Abstract
In eukaryotic organisms, histone posttranslational modifications (PTMs) are indispensable for their role in maintaining cellular physiology, often through their mediation of chromatin-related processes such as transcription. Targeted investigations of this ever expanding network of chemical moieties continue to reveal genetic, biochemical, and cellular nuances of this complex landscape. In this study, we present our findings on a novel class of histone PTMs: Serine, Threonine, and Tyrosine O-acetylation. We have combined highly sensitive nano-LC-MS/MS experiments and immunodetection assays to identify and validate these unique marks found only on histone H3. Mass spectrometry experiments have determined that several of these O-acetylation marks are conserved in many species, ranging from yeast to human. Additionally, our investigations reveal that histone H3 serine 10 acetylation (H3S10ac) is potentially linked to cell cycle progression and cellular pluripotency. Here, we provide a glimpse into the functional implications of this H3-specific histone mark, which may be of high value for further studies of chromatin.
Introduction
Histone posttranslational modifications (PTMs) play an invaluable role in the maintenance of cellular integrity. Transcription, cellular differentiation, DNA replication and repair, and even tumorigenesis have been shown to be integrally co-dependent on the “epigenetic” signatures at various cellular stages.Citation1 The “histone code” hypothesis, as proposed by Allis and colleagues, suggests that histone modifications act either sequentially or in combination to affect downstream biological processes through a complex network of “writer” and “eraser” enzymes and “reader” interacting proteins.Citation2 Several unique studies have added credence to this concept in eukaryotic model systems, but have simultaneously reiterated the necessity of understanding the contribution of individual marks in the epigenetic landscape.Citation3-Citation6 For instance, in a study in budding yeasts, H4 tail lysines (K5, K8, K12, and K16) were each mutated to arginines to mimic all possible combinations of H4 tail lysine acetylation. The gene expression profiles of these strains suggested that for ~1200 genes, K5ac, K8ac, and K12ac caused “non-specific, cumulative effects seen as increased transcription,” but also showed that H4K16ac independently acts as a regulator of the transcriptional profiles of select genes in these cells.Citation7 In vitro investigations by Shogren-Knaak revealed that H4K16ac single-handedly imposes architectural constraints on chromatin structure, which in turn limits accessibility of transcriptional activators to underlying genes.Citation8 In this vein, we have focused our studies on evaluating the potential role of individual H3S10ac in pluripotency and cell cycle regulation.
In the past decade, advancements in mass spectrometry-based proteomic methods have led to the discovery of a myriad of new and critical histone and non-histone PTMs.Citation9 Among others, new sites of acetylation, methylation, O-GlcNAcylation, crotonylation, propionylation, arginine citrullination, butyrylation, proline isomerization, malonylation, succinylation, formylation, and ADP-ribosylation have been detected and studied in a wide range of species and biological processes.Citation10-Citation16 Serine acetylation was first reported as a chemical antagonist to phosphorylation-mediated signaling pathways in eukaryotic cells, catalyzed by the Yersinia bacterial virulence factor, YopJ.Citation17 YopJ specifically acetylates a serine residue that is otherwise an acceptor site for a phosphoryl group by an upstream mitogen-activated protein kinase (MAPK) in human embryonic kidney (HEK) 293 cells. More recently, two proteomics-based studies have also detected this type of O-acetylation modification on non-histone proteins in higher eukaryotes.Citation18,Citation19 In our mass spectrometric analyses, we reveal several new low-level O-acetylation modifications on histone H3, including H3S10ac, none of which have been described elsewhere in the literature. The H3S10 residue has previously been the subject of intense investigation. In its phosphorylated state (H3S10phos), it facilitates chromosomal condensation and segregation during metaphaseCitation20,Citation21 and also sterically hinders heterochromatin protein 1 from binding the adjacent H3K9me3 during mitosis.Citation22 Based on these findings, we hypothesize that H3S10ac could function as a phospho-antagonist to H3S10phos in similar fashion to the YopJ catalyzed serine O-acetyl blockage of MAP kinase phospho-sites. Here, we describe our initial work characterizing this highly conserved class of histone PTMs and their potential biological roles within the epigenetic landscape of chromatin.
Results
Initial identification of O-acetylation on histone H3
Using the PILOT_PTM algorithm,Citation23 we mined existing HeLa and mouse embryonic stem cell MS/MS data sets generated in our lab, and manually validated any potentially interesting and novel hits. Our screens revealed previously unidentified sites of known types of PTMs such as lysine acetylation and crotonylation (data not shown) but, more significantly, novel categories of histone PTMs such as serine and threonine O-acetylation on histone H3. We initially identified an O-acetylated histone H3 peptide from mouse embryonic stem cells. This peptide species corresponded to the H3 9–17 propionylated tryptic fragment containing an O-acetyl group on Serine 10 (H3S10ac). Shown in is the MS/MS spectrum of the [M+2H]2+ peptide ion at 556.309 m/z with the sequence prKprS(OAc)TGGKprAPR, where “pr” on the peptide represents the propionyl amide group from our chemical derivatization procedure of histone proteins. We obtained adequate sequence coverage for this H3 peptide with reproducible retention times through all our runs and consistent detection of the b1 ion at 241.2 m/z and the b2 ion at 370.4, which complimented the y7 and y8 ions at 742.5 m/z and 871.4 m/z, respectively, and are indicative of an acetyl group (+ 42 Da) on the S10 residue.
Figure 1. Sequencing of H3S10ac peptide. (Left) MS/MS spectrum of the [M+2H]2+ ion at 556.309 m/z, which was generated from the CAD fragmentation of the prKprS(OAc)TGGKprAPR precursor peptide. Expected mono-isotopic b- (top row) and y-type (bottom row) ion fragment masses are observed. The observed accurate mass, [M+2H]2+(exp), corresponded precisely to the calculated accurate mass, [M+2H]2+(cal) of 556.309 m/z (0 ppm error). OAc refers to the acetyl modification on the S10 residue and pr refers to the propionyl amide group from chemical derivatization of histone proteins. (Right) Structure of an acetylated serine residue
![Figure 1. Sequencing of H3S10ac peptide. (Left) MS/MS spectrum of the [M+2H]2+ ion at 556.309 m/z, which was generated from the CAD fragmentation of the prKprS(OAc)TGGKprAPR precursor peptide. Expected mono-isotopic b- (top row) and y-type (bottom row) ion fragment masses are observed. The observed accurate mass, [M+2H]2+(exp), corresponded precisely to the calculated accurate mass, [M+2H]2+(cal) of 556.309 m/z (0 ppm error). OAc refers to the acetyl modification on the S10 residue and pr refers to the propionyl amide group from chemical derivatization of histone proteins. (Right) Structure of an acetylated serine residue](/cms/asset/c9855406-c97f-4aee-b1a5-217e0db38242/kepi_a_10926025_f0001.gif)
Histone O-acetylation is conserved across many eukaryotic species
H3S10ac was first discovered in mouse embryonic stem cell samples, and initially went undetected in several human samples tested (HeLa, Human Embryonic Kidney [HEK 293] and Human Foreskin Fibroblast [HFF]) when analyzed by the PILOT-PTM program. To account for the low levels of O-acetylated peptides, we performed hypothesis-driven nano-LC-MS/MS, targeting Serine O-acetylated precursor peptides, based on theoretical m/z values calculated for the various eukaryotic histone sequences, listed in . Serine acetylation requires the formation of an esteric bond from the hydroxyl group on its side chain. We therefore also hypothesized that threonine and tyrosine residues, also with –OH containing side chains, held the potential for this novel O-acetylation. After several rounds of targeted nano-LC-MS/MS experiments, we identified a combined total of six serine, threonine, and tyrosine O-acetylations on the N-terminal domain of histone H3, several highly conserved across prominent model eukaryotic organisms (; Fig. S1). Most significantly, in analyzing histone samples from Saccharomyces cerevisiae, we observed a distinct conservation of O-acetylation at amino acid position 22. In yeast and tetrahymena, the H3 amino acid sequence contains a serine (S) at position 22, while a threonine (T) is found at position 22 in human and Drosophila melanogaster. ( and ). Similar to the 9–17 peptide, we invariably obtain > 90% sequence coverage for this 18–26 fragment, [M+2H]2+ = 591.845 m/z and 598.854 m/z for yeast and Drosophila, respectively. The b5 ions, 682.2 m/z (yeast) and 696.7 m/z (Drosophila), and y5 ions, 630.4 m/z (yeast) and 644.3 m/z (Drosophila) denote acetyl marks on the S and T at position 22, respectively. Yeast MS/MS data also revealed a mixture of H3S28ac and H3S31ac, with the H3S31ac being slightly more abundant than the H3S28ac mark (), based on the fragment ion relative ratio seen in the tandem mass spectra (Fig. S2).
Table 1. Novel sites of histone O-acetylation
Figure 2. Yeast S22ac and Drosophila T22ac. The top panel shows the MS/MS spectrum of the [M+2H]2+ precursor ion at 591.845 m/z, the 18–26 peptide (prKprQLAS[OAc]KprAAR) of histone H3 of S. cerevisiae. Conversely, the bottom panel shows the MS/MS spectrum of the complementary 18–26 precursor peptide, extracted from Drosophila S2 cell histone samples. Both spectra show that the residue at position 22 is acetylated in a manner that is independent of the residue; serine in yeast and threonine in Drosophila.
![Figure 2. Yeast S22ac and Drosophila T22ac. The top panel shows the MS/MS spectrum of the [M+2H]2+ precursor ion at 591.845 m/z, the 18–26 peptide (prKprQLAS[OAc]KprAAR) of histone H3 of S. cerevisiae. Conversely, the bottom panel shows the MS/MS spectrum of the complementary 18–26 precursor peptide, extracted from Drosophila S2 cell histone samples. Both spectra show that the residue at position 22 is acetylated in a manner that is independent of the residue; serine in yeast and threonine in Drosophila.](/cms/asset/8eaba50d-d118-4b1d-baa4-00b261637768/kepi_a_10926025_f0002.gif)
To further characterize this new class of modification, we quantified their abundances relative to other commonly analyzed histone PTMs. Most of these marks were of very low abundance (1% or less) for any of the organisms interrogated (), which are dwarfed percentages in relation to most other well-known acetylation sites on histone H3 or H4 (). The highest levels of histone H3 O-acetylation (H3S10, H3T22, and H3S28) are consistently found in mESCs and drosophila S2 cells; both cell types of embryonic origin. In agreement with this observation, higher levels of global H4 lysine acetylation and some H3 acetyl sites are also typical in pluripotent cells that have more “structurally open” genomes. During these quantification analyses, we took extra care to avoid some common issues that can emerge while analyzing any type of low level modification, as previous work has shown in vitro artifacts to be problematic when focusing on potential protein modifications.Citation24,Citation25 For instance, (left panel) shows an MS layout of the peptide peaks used for quantification of H3S10ac. Each peak represents either the unmodified or differentially modified forms of the H3 9–17 peptide fragment (KSTGGKAPR), corresponding to their [M+2H]2+ m/z values. As annotated, several residues on this peptide, namely K9 and K14, are host to a myriad of modifications. One peak, when sequenced was shown to be an H3S10ac “false positive” peptide (closer to 46 min), which shared an identical [M+2H]2+ value of 556.309 m/z and eluted within 45 s of the actual H3S10ac peptide (near 45 min). Here, the more prominent peak at 556.309 m/z corresponds to H3S10prK14ac, as highlighted by the MS/MS peptide sequence (, right panel). At this point, we are not sure if this Serine O-propionylated peptide is an endogenous modification or an in vitro artifact resulting from the chemical derivatization procedure. To further validate H3S10ac, we compared the MS/MS spectrum of a synthetic H3 O-acetyl peptide with that of the in vivo derived peptide from . Both H3 peptides, prKprS(OAc)TGGKprAPR, had similar retention times (data not shown) and exhibited near identical parent masses and CAD MS/MS spectra ().
Table 2. Global H3 and H4 lysine acetylation
Figure 3. Extracted ion chromatogram for peptide quantification. The left panel shows extracted ion chromatograms for various modified peptides ([M+2H]2+ ions) spanning the H3 9–17 residues, KSTGGKAPR, after chemical derivatization by propionylation. Labels indicate the particular modified form eluted in that peak, as determined after inspection of the corresponding MS/MS spectra. Occasionally, non-target peptide peaks with identical precursor masses will elute slightly after the S10ac peptide. For example, the right panel shows a detailed MS/MS analysis of a prominent peptide peak (H3S10prK14ac) with an identical [M+2H]2+ value of 556.309 m/z, which eluted within 45 s of the targeted H3S10ac peptide. The S10pr and S10ac peptide species contain the same modifications in different sequences (S10pr - prKprSprTGGKacAPR, S10ac - prKprS[OAc]TGGKprAPR) and therefore the same m/z values and elute at almost identical times. For quantification, the relative abundance of each peak, expressed as percentages, is calculated by measuring the area under the XIC peak corresponding to each specifically modified form and expressing that value as a fraction of the total sum of the peak areas corresponding to all observed modified forms.
![Figure 3. Extracted ion chromatogram for peptide quantification. The left panel shows extracted ion chromatograms for various modified peptides ([M+2H]2+ ions) spanning the H3 9–17 residues, KSTGGKAPR, after chemical derivatization by propionylation. Labels indicate the particular modified form eluted in that peak, as determined after inspection of the corresponding MS/MS spectra. Occasionally, non-target peptide peaks with identical precursor masses will elute slightly after the S10ac peptide. For example, the right panel shows a detailed MS/MS analysis of a prominent peptide peak (H3S10prK14ac) with an identical [M+2H]2+ value of 556.309 m/z, which eluted within 45 s of the targeted H3S10ac peptide. The S10pr and S10ac peptide species contain the same modifications in different sequences (S10pr - prKprSprTGGKacAPR, S10ac - prKprS[OAc]TGGKprAPR) and therefore the same m/z values and elute at almost identical times. For quantification, the relative abundance of each peak, expressed as percentages, is calculated by measuring the area under the XIC peak corresponding to each specifically modified form and expressing that value as a fraction of the total sum of the peak areas corresponding to all observed modified forms.](/cms/asset/1d1d749e-ff10-4f38-a064-43d7335a81e2/kepi_a_10926025_f0003.gif)
Figure 4. Detection and validation of H3S10ac. A comparison of the MS/MS spectra of the [M+2H]2+ ion at 556.309, prKprS(OAc)TGGKprAPR for the in vivo-derived (top panel) and synthetic (bottom panel) H3 9–17 peptides.
![Figure 4. Detection and validation of H3S10ac. A comparison of the MS/MS spectra of the [M+2H]2+ ion at 556.309, prKprS(OAc)TGGKprAPR for the in vivo-derived (top panel) and synthetic (bottom panel) H3 9–17 peptides.](/cms/asset/ea2eb9f4-f845-48ab-8ae7-7897de2f391d/kepi_a_10926025_f0004.gif)
As H3S10ac levels were slightly different in the different cell types, we sought to test the hypothesis that H3S10ac levels can be modulated by the activity of histone deacetylases (HDACs). For this, we treated cells with sodium butyrate, a pan-inhibitor for class I and class II HDACs. As mentioned earlier, in untreated HeLa cells, the levels of H3S10ac were either outside the range of our detection method, or absent. However, in the presence of 8mM sodium butyrate, the levels of H3S10ac rose to a detectable level around 0.5% (). As other histone H3 and H4 acetylation sites increase 5–10 fold in abundance following HDAC inhibition (data not shown), we estimate that the global level of H3S10ac in asynchronous somatic human cells to be extremely low (< 0.05%). Most other O-acetylation sites were also found at very low abundances (1% or less), regardless of the site of modification or organism from which it originated (). However, it should again be noted that the cell types where H3S10ac was most easily observable were in mouse embryonic stem cells, and Drosophila S2 cells, which are derived from fly embryos. This observation of enhanced H3S10ac in cells of embryonic origin is further probed in the experiments described below.
Orthogonal immunoassay approaches confirm in vivo O-acetylation of histone H3
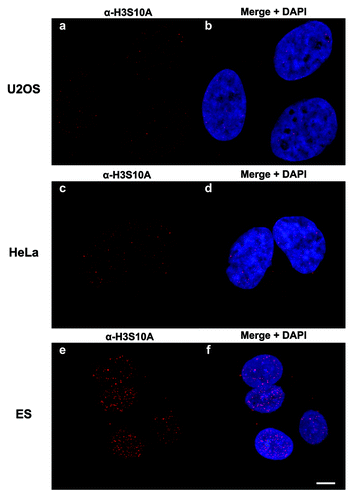
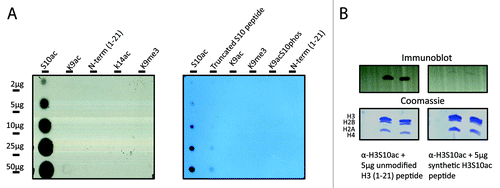
As a complementary approach to our nano-LC-MS/MS studies, we generated a site-specific H3S10ac antibody. On initial testing, the rabbit antiserum was only partially effective (data not shown). However, after several steps of column affinity chromatography with the target peptide and subsequently with the unmodified peptide to deplete any non-H3S10ac recognizing antibodies in the serum, the final affinity-purified antibody demonstrated specificity only to histone H3S10ac. Shown in are dot blots against a panel of acetylated, methylated, phosphorylated, and unmodified H3 9–17 peptides. The purified α-H3S10ac was very specific and robustly recognized only the H3S10ac peptide even at high loading amounts (). Synthetic peptides containing K9 or K14 acetylation, which are within the recognized epitope of the peptide, were not recognized indicating that the antibody specifically recognized an O-acetyl bond. To ensure that the antibody did not recognize any other Serine O-linked bond, we tested α-H3S10ac against an H3S10phos peptide; the antibody did not bind. Furthermore, the H3S10ac signal can be efficiently “competed away” by pre-incubation of the antibody with a synthetic H3S10-acetylated peptide, but not by unmodified peptides or peptides acetylated at different residues ( and data not shown). These immunoassay experiments demonstrate that our MS/MS detection of H3S10ac is a genuinely new in vivo modification on histone H3, and also now shows that we have generated a potent immune-reagent that can be used in further genomic, imaging or biological experiments. Toward that end, we performed immunofluorescence microscopy experiments using the antibody on a few selected cell types (). Immunofluorescence staining on pre-extracted U2OS, HeLa, and ESCs shows that the histone H3 serine 10 acetylation is in a diffuse and slightly punctate pattern throughout the nucleus, which is defined by DAPI staining. In agreement with our quantitative proteomics data, we find that this mark is low level and difficult to detect in non-pluripotent cell types such as U2OS and HeLa (), while the highest levels of this modification are observed in mESCs ().
Figure 5. Immunoaffinity competition assay. (A) Validation of α-H3S10ac specificity: increasing concentrations (2–50µg) of differently modified peptides were blotted on to a nitrocellulose membrane and incubated with purified polyclonal α-H3S10ac. The antibody recognized only the synthetic H3S10ac peptide. (B) Bulk iPSC histones were analyzed by western blot for detection of H3S10ac. The signal was competed away by pre-incubation of the synthetic H3S10ac peptide.
H3S10ac abundance levels linked to cellular pluripotency
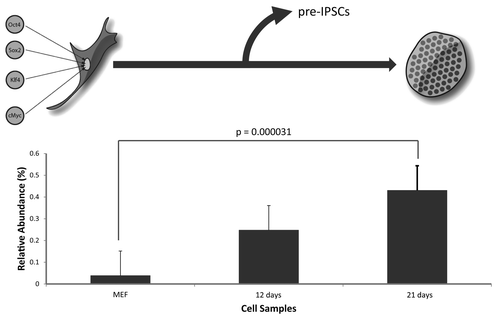
Significant increases in global histone acetylation in early embryonic stem cells have been previously reported, and HDAC inhibitors have also been shown to be effective in enhancing the yield of induced pluripotent stem cells (iPSCs) generated from somatic cell precursors.Citation26 For these reasons, we measured the abundance levels of H3S10ac during a time-course production of iPSCs. Using of the Takahashi and Yamanaka model, we reprogrammed mouse embryonic fibroblasts (MEFs) to iPSCs by overexpressing the four ‘Yamanaka transcription factors’, Oct 4, Klf4, Sox2, and c-Myc over the course of 21 d. We extracted histone samples at various time points and quantified the relative abundance of H3S10ac from somatic MEF cells, and iPSCs, by nano-LC-MS. Our data shows that H3S10ac was 10-fold higher in fully reprogrammed iPSCs (21 d) compared with the starting MEF histones (, p < 0.00003). There is also an increase in total K9ac and K14ac for the peptide but with smaller fold changes (Fig. S2). The levels of H3S10ac are also increased in the iPCSs within 12 d, but not to a significant level compared with the MEFs; however, the trend in the rise of the mark is apparent. These experiments show that H3S10ac also appears to be enriched in pluripotent iPSCs, similar to our earlier findings in mouse ESCs. This suggests that this mark may play a role in the maintenance of the pluripotent state possibly before cellular differentiation, although this idea remains to be tested in future studies.
Figure 7. Quantification of H3S10ac during reprogramming. Differentiated somatic MEFs were reprogrammed to iPSCs over the course of 21 d by overexpressing the four (4) Yamanaka transcription factors, Oct4, c-Myc, Sox2, Klf4. The levels of H3S10ac were quantified during this time course by nano-LC-MS analysis. Three independent experiments were performed and the error bars represent standard error.
Potential cell cycle regulation of H3S10ac
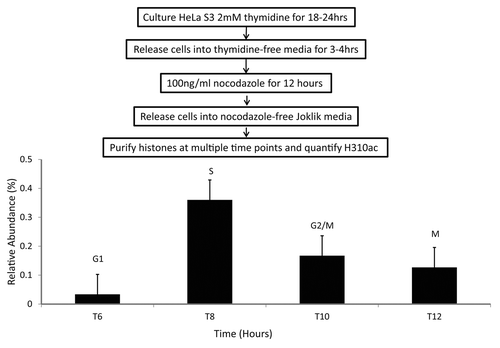
Histone H3S10phos plays a major role during mitosis, as it is highly enriched in this particular cell cycle phase and is needed for proper condensation and segregation of chromosomes. To test the possibility that H3S10ac may be cell cycle regulated and under-enriched in mitosis at the onset of H3S10phos, we performed an analysis of H3S10ac from synchronized HeLa cells using a thymidine-nocodazole block (blocks at M phase) and release. In synchronized HeLa S3 suspension cells, we observed that during S phase, H3S10ac was > 10-fold higher compared to cells in G1, and at least 2–3-fold higher relative to G2/M and M phase cells (). Comparatively, for the same samples, total levels of H3K9ac and H3K14ac do not change significantly (Fig. S3).This initial result hints at a role for H3S10ac during DNA replication, which is not entirely surprising as S phase contrasts M phase, mostly in regards to chromosome segregation and drastic changes in the levels of H3S10phos. Alternatively, H3S10ac might be implicated as a co-translational PTM, in similar fashion to N-terminal acetylation of histone H4, which is then regulated as histones progress through the cell cycle.
Figure 8. H3S10ac through HeLa cell cycle. HeLa suspension cells were synchronized at M phase of cell cycle through a thymidine-nocodazole synchronization followed by release into nocodazole-free media. The levels of H3S10ac were quantified at 6, 8, 10, and 12-h time points after release through nano-LC-MS analysis. Three independent experiments were performed and the error bars represent standard error.
Discussion
Non-histone serine and threonine O-acetylation has been reported to be present on a handful of yeast kinases that are important in pathways controlling cell shape and division.Citation19 Additionally, a very recent study has also reported the detection of a few O-acetylation sites on histones from mouse brain tissues, mostly on histone H1, H2A, and H2B.Citation27 Our investigations into the role of H3 O-acetylation began with a bioinformatics screen of histone MS/MS data generated from HeLa cells and mouse pluripotent cells. From our interrogations of histones extracted from several model organisms, we found that the initial sites discovered were only a fraction of the multiple O-acetylation sites on H3, found in both lower eukaryotes such as yeasts and tetrahymena, and metazoans, such as mice and humans. The high conservation of most of these O-acetyl marks across the different organisms (Fig. S1) signifies a potential evolutionary selection for this PTM. This also suggests that the enzymes that regulate this mark, HATs or HDACs may also be conserved across divergent organisms. In support, we found that treatment of HeLa cells with the class I and II HDAC inhibitor, sodium butyrate increases the levels of H3S10ac. However, we cannot rule out that this mark might be added to histones in a non-enzymatic process, as has been shown through investigations of different marks on proteins including histones.Citation28-Citation30 Interestingly, in yeast we found a co-occurrence of H3S28ac and nearby H3S31ac. This S31 residue is found only on the H3 variant H3.3 in mammalian cells (which is the only sequence found in yeast), and could provide an H3 variant specific PTM, which is quite rare. H3.3S31ac was not detected in any mammalian sample due to this variant being found in much lower levels compared with the other H3 variants, but this does not rule out that the mark could exist at levels below our MS detection limit. The low abundance of serine, threonine, and tyrosine acetylation (< 1%) may indicate that these marks are found on very discrete locations within the genome, similar to other modifications such as H3K4me3, found only at promoter regions,Citation31 or that they are tightly regulated to coincide with very specific biological conditions or temporal events. Due to the low level abundance, one hypothesis is that H3S10ac may serve as an activating marker to a distinct population of genes; a possibility that we are currently pursuing by first performing Chip-Seq to identify candidate genes.
Previous studies have shown that similar PTM novelties, such as Serine O-methylation are artifacts caused by a gas phase rearrangement reaction.Citation32 Specially, Zhang et al. found that a gas phase transfer of a methyl group from histone H3 K27 to S28 on mostly singly charged ions created a false positive identification of the Ser O-methylation. This methyl migration during the MS/MS fragmentation of +1 charged methylated histone peptides had also been previously noted to be a problematic issue for MALDI +1 generated ions, as opposed to ESI multiply charged ion MS/MS spectra.Citation25 In general, we never perform fragmentation on singly charged peptide ions, as they tend to not give sufficient fragmentation for peptide sequence analysis and are therefore confident that we have eliminated any potential false positives from our designations. It has also been shown that lysine, arginine, histidine, and glutamic acid residues can become artificially methylated as a result of staining and/or fixing SDS-PAGE gels with methanol.Citation33 Focusing on H3S10ac, we used two orthogonal approaches to confirm the existence of this mark as a true in vivo modification; namely by comparison to a synthetic peptide standard and the use of non-mass spectrometric immune-affinity assays. Using our affinity purified H3S10ac antibody, we showed through IF experiments that this mark was a true nuclei localized in vivo mark, and that our antibody could be utilized in imaging experiments. IF experiments were consistent with the findings that most cells exhibit low levels of H3S10ac, but that this histone modification was enriched in pluripotent cells such as mESCs.
Most histone PTMs are not cell cycle regulated,Citation34 with the exception of histone H4K20 methylation and mitotic phosphorylation H3 marks.Citation34-Citation39 One such mark, H3S10phos is invariably less than 2% in asynchronous cells but increases to approximately 60% during M phase, coupled to H3K9me2.Citation40 Although, it must be noted that even at lower levels, H3S10phos contributes to several downstream signaling, and transcriptional activities.Citation21,Citation38,Citation41 Taking advantage of cell synchronization experiments in HeLa suspension cells, we found that H3S10ac was enriched in S phase compared with other synchronized phases. One hypothesis to explain the requirement for this particular acetylation would be that O-acetylation bars kinase activity from the H3S10 residue, which is needed for proper chromosomal condensation and segregation.Citation21 This ‘blocking’ mechanism has been previously reported for conserved serine and threonine residues on MAP kinase, and TAK1 proteins, by the catalytic activity of Yersinia pestis YopJ virulence factor, which acts as a serine/threonine acetyl-transferaseCitation17,Citation42 Several key studies have similarly shown a distinct correlation between heterochromatin silencing through histone PTMs and replication timing;Citation43,Citation44 cells treated with Trichostatin A, a broad-spectrum histone deacetylase (HDAC) inhibitor, accelerates the timing of replication of hypo-acetylated heterochromatin.Citation45 Additionally, deletion of the S. cerevisiae HDAC, Rpd3 accelerates origin firing initiation at early and late origins and molecular manipulations of the Gcn5 histone acetyl-transferase (HAT) induces premature late origin firing.Citation46 Our observation that the H3S10ac levels are heightened at S phase warrants studies into the role of this mark in DNA synthesis, histone deposition, and/or origins of replication.
Many H3 and H4 acetylation sites do not increase during reprogramming from somatic cells (MEFs) to pluripotent cells (ESCs and iPSCs) as seen in , so there might be reason to expect a functional link between H3S10ac and embryonic type cells or in development, as opposed to an overall non-specific change in acetylation. We are currently probing the relationship between H3S10ac and the “bivalent domain”-associated marks of H3K4me3 and H3K27me3.Citation6 It would be interesting to determine if H3S10ac could be found on the same histone tail as the two bivalent domains, although this requires the use of advanced Middle Down proteomicsCitation47 following ChIP enrichment using our H3S10ac antibody. Although our data presents most of the O-acetyl marks independently of short-range interactions with other modified lysine residues on the same peptide, it is possible for H3S10ac to be present on the same nucleosome as other known PTMs, but not the same histone molecule, as demonstrated in recent work that shows the existence of asymmetrically modified histone PTMs on nucleosomes.Citation48
In conclusion, we have used mass spectrometry based proteomics and antibody-based approaches to identify serine, threonine, and tyrosine O-acetylations on histone H3 from different cell types, and organisms. Our results suggest a role for this new PTM during the cell cycle, possibly antagonisticto H3S10phos during DNA replication. This mark is also upregulated in pluripotent cells but whether it is needed for the maintenance of this state, or plays a role in cellular differentiation remains to be discovered. Future work will be directed at identifying the enzymes that regulate this mark, and also determining the possible function of the modification through ChIP-Seq genomic experiments to identify gene targets and their regulatory mechanisms.
Materials and Methods
Cell culture
HeLa and HEK 293 cells
HeLa S3 suspension cultures were maintained at 37 °C in 0.2 LPM CO2 in Joklik modified Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 1% Glutamax (Invitrogen), 10% newborn calf serum (Hyclone), and 1% penicillin/streptomycin at 2–6 × 105 cells/ml. HEK293 cells were grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. To harvest HeLa and HEK 293 samples, cells were pelleted at 600 rcf and washed in phosphate-buffered saline (PBS). The pellets were flash frozen in liquid nitrogen and stored at -80 °C.
HeLa cell cycle synchronization with thymidine-nocodozole
Asynchronously growing HeLa S3 cells were synchronized with thymidine and nocodazole by incubating in 2mM thymidine for 18–24 h, followed by thymidine-free media for 3–4 h, 100ng/ml nocodazole for 12 h, and finally into standard nocodazole-free Joklik medium. Following release from thymidine-nocodazole blocks, aliquots were collected and ethanol fixed for flow cytometric analysis of synchronization efficiencies. Separate aliquots were flash-frozen in liquid nitrogen and stored at -80 °C for histone acid extraction and mass spectrometric analysis.
Tetrahymena
T. thermophila was grown in super protease peptone medium (1% proteose peptone, 0.1% yeast extract, 0.2% glucose, 90 µM EDTA ferric sodium salt) at 30 °C, as previously described.Citation49 Conjugation was initiated when log-phase cells of different mating types were washed, starved (16–24 h at 30 °C), and mixed in 10 mM Tris buffer (pH 7.5) or Dryl’s phosphate buffer (1.5 mM CaCl2, 1 mM NaH2PO4, 1 mM Na2HPO4, 2 mM sodium citrate). The cells were centrifuged at maximum speed for 5 min, followed by purification of the somatic macronucleus (MAC) and the germline micronucleus (MIC), as previously described.Citation50,Citation51
Drosophila S2 cells
S2 cells were grown in CCM3 media (ThermoFisher) to reach a density of 106 to 107 cells/ml. One liter of cells was washed in PBS once, harvested by centrifugation at 600 rcf, flash frozen, and stored at -80 °C.
Saccharomyces cerevisiae
A wild type yeast strain in the BY4741 background was cultured in YEPD media to yield ~1010 cells, as previously described.Citation52 The cells were harvested by centrifugation at 3200 rcf, flash frozen, and stored at -80 °C. Nuclei were extracted from spheroplasts, as previously describedCitation53 and stored at -80 °C.
MEFs, mESCs, iPSCs
iPSCs (female line 2D4- made with Oct4, Sox2, c-Myc, and Klf4,Citation54 12-d “partially reprogrammed” pre-iPSCs (female line 1A2 and male line 12-1)Citation55-Citation58 male ES line V6.5 and male and female Nanog-GFPCitation59 and Stemcca fibroblasts (derived from d14.5 embryos) were grown in standard media. Stemcca mouse lines contained a single tet-inducible cassette with all 4 reprogramming factors, along with an Oct4-GFP reporter in a R26 M2rtTA/wt background, which were generated similar to Stadtfeld et al.Citation60 Reprogramming experiments were performed using retroviruses generated from a pMX backbone encoding Oct4, Sox2, c-Myc or Klf4 cDNAs as described in Sridharan et al.Citation56 and conducted in media containing 15% serum. Reprogramming was induced in stemcca MEFs, with doxycycline at 1µg/ml. Oct4-GFP positive colonies were isolated and used for further expansion to ensure that all cells used for histone extraction were fully reprogrammed. Cells were harvested by centrifugation at 600 rcf, flash frozen, and stored at -80 °C at 0, 12, and 21 d over the course of reprogramming.
Histone extraction, digestion and chemical derivatization
Cell pellets were thawed on ice before nuclei isolation and histone extractions as previously described.Citation61 Briefly, cells were lysed using NP-40 in nuclei isolation buffer with 5 μmol/l microcystin, 0.3 mmol/l 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF) and 10 mmol/l sodium butyrate. Histones were isolated from nuclei by extraction with 0.4 N H2SO4, precipitated with trichloroacetic acid, washed in acetone, dried overnight, and resuspended in water. Histone samples were digested in-solution with trypsin following chemical propionylation as previously described.Citation62 Briefly, histones were resuspended in 20 µl in ammonium bicarbonate (pH 8). Ten microliters of propionic anyhdyride reagent (3:1 isopropanol:propionic anhydride) was added, and the sample vortexed and pulse-centrifuged for several seconds. Ammonium hydroxide was added dropwise to ensure that the reaction maintained a pH 8 value. This reaction was incubated at 37 °C for 15 min and dried down to approximately 5 µl in a SpeedVac concentrator. This propionylation step was repeated and samples were digested with trypsin in a 20:1 (wt/wt) protein:trypsin ratio and incubated at 37 °C for 5 h. The digested peptides were subjected to two more rounds of propionic anhydride derivatization. The reconstituted samples were desalted using homemade STAGE tips, and prepared for MS analysis, as previously described.Citation34
Histone analysis by nano-LC-MS/MS
MS sequencing analysis of sample histone peptides was performed as previously described.Citation62 Briefly, samples were loaded via an Eksigent autosampler (Eksigent Technologies Inc.) onto a 75 µm C18 reverse phase capillary column packed with Magic C18 reversed-phase 5μm particles and constructed with an integrated electrospray ionization tip. Samples were resolved using an 110 min 1–100% buffer B gradient (buffer A = 0.1 mol/L acetic acid, Buffer B = 70% acetonitrile in 0.1 mol/L acetic acid at 0.070 µl/min on an Agilent HPLC system (Agilent). The peptides were directly electrosprayed into and analyzed by an LTQ-Orbitrap XL (ThermoFisher Scientific). Full scan MS spectra were acquired from over an m/z range 200–1500 at 30 000 resolution in the Orbitrap after accumulation of approximately 500 000 ions, followed by 5 to 7 data-dependent MS/MS spectra collected in the ion trap (CAD energy 40%) following accumulation of 10 000 ions. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 30 s and a relative mass window of < 1 Da. MS/MS spectra were searched using the algorithm, PILOT_PTMCitation23,Citation63 identifies potential protein modifications by permuting all known PTMs, chemical derivatives, and artifacts found in the UniMod,Citation64 RESID,Citation65 and Delta MassCitation66 databases (over 900 entries) for each residue of a peptide and comparing it to the unmodified tandem MS data for a human histone H3 template sequence (GI:1894787). Propionylation (+56.026 Da) on the N-terminus of the peptides was set as a fixed modification. For histone PTM searches, propionylation (+56.026 Da), acetylation (+42.010 Da), mono- (+70.042 Da), di- (+28.031 Da), and trimethylation (+42.046 Da) were selected as variable modifications on tryptic peptides (up to 3 missed cleavages). Parent mass tolerance was set to 0.1 Da and fragment ion tolerance was set to 0.5 Da and results filtered to a 1% FDR. All the identified novel modified peptides were manually inspected according to the rules described previously.Citation67 We also performed MS experiments where we targeted specific precursor ions for MS/MS analysis of calculated O-acetylated peptides (hypothesis driven MS) to check for the presence of very low level O-acetylated peptides that could be missed in a standard data-dependent MS/MS experiment.
MS and MS/MS analysis
A peptide which mimicked the H3 1–21 peptide with S10ac was synthesized in house (Princeton University Mass Spectrometry Facility) using standard Fmoc chemistry and was purified to > 98% analytical purity using RP-HPLC. For comparison to the endogenous H3S10ac peptide, the synthetic lyophilized peptide was reconstituted in ammonium bicarbonate, chemically derivatized with propionic anhydride, digested with trypsin and analyzed by nano-LC-MS/MS using the conditions listed above for sequencing comparison to the in vivo peptide. For these experiments, all H3 9–17 modified peaks were manually validated.
The abundance of each modified peptide was determined by manual chromatographic peak integration of full MS scans using Qual Browser software (ThermoFisher Scientific, Inc.). To quantify the relative abundance of each O-acetylated peptide, the area under the curve for each peak is expressed as a percentage of the total histone peptide (sum of all forms of a particular peptide) for 3–5 biological replicates as previously described.Citation61,Citation62 The statistical significance of the relative abundance of each histone modification as compared with the relative abundance of other existing histone PTMs was calculated using a two-tailed, unpaired Student’s t-test, and the associated P-values were generated using Microsoft Excel.Citation68
Immunoaffinity purification of anti-H3S10ac antibody
The anti-H3S10ac IgG was generated by Proteintech Group by immunizing 3 rabbits with 2 serine O-acetylated peptides mimicking the H3 7–14 sequence, in tandem (ARKS[OAc]TGGKARKS[OAc]TGGKC). The polyclonal serum was further purified by column chromatography with Affi-Gel 10 beads (Bio-Rad). The peptide H3S10ac peptide (ARTKQTARKS[OAc]TGGKAPRKQLA) was conjugated to the agarose beads as described.Citation69 The H3S10ac-specific antibodies were separated from other non-specific binders by column chromatography and then eluted from the column under high pH. This column purification step was repeated with Affi-Gel 10 beads conjugated to an unmodified histone H3 tail peptide and the flow-through was collected, analyzed by Bradford® for protein concentration and tested by Dot Blot analysis against a panel of variously modified and unmodified forms of the H3 9–17 peptide.Citation70 An amount of 5–7 µg of acid extracted bulk histone protein from iPSCs were resolved in an 18% SDS-PAGE. The H3S10ac signal was detected by western blot analysis with the α-H3S10ac antibody. The signal specificity was tested by competition experiments in which the primary antibody was pre-incubated for 2 h at 4 °C with both the aforementioned synthetic H3S10ac peptide and the complementary unmodified peptide, independently.
Propidium iodide staining and flow cytometry
After ethanol fixation, cells were washed with phosphate buffered saline. Cells were incubated with 0.08 mg/ml propidium iodide (Roche) and 0.02 mg/ml RNAase in phosphate buffered saline for over 1 h with minimal light exposure at room temperature before flow cytometric analysis, as performed by the Princeton University Flow Cytometry Core Facility.
Immunofluorescence
Immunofluorescence labeling was done on pre-extracted cells. Unless otherwise noted all steps were done at room temperature. For pre-extraction, cells grown overnight on 1.5 coverslips (Fisherbrand) were washed with 1X PBS and incubated in CSK buffer (10 mM PIPES pH 7.0, 100 mM, 300 mM Sucrose, 3 mM MgCl2) + Triton X-100 (0.5% for U2OS and HeLa cells, and 0.25% for ESCs) + protease inhibitors (1mM PMSF, 1 µg/mL Leupeptin, 1 µg/mL Pepstatin, 1 µg/mL Aprotinin) on ice. The U2OS and HeLa cells were pre-extracted (3 min), rinsed 2× with 1× PBS, fixed in 3% formaldehyde (15 min), and rinsed 2× with 1× PBS. The ESCs were pre-extracted (5 min), rinsed 2× with 1X PBS, fixed in 3% formaldehyde (15 min), washed 2× with 1X PBS, permeabilized with 1X PBS + 0.1% Triton X-100 (5 min), and rinsed in 1× PBS. All of the cell types were blocked in 1× PBS + 3% BSA (1 h), incubated with α-H3S10ac antibody (1:50 in 1X PBS + 3% BSA) in a damp chamber (1 h), washed 2× in 1× PBS, and incubated with the secondary antibody (Donkey α-Rabbit AF594; 1:2000; Invitrogen A21207), diluted in 1X PBS (1 h). Cells were washed 2X in 1X PBS, incubated with DAPI (1 µm) diluted in 1X PBS (4 min), and rinsed with 1X PBS. Coverslips were immediately mounted on slides in antifade fluorescence mounting medium (1 mg/ml p-phenylenediamine, 90% glycerol in PBS, pH 8.0–9.0 adjusted with Sodium Carbonate / Bicarbonate Buffer, pH 9.2) and the edges were sealed with nail polish.
Imaging
Images were acquired using a Leica TCS SP5 II confocal microscope with a HCX PL APO 63×/1.40–0.60 oil CS lens using the 561 DPSS and 405 diode lasers, and 6× zoom (Leica Microsystems). For U2OS and HeLa cells, 6 images were acquired at 0.5 µm intervals, and for ESCs, 10 images were acquired at 0.5 µm intervals. Maximum projections are presented in the Results above.
| Abbreviations: | ||
| PTMs | = | posttranslational modifications |
| HPLC-MS | = | high performance liquid chromatography mass spectrometry |
| MEFs | = | mouse embryonic fibroblasts |
| mESCs | = | mouse embryonic stem cells |
| iPSCs | = | induced pluripotent stem cells |
Additional material
Download Zip (124.1 KB)Acknowledgments
S.M.J and A.M. are supported by R01 GM 093000-02. Imaging was done in the Wistar Cancer Center Imaging Facility supported by The Wistar Cancer Center Core Grant (P30 CA10815). BAG acknowledges funding from a National Science Foundation (NSF) Early Faculty CAREER award, NSF grant CBET-0941143, and an NIH Innovator grant (DP2OD007447) from the Office of the Director, National Institutes of Health. We thank Sean Taverna for the Tetrahymena histone samples.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: http://www.landesbioscience.com/journals/epigenetics/article/26025
References
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet 2009; 43:559 - 99; http://dx.doi.org/10.1146/annurev.genet.032608.103928; PMID: 19886812
- Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293:1074 - 80; http://dx.doi.org/10.1126/science.1063127; PMID: 11498575
- Karlić R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci U S A 2010; 107:2926 - 31; http://dx.doi.org/10.1073/pnas.0909344107; PMID: 20133639
- Yan C, Boyd DD. Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol Cell Biol 2006; 26:6357 - 71; http://dx.doi.org/10.1128/MCB.00311-06; PMID: 16914722
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 2009; 138:1122 - 36; http://dx.doi.org/10.1016/j.cell.2009.07.031; PMID: 19766566
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315 - 26; http://dx.doi.org/10.1016/j.cell.2006.02.041; PMID: 16630819
- Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci U S A 2005; 102:5501 - 6; http://dx.doi.org/10.1073/pnas.0500136102; PMID: 15795371
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006; 311:844 - 7; http://dx.doi.org/10.1126/science.1124000; PMID: 16469925
- Britton LM, Gonzales-Cope M, Zee BM, Garcia BA. Breaking the histone code with quantitative mass spectrometry. Expert Rev Proteomics 2011; 8:631 - 43; http://dx.doi.org/10.1586/epr.11.47; PMID: 21999833
- Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci U S A 2010; 107:19915 - 20; http://dx.doi.org/10.1073/pnas.1009023107; PMID: 21045127
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011; 146:1016 - 28; http://dx.doi.org/10.1016/j.cell.2011.08.008; PMID: 21925322
- Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics 2007; 6:812 - 9; http://dx.doi.org/10.1074/mcp.M700021-MCP200; PMID: 17267393
- Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc Natl Acad Sci U S A 2007; 104:60 - 5; http://dx.doi.org/10.1073/pnas.0606775103; PMID: 17190813
- Messner S, Hottiger MO. Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol 2011; 21:534 - 42; http://dx.doi.org/10.1016/j.tcb.2011.06.001; PMID: 21741840
- Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y. Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics 2012; 11:100 - 7; http://dx.doi.org/10.1074/mcp.M111.015875; PMID: 22389435
- Nelson CJ, Santos-Rosa H, Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 2006; 126:905 - 16; http://dx.doi.org/10.1016/j.cell.2006.07.026; PMID: 16959570
- Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 2006; 312:1211 - 4; http://dx.doi.org/10.1126/science.1126867; PMID: 16728640
- Lu Z, Cheng Z, Zhao Y, Volchenboum SL. Bioinformatic analysis and post-translational modification crosstalk prediction of lysine acetylation. PLoS One 2011; 6:e28228; http://dx.doi.org/10.1371/journal.pone.0028228; PMID: 22164248
- Zhang K, Chen Y, Zhang Z, Tao S, Zhu H, Zhao Y. Unrestrictive identification of non-phosphorylation PTMs in yeast kinases by MS and PTMap. Proteomics 2010; 10:896 - 903; PMID: 20049863
- Prigent C, Dimitrov S. Phosphorylation of serine 10 in histone H3, what for?. J Cell Sci 2003; 116:3677 - 85; http://dx.doi.org/10.1242/jcs.00735; PMID: 12917355
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 1999; 97:99 - 109; http://dx.doi.org/10.1016/S0092-8674(00)80718-7; PMID: 10199406
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 2005; 438:1116 - 22; http://dx.doi.org/10.1038/nature04219; PMID: 16222246
- Baliban RC, DiMaggio PA, Plazas-Mayorca MD, Young NL, Garcia BA, Floudas CA. A novel approach for untargeted post-translational modification identification using integer linear optimization and tandem mass spectrometry. Mol Cell Proteomics 2010; 9:764 - 79; http://dx.doi.org/10.1074/mcp.M900487-MCP200; PMID: 20103568
- Zhang J, Chen Y, Zhang Z, Xing G, Wysocka J, Zhao Y. MS/MS/MS reveals false positive identification of histone serine methylation. J Proteome Res 2010; 9:585 - 94; http://dx.doi.org/10.1021/pr900864s; PMID: 19877717
- Xiong L, Ping L, Yuan B, Wang Y. Methyl group migration during the fragmentation of singly charged ions of trimethyllysine-containing peptides: precaution of using MS/MS of singly charged ions for interrogating peptide methylation. J Am Soc Mass Spectrom 2009; 20:1172 - 81; http://dx.doi.org/10.1016/j.jasms.2009.02.014; PMID: 19303795
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol 2008; 26:1269 - 75; http://dx.doi.org/10.1038/nbt.1502; PMID: 18849973
- Tweedie-Cullen RY, Brunner AM, Grossmann J, Mohanna S, Sichau D, Nanni P, Panse C, Mansuy IM. Identification of combinatorial patterns of post-translational modifications on individual histones in the mouse brain. PLoS One 2012; 7:e36980; http://dx.doi.org/10.1371/journal.pone.0036980; PMID: 22693562
- Healy S, Heightman TD, Hohmann L, Schriemer D, Gravel RA. Nonenzymatic biotinylation of histone H2A. Protein Sci 2009; 18:314 - 28; http://dx.doi.org/10.1002/pro.37; PMID: 19160459
- Mano N, Kasuga K, Kobayashi N, Goto J. A nonenzymatic modification of the amino-terminal domain of histone H3 by bile acid acyl adenylate. J Biol Chem 2004; 279:55034 - 41; http://dx.doi.org/10.1074/jbc.M409205200; PMID: 15465822
- Jaisson S, Gillery P. Evaluation of nonenzymatic posttranslational modification-derived products as biomarkers of molecular aging of proteins. Clin Chem 2010; 56:1401 - 12; http://dx.doi.org/10.1373/clinchem.2010.145201; PMID: 20562349
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 2008; 40:897 - 903; http://dx.doi.org/10.1038/ng.154; PMID: 18552846
- Zhang J, Chen Y, Zhang Z, Xing G, Wysocka J, Zhao Y. MS/MS/MS reveals false positive identification of histone serine methylation. J Proteome Res 2010; 9:585 - 94; http://dx.doi.org/10.1021/pr900864s; PMID: 19877717
- Jung SY, Li Y, Wang Y, Chen Y, Zhao Y, Qin J. Complications in the assignment of 14 and 28 Da mass shift detected by mass spectrometry as in vivo methylation from endogenous proteins. Anal Chem 2008; 80:1721 - 9; http://dx.doi.org/10.1021/ac7021025; PMID: 18247584
- Zee BM, Britton LM, Wolle D, Haberman DM, Garcia BA. Origins and formation of histone methylation across the human cell cycle. Mol Cell Biol 2012; 32:2503 - 14; http://dx.doi.org/10.1128/MCB.06673-11; PMID: 22547680
- Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc Natl Acad Sci U S A 1998; 95:7480 - 4; http://dx.doi.org/10.1073/pnas.95.13.7480; PMID: 9636175
- Polioudaki H, Markaki Y, Kourmouli N, Dialynas G, Theodoropoulos PA, Singh PB, Georgatos SD. Mitotic phosphorylation of histone H3 at threonine 3. FEBS Lett 2004; 560:39 - 44; http://dx.doi.org/10.1016/S0014-5793(04)00060-2; PMID: 14987995
- Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science 2010; 330:239 - 43; http://dx.doi.org/10.1126/science.1194498; PMID: 20929775
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell 2000; 5:917 - 26; http://dx.doi.org/10.1016/S1097-2765(00)80257-9; PMID: 10911986
- Eberlin A, Grauffel C, Oulad-Abdelghani M, Robert F, Torres-Padilla ME, Lambrot R, Spehner D, Ponce-Perez L, Würtz JM, Stote RH, et al. Histone H3 tails containing dimethylated lysine and adjacent phosphorylated serine modifications adopt a specific conformation during mitosis and meiosis. Mol Cell Biol 2008; 28:1739 - 54; http://dx.doi.org/10.1128/MCB.01180-07; PMID: 18180282
- Zee BM, Levin RS, Dimaggio PA, Garcia BA. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin 2010; 3:22; http://dx.doi.org/10.1186/1756-8935-3-22; PMID: 21134274
- Johansen KM, Johansen J. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res 2006; 14:393 - 404; http://dx.doi.org/10.1007/s10577-006-1063-4; PMID: 16821135
- Paquette N, Conlon J, Sweet C, Rus F, Wilson L, Pereira A, Rosadini CV, Goutagny N, Weber AN, Lane WS, et al. Serine/threonine acetylation of TGFβ-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc Natl Acad Sci U S A 2012; 109:12710 - 5; http://dx.doi.org/10.1073/pnas.1008203109; PMID: 22802624
- Rampakakis E, Di Paola D, Chan MK, Zannis-Hadjopoulos M. Dynamic changes in chromatin structure through post-translational modifications of histone H3 during replication origin activation. J Cell Biochem 2009; 108:400 - 7; http://dx.doi.org/10.1002/jcb.22266; PMID: 19585526
- Casas-Delucchi CS, van Bemmel JG, Haase S, Herce HD, Nowak D, Meilinger D, Stear JH, Leonhardt H, Cardoso MC. Histone hypoacetylation is required to maintain late replication timing of constitutive heterochromatin. Nucleic Acids Res 2012; 40:159 - 69; http://dx.doi.org/10.1093/nar/gkr723; PMID: 21908399
- Kemp MG, Ghosh M, Liu G, Leffak M. The histone deacetylase inhibitor trichostatin A alters the pattern of DNA replication origin activity in human cells. Nucleic Acids Res 2005; 33:325 - 36; http://dx.doi.org/10.1093/nar/gki177; PMID: 15653633
- Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol 2004; 24:4769 - 80; http://dx.doi.org/10.1128/MCB.24.11.4769-4780.2004; PMID: 15143171
- Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics 2009; 8:2266 - 84; http://dx.doi.org/10.1074/mcp.M900238-MCP200; PMID: 19654425
- Voigt P, LeRoy G, Drury WJ 3rd, Zee BM, Son J, Beck DB, Young NL, Garcia BA, Reinberg D. Asymmetrically modified nucleosomes. Cell 2012; 151:181 - 93; http://dx.doi.org/10.1016/j.cell.2012.09.002; PMID: 23021224
- Gorovsky MA, Yao MC, Keevert JB, Pleger GL. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol 1975; 9:311 - 27; http://dx.doi.org/10.1016/S0091-679X(08)60080-1; PMID: 805898
- Allis CD, Dennison DK. Identification and purification of young macronuclear anlagen from conjugating cells of Tetrahymena thermophila. Dev Biol 1982; 93:519 - 33; http://dx.doi.org/10.1016/0012-1606(82)90139-7; PMID: 7141113
- Taverna SD, Ueberheide BM, Liu Y, Tackett AJ, Diaz RL, Shabanowitz J, Chait BT, Hunt DF, Allis CD. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc Natl Acad Sci U S A 2007; 104:2086 - 91; http://dx.doi.org/10.1073/pnas.0610993104; PMID: 17284592
- Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci U S A 2006; 103:6988 - 93; http://dx.doi.org/10.1073/pnas.0601676103; PMID: 16627621
- Gardner KE, Zhou L, Parra MA, Chen X, Strahl BD. Identification of lysine 37 of histone H2B as a novel site of methylation. PLoS One 2011; 6:e16244; http://dx.doi.org/10.1371/journal.pone.0016244; PMID: 21249157
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663 - 76; http://dx.doi.org/10.1016/j.cell.2006.07.024; PMID: 16904174
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature 2008; 454:49 - 55; http://dx.doi.org/10.1038/nature07056; PMID: 18509334
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell 2009; 136:364 - 77; http://dx.doi.org/10.1016/j.cell.2009.01.001; PMID: 19167336
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol 2008; 6:e253; http://dx.doi.org/10.1371/journal.pbio.0060253; PMID: 18942890
- Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 2009; 5:491 - 503; http://dx.doi.org/10.1016/j.stem.2009.09.012; PMID: 19818703
- Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka IR, Paddison PJ. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells 2009; 27:2979 - 91; PMID: 19785031
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2008; 2:230 - 40; http://dx.doi.org/10.1016/j.stem.2008.02.001; PMID: 18371448
- Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. J Biol Chem 2010; 285:3341 - 50; http://dx.doi.org/10.1074/jbc.M109.063784; PMID: 19940157
- Plazas-Mayorca MD, Zee BM, Young NL, Fingerman IM, LeRoy G, Briggs SD, Garcia BA. One-pot shotgun quantitative mass spectrometry characterization of histones. J Proteome Res 2009; 8:5367 - 74; http://dx.doi.org/10.1021/pr900777e; PMID: 19764812
- Baliban RC, Dimaggio PA, Plazas-Mayorca MD, Garcia BA, Floudas CA. PILOT_PROTEIN: identification of unmodified and modified proteins via high-resolution mass spectrometry and mixed-integer linear optimization. J Proteome Res 2012; 11:4615 - 29; http://dx.doi.org/10.1021/pr300418j; PMID: 22788846
- Creasy DM, Cottrell JS. Unimod: Protein modifications for mass spectrometry. Proteomics 2004; 4:1534 - 6; http://dx.doi.org/10.1002/pmic.200300744; PMID: 15174123
- Garavelli JS. The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics 2004; 4:1527 - 33; http://dx.doi.org/10.1002/pmic.200300777; PMID: 15174122
- Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods 2007; 4:798 - 806; http://dx.doi.org/10.1038/nmeth1100; PMID: 17901869
- Chen Y, Kwon SW, Kim SC, Zhao Y. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J Proteome Res 2005; 4:998 - 1005; http://dx.doi.org/10.1021/pr049754t; PMID: 15952748
- Leroy G, Chepelev I, Dimaggio PA, Blanco MA, Zee BM, Zhao K, Garcia BA. Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol 2012; 13:R68; http://dx.doi.org/10.1186/gb-2012-13-8-r68; PMID: 22897906
- Laboratories B-R. Instruction Manual. Affi-Gel 10 Activated Immunoaffinity Supports. Hercules, CA, 2012.
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 10 12 Forthcoming 2011