem 2011; 286:44788 - 98; http://dx.doi.org/10.1074/jbc.M111.303081; PMID: 22084246
- Abudara V, Jiang RG, Eyzaguirre C. Behavior of junction channels between rat glomus cells during normoxia and hypoxia. J Neurophysiol 2002; 88:639 - 49; PMID: 12163517
- Jiang RG, Eyzaguirre C. Calcium channels of cultured rat glomus cells in normoxia and acute hypoxia. Brain Res 2005; 1031:56 - 66; http://dx.doi.org/10.1016/j.brainres.2004.10.021; PMID: 15621012
- Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet 2011; 7:e1002154; http://dx.doi.org/10.1371/journal.pgen.1002154; PMID: 21731508
- Weiss M, Elsner M, Kartberg F, Nilsson T. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys J 2004; 87:3518 - 24; http://dx.doi.org/10.1529/biophysj.104.044263; PMID: 15339818
- Banks DS, Fradin C. Anomalous diffusion of proteins due to molecular crowding. Biophys J 2005; 89:2960 - 71; http://dx.doi.org/10.1529/biophysj.104.051078; PMID: 16113107
- Gendron PO, Avaltroni F, Wilkinson KJ. Diffusion coefficients of several rhodamine derivatives as determined by pulsed field gradient-nuclear magnetic resonance and fluorescence correlation spectroscopy. J Fluoresc 2008; 18:1093 - 101; http://dx.doi.org/10.1007/s10895-008-0357-7; PMID: 18431548
- Kapusta P, Macháň R, Benda A, Hof M. Fluorescence Lifetime Correlation Spectroscopy (FLCS): Concepts, Applications and Outlook. Int J Mol Sci 2012; 13:12890 - 910; http://dx.doi.org/10.3390/ijms131012890; PMID: 23202928
- Sengupta P, Garai K, Balaji J, Periasamy N, Maiti S. Measuring size distribution in highly heterogeneous systems with fluorescence correlation spectroscopy. Biophys J 2003; 84:1977 - 84; http://dx.doi.org/10.1016/S0006-3495(03)75006-1; PMID: 12609900