Abstract
Disorders of human communication abilities can be classified into speech and language disorders. Speech disorders (e.g., dyspraxia) affect the sound generation and sequencing, while language disorders (e.g., dyslexia and specific language impairment, or SLI) are deficits in the encoding and decoding of language according to its rules (reading, spelling, grammar). The diagnosis of such disorders is often complicated, especially when a patient presents more than one disorder at the same time. The present review focuses on these challenges. We have combined data available from the literature with an in silico approach in an attempt to identify putative miRNAs that may have a key role in dyspraxia, dyslexia and SLI. We suggest the use of new miRNAs, which could have an important impact on the three diseases. Further, we relate those miRNAs to the axon guidance pathway and discuss possible interactions and the role of likely deregulated proteins. In addition, we describe potential differences in expressional deregulation and its role in the improvement of diagnosis. We encourage experimental investigations to test the data obtained in silico.
Keywords: :
Introduction
Disorders of human communication abilities are generally classified into speech and language disorders. Speech disorders (e.g., dyspraxia) affect the sound generation and sequencing, while language disorders (e.g., dyslexia and specific language impairment [SLI] are deficits in the encoding and decoding of language according to its rules (reading, spelling, and grammar). The diagnosis of such disorders often presents significant challenges in how often the affected children present otherwise a normal development. The situation is further complicated in distinguishing speech from language disorders in individuals affected by both speech and language disorders. To date, several target genes associated with dyslexia, SLI and dyspraxia have been identified by a number of studies.Citation1,Citation2 Common genes associated with dyslexia include GC-Rich Sequence DNA-Binding Factor 2 (C2orf3), Doublecortin Domain Containing 2 (DCDC2), Dyslexia Susceptibility 1 Candidate 1 (DYX1C1), Dyslexia-Associated Protein KIAA0319 (KIAA0319), Dyslexia-Associated Protein KIAA0319-Like Protein (KIAA0319L), Mitochondrial Ribosomal Protein L19 (MRPL19), and Roundabout Axon Guidance Receptor Homolog 1 (ROBO1). Genes associated with dyspraxia include N-Acetylglucosamine-1-Phosphodiester Alpha-N-Acetylglucosaminidase (NAGPA), Forkhead box protein P2 (FOXP2). Genes associated with SLI include Contactin-associated protein-like 2 (CNTNAP2), ATPase Ca2+ Transporting Type 2C Member 2 (ATP2C2) and C-Maf Inducing Protein (CMIP).Citation2 The identification of novel biomarkers, however, would be crucial for a better understanding of the affected molecular pathways and for improvement of dyslexia, dyspraxia, and SLI diagnosis in affected individuals. The present review focuses on this problem. Using data concerning deregulated proteins in dyslexia, dyspraxia, and SLI, we aim at identifying miRNAs associated with dyslexia and/or dyspraxia, and SLI and finding possible new biomarkers useful for diagnosis and distinction of these disorders. In recent years, research throughout the world focused much on the roles of miRNAs in the most different physiological processes. miRNAs are a group of small non-coding RNAs (18–25 bp after maturation). They are known to perform a unique task of post transcriptional gene regulation by causing, depending on various grades of complementarities, protein synthesis blocking or mRNA degradation.Citation3 Since their discovery, miRNAs have been known to regulate the expression of a large number of proteins; it is now thought that they could regulate up to 30% (or even more) of the human genome. Despite their important physiological and pathological roles, miRNAs could play a relevant role in diagnosis and prognosis of a large number of diseases, as the qualitative and quantitative miRNA composition varies in different tissues and depends on the health status. A great advantage in the research of miRNAs for prognostic and diagnostic purposes comes from a variety of software available online. In recent years, different algorithms have been developed to predict the role of miRNAs expressed in humans, Drosophila, and plants, among other organisms. In order to identify mRNA targets common to more than one miRNA in faster and more efficient ways, and to identify new miRNAs modulated in specific pathways, we have developed a computer program named SID1.0 (simple String Identifier) and successfully applied it in the identification of deregulated miRNAs.Citation3 This simple program of string identification has proven to be a very useful tool for predicting miRNAs involved in pathogenesis, thereby representing a potential tool in diagnostics and clinical monitoring. From a general stand point, SID1.0 simply implements an exhaustive search strategy. Its merit relies upon the biological application. As far as we know, this is the first software designed to filter data retrieved from available databases (e.g., TargetScan, miRanda, RNAhybrid, PicTar, DIANA-MicroT 3.0), allowing the recovery of additional information not directly available from the providers.
Genes Known for their Association with Speech and Language Disorders
From the literature, several genes appear to be altered in dyslexia (C2orf3, DCDC2, DYX1C1, KIAA0319, KIAA0319L, MRPL19, ROBO1), dyspraxia (NAGPA, FOXP2), and SLI (CNTNAP2, ATP2C2, CMIP). The function of C2orf3 is still not known; however, it is highly expressed in the brain. MRPL19, a gene located in the same locus as C2orf3 on chromosome 2, seems to participate in mitochondrial energy metabolism and its expression is associated to dyslexia as well.Citation4 DCDC2 and KIAA0319 are located on chromosome 6. DCDC2 has recently been shown to affect ciliary signaling and, when overexpressed, to alternate the morphology of ciliated neurons.Citation5 KIAA0319 has been shown to be under expressed in dyslexia and seems to be involved in neuronal migration, so that its decreased levels cause a morphology change in migrating neurons.Citation6 KIAA0319L, a homolog of KIAA0319, could play a similar function in dyslexia; however, its role remains unclear.Citation7 DYX1C1 is one of the main candidate genes to be altered in dyslexia.Citation8 Similarly to other candidate genes involved in dyslexia, DYX1C1 plays a role in neuronal migration, being required for the transition out of the multipolar stage of migration.Citation9 ROBO1 has also been described as a candidate dyslexia gene; however, its involvement might be restricted to specific genotypes and isolated cases. ROBO1 is a member of the immunoglobulin gene superfamily. Its function is related to axon guidance and neuronal precursor cell migration, as it is activated by SLIT proteins, resulting in a repulsive effect on glioma cell guidance in the developing brain.Citation10 Concerning dyspraxia, two main candidate genes have been described. NAGPA encodes a N-acetylglucosamine-1-phosphodiester α-N-acetylglucosaminidase that is a component of the lysosomal enzyme-targeting pathway and cleaves α-N-acetylglucosamine moieties uncovering mannose-6-phosphate moieties and targeting the processed enzymes to the lysosomes.Citation11 FOXP2, mostly known as a marker for dyspraxia, encodes for a transcriptional regulator controlling neuron growth and differentiation.Citation12 Three candidate genes, CNTNAP2, ATP2C2, and CMIP, have been described for SLI. CNTNAP2 encodes a contactin-associated protein-like 2 with functions in cell adhesion. Interestingly, CNTNAP2 is known to be downregulated by FOXP2.Citation13 CMIP and ATP2C2 are located on chromosome 16. The precise functions of CMIP in the brain, however, are not clear, as CMIP presents different splicing isoforms and is predominantly known as part of the T-cell signaling pathway.Citation14 ATP2C2 encodes the secretory pathway Ca2+, Mn2+ transporting ATPase (SPCA2) and its alteration could affect the translocation of enzymes to the lysosomes.Citation15
Identification of Putative miRNAs Associated with Speech and Language Disorders by SID1.0 Analysis
From the TargetScan database, we obtained the predicted miRNAs for the above-described target genes.Citation16 The predicted miRNAs are indicated with a specific gene ID system (Refseq ID). For each target gene, a data set (i.e., a group list of Refseq IDs) of the predicted targeted miRNAs was created. Since a visual inspection of the IDs would be unpractical, due to their large number (in some cases IDs could be in the thousands), they have been automatically indexed using a program written in Fortran that looks for Refseq IDs shared by the predicted target genes of the different data sets. SID1.0 is based on an algorithm of sequential exhaustive search that has been implemented in Fortran 90 using a straightforward approach.Citation3 SID1.0 performs an exhaustive search within each individual one-column ASCII input file and reports the result (i.e., the number of common targets) on an ASCII output file in the form of a table that summarizes the common IDs. Thus, the main advantage of SID1.0, which works as a filter on the information provided by the web pages hosting the miRNA databases, is that it is completely independent from the algorithms on which the databases rely upon. In this way, our procedure builds upon the prediction algorithms used in the databases, whose outputs are scrutinized by SID1.0. This latter, has been developed and tested in a Mac OS X environment, and is currently compiled using the Gnu Fortran compiler. For each gene, a data set of the miRNAs predicted to target a gene was created. Furthermore, for a defined miRNA name, target genes can be automatically retrieved from the DIANA-microT 3.0 database. A list of gene names or a list of RefSeq IDs are provided and the program translates them into Ensembl IDs.Citation17
Common miRNAs Associated with Speech and Language Disorders
By using our SID1.0 predicting tool, we identified the common miRNAs corresponding to the above described target genes. These miRNAs are shown in .Citation18 hsa-miR-548c-3p was the only miRNA found in common to all the dyslexia genes. Different common miRNAs were found in dyspraxia (hsa-miR-182; hsa-miR-34c-5p; hsa-miR-34a; hsa-miR-449a; hsa-miR-449b; hsa-miR-1271; hsa-miR-96; hsa-miR-9; hsa-miR-647; hsa-miR-604; hsa-miR-214; hsa-miR-657) and in SLI (hsa-miR-1207-5p; hsa-miR-188-3p; hsa-miR-1225-3p; hsa-miR-299-3p). As indicated by these results, there were no common miRNAs between all the genes analyzed. When we tried to combine the analysis of all these groups we found that some of the previous miRNAs were common to most of the genes analyzed. Combining dyslexia and dyspraxia genes, hsa-miR-548c-3p was the most common miRNA. The same miRNA was obtained by combining dyslexia and SLI genes, but also by combining all the groups. Differently, hsa-miR-1207-5p was the most common miRNA in the combination of dyspraxia and SLI genes. These results suggest that hsa-miR-548c-3p is specifically modulated in dyslexia with speech and language disorders and that hsa-miR-1207-5p is only modulated in speech and language disorders.
Table 1. Common miRNAs (RefSeq ID) of dyslexia, dyspraxia, and SLI genes
New Putative Genes Involved in Speech and Language Disorders and Related to the Axon Guidance Pathway
The identification of novel biomarkers is crucial to understanding the molecular pathways affected in dyslectic individuals and to diagnose this disorder. Many of the already known genes involved in speech and language disorders (such as ROBO-1, KIAA0319, and FOXP2, among others) are related to neuronal development and axon guidance. Apparently of central importance, the axon guidance pathway could include other key genes also important in language and speech disorders. Having identified a new set of miRNAs possibly involved in dyslexia, dyspraxia, and SLI, we wanted to identify new candidate genes involved in specific language and speech disorders. Using the DIANA-mirPath database (DIANA LAB), we identified axon guidance pathway genes that could be involved in dyslexia, dyspraxia, and SLI (). Among them, two miRNAs were chosen for their representative role. The first, hsa-miR-548c-3p, was the only miRNA common to all the dyslexia genes, and also common to the combination of dyspraxia and SLI genes. The second, hsa-miR-1207–5p, was the most common miRNA in the combination of dyspraxia and SLI genes. The miRNAs identified above could be mostly related to different target genes of the axon guidance pathway, which are not targeted by all the diseases in the same way ().
Table 2. Axon guidance pathway modulated by the identified miRNAs
Figure 1. Targets of the axon guidance pathway, possibly modulated by miRNAs in dyslexia (green bar), dyspraxia (red bar), and SLI (blue bar). The interactions are based on hsa04360 KEGG pathway description (http://www.genome.jp/dbget-bin/www_bget?pathway+hsa04360) and literature (see section 5).
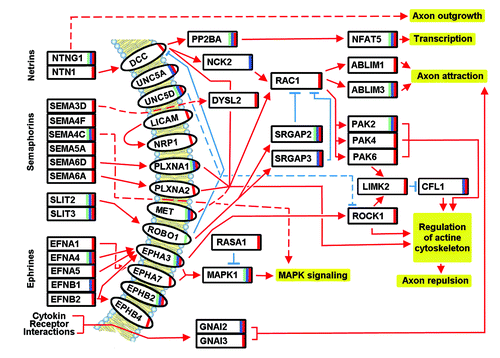
Worth mentioning are the SLIT (Slit Homolog) proteins, SLIT 2, and SLIT 3, whose expression appears to be altered in all three diseases. SLIT 2 seems to act as molecular guidance cue in cellular migration and to be essential for midline guidance by acting as a repulsive signal, preventing inappropriate midline crossing of axons.Citation19 The same seems to be valid for SLIT3.Citation20 The SLIT proteins further activate ROBO1 (a member of the immunoglobulin gene superfamily that has already described to be modulated in dyslexia). The activation of this receptor results in a repulsive effect on glioma cell guidance in the developing brain. ROBO1 may silence the effects of NTN1 (Netrin 1) by formation of a complex with DCC (deleted in colorectal carcinoma). On the other hand, it may also activate srGAPs. Two of the srGAP proteins (GTPase activating proteins) are in fact modulated by the identified miRNAs. SrGAP2 seems to be implicated in all the three diseases, while srGAP3 seems to be specific only to dyspraxia and SLI. Both may attenuate RAC1 (Ras-Related C3 Botulinum Toxin Substrate 1) signaling and regulate actin dynamics for cell migration and differentiation. They also play an important role in both axons and dendrites outgrowth, and in the maturation of dendritic spines. In addition, they stimulate the branching of the leading process, negatively regulate neuron radial migration in the cerebral cortex, and may have implications for cognition, learning, and memory.Citation21-Citation23
Another important group of target genes include netrins and their related target proteins. NTNG1 (Netrin G1) is important for the three diseases and implicated in axon and dendrite outgrowth. NTN1 seems to be deregulated only in dyspraxia. NTN1 in association with either DCC or UNC5 receptors leads to axon attraction or repulsion, respectively, and serves as a survival factor via its association with its receptors, which prevent the initiation of apoptosis.Citation24 Of the UNC5 family (Unc-5 Homolog), two receptors, UNC5A (dyspraxia and SLI) and UNC5D (all three diseases) are targeted. They both mediate axon repulsion in growth cones, caused by their association with DCC that may trigger signaling for repulsion.Citation25 DCC, another netrin receptor protein, targeted probably only in dyspraxia, mediates axon attraction of neuronal growth cones in the developing nervous system via RAC1 (targeted in dyspraxia), NCK2 (NCK Adaptor Protein 2; targeted in SLI), and the ABLIM (Actin Binding LIM) proteins. ABLIM1 may be targeted only in dyspraxia, while ABLIM3 appears to be targeted in all three diseases; both mediate interactions between actin filaments and cytoplasmic targets important in axon attraction.Citation26 Furthermore, DCC, via PP2BA (Protein Phosphatase 3, Catalytic Subunit, Alpha Isozyme) (all three diseases) induces NFAT5 (all three diseases), a member of the nuclear factors of activated T-cell family of transcription factors.
Semaphorins and their related pathways seem to be modulated predominantly in dyspraxia. In fact, only the semaphorin SEMA4C may be modulated in all three diseases, while the semaphorins likely modulated in dyspraxia include SEMA3D, SEMA4F, SEMS5A, SEMA6A, and SEMA6D. Semaphorins act through various semaphorin co-receptors, also supposedly modulated in the three diseases. While PLXNA1 (Plexin A1) seems to be modulated in all the three diseases, PLXNA2 (Plexin A2) seems to be modulated only in dyspraxia, both play a role in axon guidance, invasive growth and cell migration. Importantly, MET (Mesenchymal epithelial transition factor), NRP1 (Neutrophilin 1), L1CAM (L1 Cell Adhesion Molecule), and RAC1, implicated in the semaphorin signaling may also be modulated exclusively in dyspraxia. MET is a receptor tyrosine kinase that transduces signals from the extracellular matrix into the cytoplasm, regulating many physiological processes including proliferation, scattering, morphogenesis, and survival. MET may interact with the PLXNB receptors and activate RAC signaling.Citation27 NRP1 (neuropilin 1) may bind many ligands and co-receptors, semaphorins being one group of those. It may mediate the chemorepulsant activity of semaphorins (semaphorin 3A).Citation28 L1CAM is an axonal glycoprotein belonging to the immunoglobulin family. It has an important role as a cell adhesion molecule and plays an important role in neuronal migration and differentiation.Citation29 RAC1 is a GTPase that belongs to the RAS superfamily of small GTP-binding proteins, as mentioned above, and plays a central role in axon guidance, as it may regulate cytoskeletal reorganization, axon attraction, and repulsion. Some of its targets have been already described above, and include the ABLIM proteins, leading to axon attraction. Another group of proteins activated by RAC1 includes the PAK proteins, a family of serine/threonine p21-activating kinases, which are critical effectors that link RAC1 to cytoskeleton reorganization and nuclear signaling. Two PAK (p21 activated kinase) proteins, however, are likely additionally modulated by miRNAs in dyspraxia (PAK 4 and PAK 6) and one in all the three diseases (PAK2). PAK2 plays a role in a variety of different signaling pathways including cytoskeleton regulation, cell motility, cell cycle progression, apoptosis or proliferation.Citation30 PAK4 is a mediator of filopodia formation and may play a role in the reorganization of the actin cytoskeleton. It phosphorylates the protein phosphatase SSH1 (inactivation) and the LIMK kinases (activation), leading to increased inhibitory phosphorylation of the actin binding/depolymerizing factor cofilin and to stabilization of actin filaments. PAK4 further regulates cell motility, the assembly of focal adhesions, and actin stress fibers.Citation31 PAK6 interacts with the androgen receptor and inhibits androgen mediated gene transcription.Citation32 The kinase LIMK2 itself seems to be modulated in dyspraxia. As mentioned previously, it is phosphorylated by PAK or ROCK and, in turn, it phosphorylates cofilin (CFL1), inhibiting its actin-depolymerizing activity. It is thought that this pathway contributes to the reorganization of the actin cytoskeleton.Citation33 Cofilin may be modulated in dyspraxia and SLI. Cofilin is a widely distributed intracellular actin-modulating protein that binds and depolymerizes filamentous F-actin and inhibits the polymerization of monomeric G-actin in a pH-dependent manner. It is involved in the translocation of the actin-cofilin complex from cytoplasm to nucleus.Citation34 DPYSL2, a member of the collapsin response mediator protein family, may be altered in syspraxia and is already known to be altered in various neurological diseases (e.g., Alzheimer disease). This protein facilitates neuron guidance, growth, and polarity, and promotes microtubule assembly. It is part of semaphorin class 3 signaling and is required for growth cone collapse and also synaptic signaling through interactions with calcium channels.Citation35
Ephrins are a group of proteins that seem to show more variations in expression among the three disorders. Ephrins and ephrin-related receptors comprise the largest subfamily of receptor protein-tyrosine kinases and are implicated in nervous system development. Based on their structures, ephrins are divided into EFNA, which are anchored to the membrane by a glycosyl-phosphatidyl-inositol linkage, and EFNB, which are transmembrane proteins. EFNA4 may be deregulated in all three disorders, and is crucial for migration, repulsion and adhesion during neuronal development.Citation36 EFNB1 (Ephrin B1) plays a role in constraining the orientation of longitudinally projecting axons and may be deregulated in SLI and dyspraxia, while EFNA1, EFNA5, and EFNB2 seem to be modulated in their expression only in dyspraxia.Citation37 Further targets in the ephrin signaling include the ephrin receptors. The ephrin receptors are divided into two groups based on the similarity of their extracellular domains and their affinities for EFNA or EFNB. In our analysis, four ephrin receptors seem to be modulated in the disorders, two of them, EPHA3 and EPHB2, in all three disorders and two of them, EPHA7 and EPHB4, only in dyspraxia. EPHA3 binds predominantly EFNA5 and plays a role in the segregation of motor and sensory axons during neuromuscular circuit development.Citation38 EPHB2 is involved in the guidance of commissural axons that form a major interhemispheric connection of the cerebral cortex.Citation39 EPHA7 binds predominantly EFNA5 and their interaction regulates brain development, having a repellent activity on axons and being able to induce caspase 3 dependent apoptosis. EPHA7 may induce different MAPK signaling pathways.Citation40 EPHB4 binds predominantly EFNB2. EPHB4-mediated signaling controls cellular repulsion and segregation form EFNB2-expressing cells.Citation41 As it has been seen before, EPHA receptors may activate MAPK signaling. However, MAPK1 is likely modulated by miRNAs in all three disorders. MAPK1 is a member of the MAP kinase family and may act as an integration point for multiple biochemical signals. The activation of this kinase requires its phosphorylation by upstream kinases. After activation, this kinase moves to the nucleus and phosphorylates nuclear targets. Serine/threonine kinases act as essential components of the MAP kinase signal transduction pathway and may affect different cellular functions such as cell growth, adhesion, and cytoskeletal rearrangements.Citation42 Further targets, not directly included in the previously described networks, are RASA1, a GTPase activating protein modulated in dyspraxia, and two G proteins, GNAI2 and GNAI3, the first modulated in SLI and dyspraxia and the second only in dyspraxia.
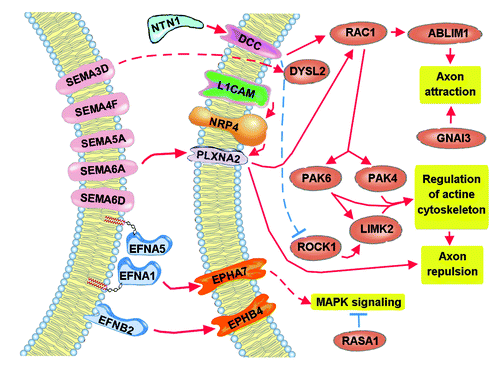
In summary, many targets of the axon guidance pathway, specifically of the netrin, semaphorin, and ephrin signaling, may be modulated in dyslexia, dyspraxia, and SLI. Furthermore, it should be noted that target distribution is not uniform among the three diseases. In fact, our investigation has identified a good number of targets that appear to be specifically modulated in dyspraxia, which could be further investigated and validated and could be of great interest in better understanding the differences between the different speech and language disorders and in the development of better and more specific diagnostic tools. summarizes the target genes (and their interactions) that could be specifically altered in dyspraxia. As it can be seen, semaphorin signaling and, partly, ephrin signaling seem to form the core of alterations, suggesting that a variety of semaphorins are altered in dyspraxia, an assumption that needs to be further confirmed by comparing with other speech and language disorders, and that could be of great importance for a better understanding of the specific pathophysiology of dyspraxia. Future target validation of additional putative miRNA targets could hopefully clear the remaining question marks and missing connections and enable a better understanding of speech and language disorders.
Figure 2. Targets of the axon guidance pathway likely altered in dyspraxia. The comparison of with denotes the possible importance of semaphorin signaling in the pathology of dyspraxia, while ephrin and netrin signaling are present too. As can be seen, the pathways lead to a favoring of axon repulsion through the PAK kinases and to axon attraction through RAC1; therefore, possible loss of balance between axon repulsion and axon attraction could be the result of deregulated expression of the semaphorin signaling.
Conclusions
In the present review we aimed to identify miRNAs that may have a key role in dyspraxia, dyslexia, and SLI, using miRNA predictions of TargetScan database and SID1.0, a software specifically developed to interrelate database data. We were able to identify new miRNAs with putative important roles in the alteration of specific target gene expression, for the three disorders. Further experimental validation of these new miRNAs will be of great interest to further understand the pathophysiology and epigenetics of speech and language disorders and the potential differences in miRNA profiles that could lead to a better diagnostic approach. The new miRNAs have been further investigated in an in silico analysis of targets involved in the axon guidance pathway. A number of targets could be identified that are involved mostly in netrin, ephrin, and semaphorin signaling. A clear involvement of these targets in axon guidance should be of big interest to further investigate and validate the alteration of these targets in specific speech and language disorders. Our analysis suggests differences in expression alterations among dyslexia, dyspraxia, and SLI. Our data suggest that semaphorin signaling seems predominantly altered only in dyspraxia. The data obtained are now to be validated experimentally. A better understanding of miRNA alterations would allow the development of better diagnostic tests, considering that diagnosis and distinction among speech and language disorders is quite challenging; the identification of specific markers would be of great value. Validation of the predicted miRNAs in plasma from dyspraxia, dyslexia, and SLI affected patients would be of great importance. Furthermore, our data on putative modulated targets belonging to the axon guidance pathway points to additional examination of the role of the axon guidance pathway and its alteration in dyslexia, dyspraxia, and SLI. Possible differences among the three disorders (e.g., involvement of semaphorin signaling) should be validated and further investigated and may lead to a better understanding of pathophysiological mechanisms and differences among the three disorders.
Acknowledgments
We wish to express our gratitude to Grande Oriente d’Italia (GOI), Massoneria Italiana, Collegio delle Marche, for their guidance and useful suggestions which helped us in completing this review.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Kang C, Drayna D. Genetics of speech and language disorders. Annu Rev Genomics Hum Genet 2011; 12:145 - 64; http://dx.doi.org/10.1146/annurev-genom-090810-183119; PMID: 21663442
- Newbury DF, Monaco AP. Genetic advances in the study of speech and language disorders. Neuron 2010; 68:309 - 20; http://dx.doi.org/10.1016/j.neuron.2010.10.001; PMID: 20955937
- Albertini MC, Olivieri F, Lazzarini R, Pilolli F, Galli F, Spada G, Accorsi A, Rippo MR, Procopio AD. Predicting microRNA modulation in human prostate cancer using a simple String IDentifier (SID1.0). J Biomed Inform 2011; 44:615 - 20; http://dx.doi.org/10.1016/j.jbi.2011.02.006; PMID: 21334455
- Anthoni H, Zucchelli M, Matsson H, Müller-Myhsok B, Fransson I, Schumacher J, Massinen S, Onkamo P, Warnke A, Griesemann H, et al. A locus on 2p12 containing the co-regulated MRPL19 and C2ORF3 genes is associated to dyslexia. Hum Mol Genet 2007; 16:667 - 77; http://dx.doi.org/10.1093/hmg/ddm009; PMID: 17309879
- Massinen S, Hokkanen ME, Matsson H, Tammimies K, Tapia-Páez I, Dahlström-Heuser V, Kuja-Panula J, Burghoorn J, Jeppsson KE, Swoboda P, et al. Increased expression of the dyslexia candidate gene DCDC2 affects length and signaling of primary cilia in neurons. PLoS One 2011; 6:e20580; http://dx.doi.org/10.1371/journal.pone.0020580; PMID: 21698230
- Levecque C, Velayos-Baeza A, Holloway ZG, Monaco AP. The dyslexia-associated protein KIAA0319 interacts with adaptor protein 2 and follows the classical clathrin-mediated endocytosis pathway. Am J Physiol Cell Physiol 2009; 297:C160 - 8; http://dx.doi.org/10.1152/ajpcell.00630.2008; PMID: 19419997
- Couto JM, Gomez L, Wigg K, Cate-Carter T, Archibald J, Anderson B, Tannock R, Kerr EN, Lovett MW, Humphries T, et al. The KIAA0319-like (KIAA0319L) gene on chromosome 1p34 as a candidate for reading disabilities. J Neurogenet 2008; 22:295 - 313; http://dx.doi.org/10.1080/01677060802354328; PMID: 19085271
- Paracchini S, Ang QW, Stanley FJ, Monaco AP, Pennell CE, Whitehouse AJ. Analysis of dyslexia candidate genes in the Raine cohort representing the general Australian population. Genes Brain Behav 2011; 10:158 - 65; http://dx.doi.org/10.1111/j.1601-183X.2010.00651.x; PMID: 20846247
- Wang Y, Paramasivam M, Thomas A, Bai J, Kaminen-Ahola N, Kere J, Voskuil J, Rosen GD, Galaburda AM, Loturco JJ. DYX1C1 functions in neuronal migration in developing neocortex. Neuroscience 2006; 143:515 - 22; http://dx.doi.org/10.1016/j.neuroscience.2006.08.022; PMID: 16989952
- Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, Eklund R, Nopola-Hemmi J, Kääriäinen H, Kere J. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet 2005; 1:e50; http://dx.doi.org/10.1371/journal.pgen.0010050; PMID: 16254601
- Kornfeld S. Trafficking of lysosomal enzymes. FASEB J 1987; 1:462 - 8; PMID: 3315809
- Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, Peng S, Preuss TM, Wohlschlegel JA, Geschwind DH. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature 2009; 462:213 - 7; http://dx.doi.org/10.1038/nature08549; PMID: 19907493
- Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcón M, Oliver PL, Davies KE, Geschwind DH, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med 2008; 359:2337 - 45; http://dx.doi.org/10.1056/NEJMoa0802828; PMID: 18987363
- Nagase T, Kikuno R, Hattori A, Kondo Y, Okumura K, Ohara O. Prediction of the coding sequences of unidentified human genes. XIX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res 2000; 7:347 - 55; http://dx.doi.org/10.1093/dnares/7.6.347; PMID: 11214970
- Xiang M, Mohamalawari D, Rao R. A novel isoform of the secretory pathway Ca2+,Mn(2+)-ATPase, hSPCA2, has unusual properties and is expressed in the brain. J Biol Chem 2005; 280:11608 - 14; http://dx.doi.org/10.1074/jbc.M413116200; PMID: 15677451
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120:15 - 20; http://dx.doi.org/10.1016/j.cell.2004.12.035; PMID: 15652477
- Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics 2009; 25:1991 - 3; http://dx.doi.org/10.1093/bioinformatics/btp299; PMID: 19435746
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol 2011; 18:1139 - 46; http://dx.doi.org/10.1038/nsmb.2115; PMID: 21909094
- Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 1999; 96:795 - 806; http://dx.doi.org/10.1016/S0092-8674(00)80590-5; PMID: 10102268
- Schubert T, Denk AE, Ruedel A, Kaufmann S, Hustert E, Bastone P, Bosserhoff AK. Fragments of SLIT3 inhibit cellular migration. Int J Mol Med 2012; 30:1133 - 7; PMID: 22922792
- Bristow JM, Sellers MH, Majumdar D, Anderson B, Hu L, Webb DJ. The Rho-family GEF Asef2 activates Rac to modulate adhesion and actin dynamics and thereby regulate cell migration. J Cell Sci 2009; 122:4535 - 46; http://dx.doi.org/10.1242/jcs.053728; PMID: 19934221
- Hamill KJ, Hopkinson SB, DeBiase P, Jones JC. BPAG1e maintains keratinocyte polarity through beta4 integrin-mediated modulation of Rac1 and cofilin activities. Mol Biol Cell 2009; 20:2954 - 62; http://dx.doi.org/10.1091/mbc.E09-01-0051; PMID: 19403692
- Li X, Lee AY. Semaphorin 5A and plexin-B3 inhibit human glioma cell motility through RhoGDIalpha-mediated inactivation of Rac1 GTPase. J Biol Chem 2010; 285:32436 - 45; http://dx.doi.org/10.1074/jbc.M110.120451; PMID: 20696765
- Mazelin L, Bernet A, Bonod-Bidaud C, Pays L, Arnaud S, Gespach C, Bredesen DE, Scoazec JY, Mehlen P. Netrin-1 controls colorectal tumorigenesis by regulating apoptosis. Nature 2004; 431:80 - 4; http://dx.doi.org/10.1038/nature02788; PMID: 15343335
- Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SS, Culotti JG, Tessier-Lavigne M. Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell 1996; 87:175 - 85; http://dx.doi.org/10.1016/S0092-8674(00)81336-7; PMID: 8861902
- Roof DJ, Hayes A, Adamian M, Chishti AH, Li T. Molecular characterization of abLIM, a novel actin-binding and double zinc finger protein. J Cell Biol 1997; 138:575 - 88; http://dx.doi.org/10.1083/jcb.138.3.575; PMID: 9245787
- Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, Aaronson SA. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 1991; 251:802 - 4; http://dx.doi.org/10.1126/science.1846706; PMID: 1846706
- Appleton BA, Wu P, Maloney J, Yin J, Liang WC, Stawicki S, Mortara K, Bowman KK, Elliott JM, Desmarais W, et al. Structural studies of neuropilin/antibody complexes provide insights into semaphorin and VEGF binding. EMBO J 2007; 26:4902 - 12; http://dx.doi.org/10.1038/sj.emboj.7601906; PMID: 17989695
- Bearer CF. L1 cell adhesion molecule signal cascades: targets for ethanol developmental neurotoxicity. Neurotoxicology 2001; 22:625 - 33; http://dx.doi.org/10.1016/S0161-813X(01)00034-1; PMID: 11770884
- Shin EY, Shim ES, Lee CS, Kim HK, Kim EG. Phosphorylation of RhoGDI1 by p21-activated kinase 2 mediates basic fibroblast growth factor-stimulated neurite outgrowth in PC12 cells. Biochem Biophys Res Commun 2009; 379:384 - 9; http://dx.doi.org/10.1016/j.bbrc.2008.12.066; PMID: 19103160
- Wells CM, Jones GE. The emerging importance of group II PAKs. Biochem J 2010; 425:465 - 73; http://dx.doi.org/10.1042/BJ20091173; PMID: 20070256
- Schrantz N, da Silva Correia J, Fowler B, Ge Q, Sun Z, Bokoch GM. Mechanism of p21-activated kinase 6-mediated inhibition of androgen receptor signaling. J Biol Chem 2004; 279:1922 - 31; http://dx.doi.org/10.1074/jbc.M311145200; PMID: 14573606
- Foletta VC, Moussi N, Sarmiere PD, Bamburg JR, Bernard O. LIM kinase 1, a key regulator of actin dynamics, is widely expressed in embryonic and adult tissues. Exp Cell Res 2004; 294:392 - 405; http://dx.doi.org/10.1016/j.yexcr.2003.11.024; PMID: 15023529
- Sparrow N, Manetti ME, Bott M, Fabianac T, Petrilli A, Bates ML, Bunge MB, Lambert S, Fernandez-Valle C. The actin-severing protein cofilin is downstream of neuregulin signaling and is essential for Schwann cell myelination. J Neurosci 2012; 32:5284 - 97; http://dx.doi.org/10.1523/JNEUROSCI.6207-11.2012; PMID: 22496574
- Cole AR, Knebel A, Morrice NA, Robertson LA, Irving AJ, Connolly CN, Sutherland C. GSK-3 phosphorylation of the Alzheimer epitope within collapsin response mediator proteins regulates axon elongation in primary neurons. J Biol Chem 2004; 279:50176 - 80; http://dx.doi.org/10.1074/jbc.C400412200; PMID: 15466863
- Zhou R. The Eph family receptors and ligands. Pharmacol Ther 1998; 77:151 - 81; http://dx.doi.org/10.1016/S0163-7258(97)00112-5; PMID: 9576626
- Arvanitis DN, Béhar A, Tryoen-Tóth P, Bush JO, Jungas T, Vitale N, Davy A. Ephrin B1 maintains apical adhesion of neural progenitors. Development 2013; 140:2082 - 92; http://dx.doi.org/10.1242/dev.088203; PMID: 23578932
- Lawrenson ID, Wimmer-Kleikamp SH, Lock P, Schoenwaelder SM, Down M, Boyd AW, Alewood PF, Lackmann M. Ephrin-A5 induces rounding, blebbing and de-adhesion of EphA3-expressing 293T and melanoma cells by CrkII and Rho-mediated signalling. J Cell Sci 2002; 115:1059 - 72; PMID: 11870224
- Srivastava N, Robichaux MA, Chenaux G, Henkemeyer M, Cowan CW. EphB2 receptor forward signaling controls cortical growth cone collapse via Nck and Pak. Mol Cell Neurosci 2013; 52:106 - 16; http://dx.doi.org/10.1016/j.mcn.2012.11.003; PMID: 23147113
- Nakanishi H, Nakamura T, Canaani E, Croce CM. ALL1 fusion proteins induce deregulation of EphA7 and ERK phosphorylation in human acute leukemias. Proc Natl Acad Sci U S A 2007; 104:14442 - 7; http://dx.doi.org/10.1073/pnas.0703211104; PMID: 17726105
- Füller T, Korff T, Kilian A, Dandekar G, Augustin HG. Forward EphB4 signaling in endothelial cells controls cellular repulsion and segregation from ephrinB2 positive cells. J Cell Sci 2003; 116:2461 - 70; http://dx.doi.org/10.1242/jcs.00426; PMID: 12734395
- Mitsushima M, Suwa A, Amachi T, Ueda K, Kioka N. Extracellular signal-regulated kinase activated by epidermal growth factor and cell adhesion interacts with and phosphorylates vinexin. J Biol Chem 2004; 279:34570 - 7; http://dx.doi.org/10.1074/jbc.M402304200; PMID: 15184391