Abstract
Psychotropic agents are notorious for their ability to increase fat mass in psychiatric patients. The two determinants of fat mass are the production of newly differentiated adipocytes (adipogenesis), and the volume of lipid accumulation. Epigenetic programs have a prominent role in cell fate commitments and differentiation required for adipogenesis. In parallel, epigenetic effects on energy metabolism are well supported by several genetic models. Consequently, a variety of psychotropics, often prescribed in combinations and for long periods, may utilize a common epigenetic effector path causing an increase in adipogenesis or reduction in energy metabolism. In particular, the recent discovery that G protein coupled signaling cascades can directly modify epigenetic regulatory enzymes implicates surface receptor activity by psychotropic medications. The potential therapeutic implications are also suggested by the effects of the clinically approved antidepressant tranylcypromine, also a histone demethylase inhibitor, which has impressive therapeutic effects on metabolism in the obese phenotype.
Obesity and Epigenetic Regulation
Obesity in psychiatric patients is a major public health concern that has multiple antecedents. These include sedentary lifestyles, poor socioeconomic status and nutrition, and psychological stress leading to increased blood cortisol levels, among other factors. However, the single strongest predictor of obesity in this population is the concurrent use of psychotropics. All classes of psychotropic agents have been implicated, most notoriously antipsychotics, but mood stabilizers and antidepressants as well.Citation1-Citation3 For example, although clozapine and olanzapine exhibit the greatest potential for weight gain, even haloperidol, abilify, and ziprasidone are associated with increased risk.Citation4,Citation5 The normal increase in weight across the lifespan of an individual is 0.4 to 1.2 kg/y.Citation6 Psychotropic induction of weight gain is 10-fold higher than normal aging and, from this perspective, these medications can be considered a potent environmental toxin (). Beyond the effects of individual drugs, the obesogenic effects of psychotropic combinations, such as an antipsychotic and a mood stabilizer, has been poorly studied.
Fat mass: proliferation/differentiation vs. metabolism/storage
Fat mass is determined by two processes: increase in lipid accumulation and adipocyte size (hypertrophy) as well as a steady replacement by newly differentiated adipocytes from proliferating stem cells (hyperplasia). Studies on the fat mass indicate that certain parameters are set in childhood and adolescence; these are fat cell number and turnover rate.Citation8 In these same studies, the rate of adipogenesis is increased in obese individuals, although the proportion of new cells to old cells is stable (approximately 10%).
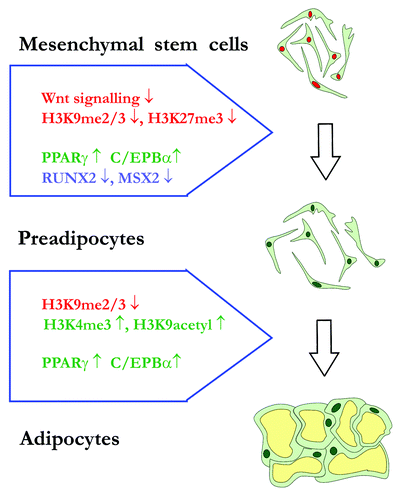
If newly proliferated cells are an important contributor to fat mass, then it is a reasonable hypothesis that the proliferation rate could vary in response to obesogenic inputs. Hyperplasia is inherently a developmental/quasi-embryological process, with sequential progression from cell-fate determination followed by terminal differentiation. This developmental sequence starts with the commitment of precursor cells, commonly derived from multipotent mesenchymal stem cells resident in adipose tissue itself, but also in bone, cartilage, adipose tissue, or blood. Mature adipose tissue retains among the highest concentrations of stem cells across any tissue, and adipose stem cells are characterized by vigorous proliferative capacity.Citation9 Pre-adipocytes largely relinquish their multipotent ability, but are phenotypically differentiable from stem cells only in the nature of a molecular signature, as both cell types retain their proliferative capabilities ().Citation10 Epigenetic gene regulation is a significant regulator of these embryological routines.
Figure 1. Developmental trajectory leading to fat containing adipocytes. Signaling cascades are presented between each stage and presented as either increasing (up arrows) or decreasing (down arrows). Molecular events that facilitate progression along the downward path to adipocytes are presented in green. Stem cells and pre-adipocytes are phenotypically similar but distinguishable by changes in their molecular signature as presented by different colored nuclei.
In parallel, there are now several genetic models indicating that energy metabolism and fat utilization, especially in the adult animal model, is influenced by activity of enzymes targeting chromatin histone proteins. Deletion knockouts of these epigenetic enzymes results in abnormal energy homeostasis, fat storage, and glucose trafficking. One of these enzymes, Jhdm2a, is minimally expressed in the brain, making it an attractive systemic target.Citation11
Antipsychotics induce both adipose hyperplasia and hypertrophy
Several antipsychotics can induce either/both the proliferation (or hyperplasia) of multipotent mesenchymal stem cells within adipose tissue, as well as the differentiation of pre-adipocytes into fat accumulating adipocytes (hypertrophy).Citation12 Thus, olanzapine, quetiapine, and risperidone can induce proliferation, as measured by DNA synthesis (hyperplasia), while clozapine and aripiprazole act by increasing fat accumulation (hypertrophy).Citation12 Clozapine will induce the differentiation of pre-adipocytes (obtained from living human subjects and differentiated in vitro) into adipocytes with enhanced lipid accumulation.Citation13
SREBP1 is a transcription factor that promotes cholesterol homeostasis (increased production in a cholesterol depleted environmentCitation14) and directly upregulates PPARγ mRNA expression (direct promoter bindingCitation14) as well as functional activity (synthesis of endogenous PPARγ ligandsCitation15). Olanzapine, clozapine, quetiapine, and risperidone (but not aripiprazole) will enhance the expression of the SREBP1 gene in in vitro cell models, increase expression of PPARγ, and hasten the differentiation of pre-adipocytes toward the mature adipocyte as manifested by lipid accumulation.Citation12,Citation16,Citation17 A single injection of clozapine can clearly perturb the SREBP1 system, with an initial increase followed by a sustained decrease of downstream lipogenic target genes and coordinated by liver accumulation of triacylglycerols, cholesterol, and phospholipids within 24–48 h.Citation18
Epigenetic modifications also regulate adipogenesis: an effector path?
Epigenetic gene regulation is the basis for the genome-wide transcriptional programming that underlies any developmental progression. The role of epigenetics in adipogenesis can be illustrated by focusing on two principal adipogenic transcription factors: PPARγ and C/EBPα. PPARγ (expressed as variants PPARγ1 and PPARγ2) is a nuclear ligand-receptor and is a master regulator of the genome-wide programs that lead to adipogenesis, during which it partners with CCAAT/enhancer binding protein (C/EBPα). PPARγ appears to be the primary initiator and is both necessary and sufficient for the successful differentiation of pre-adipocytes to adipocytes.Citation19 It is capable of inducing adipocyte-determination in C/EBPα (−/−) cells, while the converse is not true.Citation19,Citation20 PPARγ overexpression is even capable of forcibly inducing adipogenesis in fibroblasts.Citation19 Finally, its expression is also critical for sustaining the fully differentiated adipocyte state, and a PPARγ ‘knockdown’ will cause a de-differentiation of the mature adipocyte accompanied with a loss of accumulated neutral fats,Citation21 or even adipocyte-death.Citation22 C/EBPα is a powerful antimitotic transcription factorCitation23 and consolidates the cell-fate decision from a proliferating pre-adipocyte cell to a committed post-mitotic adipocyte.Citation24 While C/EBPα and PPARγ expression is enough to promote adipogenesis, other C/EBP factors (C/EBPβ and C/EBPδ) may improve efficiency. A separate protein relevant to effects of antipsychotics medications is SREBP1, which is a master regulator of lipogenic genes primarily related to either the triglyceride or cholesterol pathways. SREBP1 is a transcription factor that binds to its response element SRE and can directly upregulate PPARγ gene expression.Citation19
There are two ways in which epigenetic modifications can facilitate or restrict adipogenic programs. The first is through increasing or decreasing levels of a primary transcription factor such as PPARγ or C/EBPα through direct epigenetic regulation of their individual promoters. The second is through the assembly of either activator or repressor multi-protein complexes by the primary transcription factor. These assemblies are positioned on downstream promoter regions possessing the recognition sequences (PPRE in the case of PPARγ), and almost always operate by the inclusion or exclusion of epigenetic modifying enzymes. This genomewide network of DNA promoter sequences targeted by a transcription factor is called a “cistrome.” PPARγ will bind to its consensus sequence, PPRE, and in the absence of ligand, will recruit and assemble a variety of co-repressors, such as histone deacetylases (e.g., HDAC3), as well as SMRT and NCoR, thereby focusing these repressive proteins onto the targeted promoter.Citation25 However, when an agonist binds the positioned ligand-receptor, the resulting change in receptor conformation facilitates the replacement of co-repressors with co-activators, such as p300/CREB proteins possessing histone acetyltransferase activity (HAT) capable of opening chromatin. In fact, histone modifying proteins associated with PPARγ can induce a wide range of both “open” (H3k4me2/3, H3k9ac, H3k27ac) or “closed” modifications (H3K9me2/3, H3k27me2/3).
Selected epigenetic modifications relevant to adipogenic programs
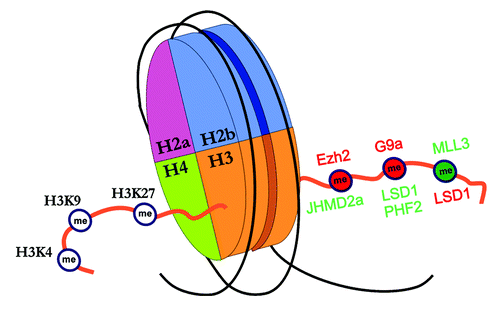
Chromatin, a complex of proteins and DNA sequence, is the platform upon which the epigenetic modifications are written, with the nucleosome as the fundamental repeating unit of chromatin.Citation26-Citation28 Epigenetic modifications work to either facilitate (“open chromatin” allowing free access of transcription proteins to the DNA sequence) or restrict gene transcription (“closed chromatin” restricting access of transcription proteins). Acetylation is always a permissive modification and is catalyzed by two opposing histone modifying enzymes: histone acetyltransferases (HATs are facilitative) and deacetylases (HDACs are restrictive). Histone methylation on the other hand is complex, and can be either facilitative or restrictive to gene expression, depending on the address. The enzymes responsible for histone methylation/demethylation are numerous and site specific to the address of the targeted lysine residue (). Thus, SETDB1 is a histone methyltransferase (HMT) targeting H3K9 for methylation, while MLL3 prefers to methylate H3K4. Finally as a generalization, DNA methylation, H3K9-methylation, and H3K27-methylation are commonly restrictive, while H3K4 methylation is permissive.
Figure 2. Nucleosome assembly of eight separate histone proteins (two each of H2a, H2b, H3, and H4), and various restrictive/permissive modifications. H3 tail on left: Lysine sites on the left H3 tail are addressed and serve as a regulatory site for the attachment of methyl groups. H3 tail on right: Methyl groups in red color represent restrictive chromatin assemblies, while methyl groups in green color represent facilitative chromatin. Histone methyltransferases are presented above the tail at their preferential catalytic site, while histone demethylases are presented below the tail. Enzymes in red indicate catalytic activity serving to create a restrictive modification, while enzymes in green represent catalytic activity serving to open chromatin. Note that LSD1 is restrictive at H3K4 and facilitative at H3K9.
H3K9: methylation and acetylation exert opposing effects on adipogenesis
The lysine at position 9 on the histone 3 tail is an epigenetic switch; acetylation (H3K9acetyl) creates an open chromatin assembly, while methylation (2 or 3 methyl groups leading respectively to H3K9me2 or H3K9me3) results in a particularly durable and resistant closed chromatin.Citation29 The transition from pre-adipocyte to mature adipocyte is broadly characterized by a change from a restrictive “closed” chromatin state in the pre-adipocyte to an “open” state in the mature adipocyte.Citation30
H3K9acetyl
The association between increased H3K9 acetylation and adipogenesis is supported by molecular changes in a cell-line model for adipocyte differentiation, 3T3-L1 cells. This cell line demonstrates a reduction in HDAC -1, -2, -5, and -6 as they differentiate from a pre-adipocyte state to differentiated fat containing adipocytes. These changes in enzyme levels are coordinated with changes in global levels of H3K9acetyl (increased in mature adipocytes) and H3K9me2 (decreased in mature adipocytes).Citation31 PPARγ expression is very low in pre-adipocytes prior to differentiation.Citation32 Experimentally induced reduction in HDAC enzyme activity increases expression of PPARγ, C/EPBα, and SREBP1c, and initiates the termination of the pre-adipocyte state and the start of differentiation toward a mature adipocyte.Citation31
H3K9me2/3
In pre-adipocytes, the promoter of Pparγ (and major sections of the entire gene locus) is initially restricted by closed chromatin modifications such as H3K9me2, which shifts toward an open state as the pre-adipocyte proceeds toward differentiation.Citation33,Citation34 H3K9me2 occupancy decreases during adipogenesis, as do levels of the methyltransferase, G9a, a histone methyltransferase catalyzing H3K9me2 ().Citation34 Moreover, knockout of G9a will cause a reduction of H3K9me2 on the Pparγ promoter, increasing its expression and advancing adipogenesis.Citation34 In addition, molecular or pharmacological inhibition of G9a will release PPARγ expression, which in turn will ramp up an adipogenic program powerful enough to induce transdifferentiation of myofibroblasts toward adipogenesis.Citation33,Citation35 Conversely, reduction in PPARγ expression induced by SETDB1 (another histone methyltransferase catalyzing H3K9me2) in bone marrow mesenchymal stem cells will inhibit adipogenesis and redirect development toward osteogenesis.Citation36
H3K9 chromatin along the PPARγ cistrome
Besides direct regulation of the Pparγ promoter, histone methyltransferases responsible for the formation of H3K9me2, such as SETDB1, can also be recruited by ligand-free PPARγ receptors onto cis-elements of its target gene promoters (or the PPARγ cistrome). This recruitment can be coordinated by signaling along the Wnt cascade, which inhibits adipogenesis. Wnt5a, a Wnt ligand, will activate a non-canonical Wnt pathway (i.e., not signaling through β-catenin), which in turn will activate Setdb1 and relocate it to a Pparγ multi-protein complex located on a targeted promoter.Citation36 Subsequently, Setdb1 will catalyze the formation of H3K9me2 on site.Citation36 Ligand-free PPARγ can also recruit members of the HDAC family of restrictive epigenetic enzymes as catalytic units of the PPARγ multi-protein complex.Citation31 HDAC enzymes, thus incorporated, can be antagonized by small molecule HDAC enzyme inhibitors to reverse the anticipated termination of the adipogenic program.Citation25 Of note, HDAC inhibitors are known to arrest cell division in cancer cells, an ability possibly applied to proliferating pre-adipocytes, forcing them toward terminal differentiation as fat containing adipocytes.
Conversely, increased H3K9acetyl levels (complemented by a decrease in levels of H3K9me2) are also found on the promoters of C/EBPα and ADD1/SREBP1c in differentiated adipocytes. Expression of these transcription factors is thus coordinated with a decrease in precursor (stem cell or pre-adipocyte) proliferation, and an increase of post-mitotic differentiation ().Citation31 Histone modifications along the C/EBPα promoter are additionally regulated by nuclear ligand-receptors, such as the glucocorticoid receptor, which in a ligand-free state (no steroids) will focus HDAC1 enzyme activity at the C/EBPα promoter causing deacetylation and chromatin closing. Upon steroid binding, glucocorticoid receptors will initiate the degradation and depletion of HDAC1 at this promoter, and facilitate C/EBPα expression, leading to pre-adipocyte differentiation into fully mature adipocytes.Citation31,Citation37 Further, HDAC inhibitors (increasing H3K9 acetylation) will accelerate differentiation and accumulation of triglycerides.Citation31
H3K9 demethylation
This overall scheme is supported by studies of the histone demethylase (HDM) enzyme, Jhdm2a, responsible for the removal of the restrictive H3K9me2 modification (). In Jhdm2a knockout mice, there is greater H3K9me2 accumulation along the PPARγ cistrome, coordinated with a downregulation of genes involved in energy metabolism leading to adult obesity and insulin resistance.Citation11,Citation38 Tateishi et al., (2009) demonstrate that in Jhdm2a knockout mice, there is increased obesity without increased food intake. This is accompanied by higher levels of free fatty acids, triglycerides, and cholesterol, as well as reduced metabolic response in brown adipose tissue to cold stress and β-adrenergic stimulation.Citation11,Citation38 In this genetic model, the ability of PPARγ to enhance expression of target genes is obstructed by H3K9me2 at the PPRE (PPAR response element) on target gene promoters.Citation11
H3K4 methylation signals onset of adipogenesis
The lysine at position 4 on the histone 3 tail when multiply methylated (2 or 3 methyl groups leading respectively to H3K4me2 or H3K4me3) establishes a mark of open chromatin. The H3K4me3 mark is the earliest occupant of adipogenic gene promoters in pre-adipocytes that are beginning to differentiate into adipocytes and erasure of this mark results in reduced adipogenesis.Citation30 Operationally, increasing H3K4me3 levels at the C/EBPα promoter will directly enhance C/EBPα expression and encourage adipogenesis.Citation24
H3K4 methylation and the PPARγ cistrome
H3K4 is methylated to H3K4me3 by the SET domain containing enzymes, including the MLL family (particularly MLL3) (). Homozygous knockouts of MLL3 demonstrate a 30–40% reduction in the mass of white adipose tissue (WAT) but not in brown adipose tissue (BAT).Citation39 These MLL mutants also show less triglyceride accumulation, lower glucose and insulin levels and improved insulin tolerance.Citation39 Because H3K4me3 is a mark of open chromatin, it is not surprising that MLL3 is recruited by ligand-bound PPARγ into its multi-protein complexes on targeted promoters, which in turn will increase H3K4me3 at these networked locations.Citation39 At the genome-wide scale, H3K4me2/3, besides directly regulating the PPARγ and CEPBα promoters, is also a component of the multi-protein assemblies recruited by PPARγ across its cistrome.Citation40 Conversely, a MLL3 knockout will reduce levels of H3K4me3 chromatin at PPARγ target genes, resulting in reduced open chromatin and decreased adipogenesis even under conditions of a fat diet.Citation39
H3K4 demethylation and energy metabolism
Levels of H3K4 methylation during the process of differentiation from pre-adipocyte to mature adipocyte are regulated by the balance of activity of H3K4 specific genetic loci.Citation24 LSD1 is a histone demethylase (HDM) responsible for removing (demethylating) both the “open” H3K4me2 mark as well the “closed” H3K9me2 mark (). LSD1 is recruited directly to the CEBPα promoter at the pre-adipocyte stage when constituent C/EBPα levels are low. Knockdown of LSD1 results in an increase in H3K9me2 occupancy at the C/EBPα promoter, preemptively blocking changes at H3K4, and thereby resulting in restricted C/EBPα expression and decreased differentiation.Citation24 It should be noted that LSD1 is highly expressed in WAT.Citation41
Given that LSD1 is a repressive enzyme in adipose tissue (by reducing levels of the open chromatin mark H3K4me2), Hino et al. demonstrate that an LSD1-knockdown will de-repress expression of genes that increase energy expenditure.Citation41 A similar effect can be seen with pharmacological inhibition of LSD1 using the antidepressant tranylcypromine, which will decrease lipid accumulation by increased lipolysis in response to insulin stimulation.Citation24 Conversely, a high fat diet will increase LSD1 gene expression, reduce the H3K4me2 open chromatin mark, and thus reduce expression of LSD1-target genes which include those engaged in oxidative metabolism.Citation41
In principle, LSD1 can repress energy expenditure genes in adipose tissue and facilitate fat accumulation. LSD1 inhibition by tranylcypromine will block the effects of a high fat diet and will markedly lower body weight under a high-fat diet condition. Notably, tranylcypromine has little effect under normal metabolic conditions. This reveals attractive properties for tranylcypromine as a method to control levels of adipose tissue by enhancing energy metabolism in patients experiencing the metabolic syndrome.
H3K27 methylation advances adipogenesis by repression of Wnt signaling
The lysine at position 27 on the histone 3 tail, when multiply methylated (2 or 3 methyl groups leading respectively to H3K27me2 or H3K27me3), establishes a mark of closed chromatin. H3K27me3 is a restrictive histone mark catalyzed by enzymes associated with the polycomb PRC2 repressive complex, with the catalytic unit of this polycomb complex being the enzyme Ezh2 ().Citation42 The Wnt/β-catenin signaling pathway works to inhibit adipogenesis and maintain a more pluripotent developmental state in the pre-adipocyte and consequently Wnt is a principal inhibitor of adipocyte differentiation.Citation43 Deletions of Ezh2 will reduce H3K27me3 on Wnt promoters, thereby de-repressing these promoters. De-repressed Wnt proteins will, in turn increase b-catenin accumulation and nuclear signaling. This Wnt/β-catenin cascade will repress downstream promoters such as Pparγ and C/Ebpα, and as a final result will inhibit adipogenesis.Citation44 By the same token, the effects of Wnt expression can be reversed by overexpression of PPARγ and C/EBPα.Citation43 In principle, H3K27me2/3 will advance adipogenesis by facilitating cell-fate determination (indirectly through suppression of Wnt proteins), while H3K9me2/3, the other restrictive modification considered in a previous section here, will inhibit adipogenesis by suppressing PPARγ and C/EBPα and repressing cell-fate determination. Incidentally, the Wnt proteins may additionally work to inhibit adipogenesis by targeting restrictive H3K9me2 modification to the PPARγ promoter via their interactions with the Setdb1 methyltransferase.Citation36
DNA methylation secures lineage commitment
Mesenchymal stem cells derived from adipose tissue are capable of multi-lineage differentiation into osteogenic, hepatic, immune, and adipose lineages. Commitment to an adipose lineage appears contingent on reduced DNA methylation along adipogenic promoters in these mesenchymal cells.Citation45 However, differentiation does not result in further hypomethylation, and the level of methylation is not correlated with increasing adipogenic mRNA transcription.Citation45 The success of terminal differentiation and the level of adipogenic-related mRNA expression may be a function of histone modifications noted above. In the fully differentiated obese state, the Pparγ promoter is again repressed by DNA methylation.Citation33 In 3T3-L1 cells used as a pre-adipocyte model, treatment with an inhibitor of DNA methyltransferase will release the Pparγ promoter and expression. A methylated Pparγ promoter can further recruit methylated DNA binding proteins such as MeCP2 and other proteins with affinity to heterochromatin such as heterochromatin protein 1 (HP1). Disengagement of this assembly from a hypermethylated DNA promoter sequence begins with the initiation of differentiation.Citation33
Effects of psychotropics on selected epigenetic enzymes and modifications
HDAC inhibition and valproic acid
Efforts to develop therapeutics targeted at the epigenetic machinery are furthest along in the category of HDAC inhibitors. In general, reduction in the enzymatic activity of histone deacetylases will establish open chromatin assemblies at the promoters of adipogenic genes, and stimulate terminal differentiation into adipocytes.Citation31 In particular, HDAC1 enzyme knockdown or overexpression will result in coordinate increase or decrease of adipogenesis, respectively.Citation31 The process of adipocyte differentiation is itself accompanied by a decline in the levels of HDAC enzymes and enzymatic activity by 50%.Citation31 These molecular effects are replicated with the use of pharmacological HDAC inhibitors, including valproic acid and Trichostatin A.
HDAC enzymes are one component of the protein assembly recruited by PPARγ across the cistrome. Direct inhibition of the embedded HDAC enzyme by small molecule pharmacology will relieve this repression, and facilitate gene transcription. In this situation, HDAC inhibitors will inhibit the repressive activity of the PPARγ-HDAC complex and release expression of adipogenic genes.Citation25 A similar but opposite dynamic is involved in the protein complex associated with the glucocorticoid nuclear receptor. In the absence of this steroid, the ligand free receptor recruits HDAC enzymes. Upon binding to its steroid ligand, this HDAC enzyme-containing complex is disassembled, thus abrogating HDAC activity. This sequence of events relaxes chromatin and advances adipogenic programs. Not surprisingly, steroids medications have the propensity to increase obesity. Small molecule HDAC inhibitors, in principle, will act in a method similar to a steroid by enhancing the adipogenic routine.Citation37 These observations are consistent with the clinical effects of valproic acid, which will induce weight gain in patients.
Valproic acid is the most widely used HDAC inhibitor in the pharmacopeia, and has been reviewed extensively elsewhere.Citation46 Of particular interest is the supplemental effects on chromatin remodeling of certain atypical antipsychotics when administered conjointly with VPA.Citation47 Clozapine and sulpiride, but not haloperidol or olanzapine, will accelerate the effects of VPA on DNA methylation and gene expression.Citation47 Beyond these initial investigations, a systematic study of other (not VPA) psychotropic drug actions on chromatin modifications in either post-mitotic brain or mitotic cells, such as the mesenchymal stem cells and pre-adipocytes, is limited. In particular, the effects of psychotropics on epigenetic regulation of mitotic cells are virtually unknown. For example, haloperidol is capable of inducing DNA demethylation in mitotic cancer cells (by 15 to 20% at specific CpG sites) with effects comparable to the prototypic DNA demethylating agent, 5-azacytidine.Citation48 Further, rats treated with haloperidol will manifest decreased methylation in peripheral lymphocytes but increased global DNA methylation in the liver.Citation49
Wnt pathway and H3K27me3: effects of lithium and antipsychotics
Lithium, valproic acid, antidepressants, and antipsychotics will all independently enhance the Wnt signaling cascade by increasing levels of phosphorylated-GSK-3b and unphosphorylated β-catenin accumulation;Citation50 both these molecular events indicate an activation of the Wnt/β-catenin signaling cascade, which is anti-adipogenic.Citation50,Citation51 Lithium acts to directly inhibit GSK-3b, while antipsychotics increase the expression of several Wnt ligands and signaling molecules such as Dishevelled-3.Citation51 It is useful to note here that adipose tissue expresses dopamine receptors and produces prolactin, making it a good target for the action of antipsychotics.Citation52 Stabilization of transmission along the Wnt pathway could in theory sustain a proliferative state of mesenchymal stem cell precursors. As noted above, accumulation of β-catenin is also a consequence of Ezh2 deletions as a consequence of reduced H3K27me3 deposition along Wnt promoters.Citation34 Increased levels of β-catenin inhibit the transcription of PPARγ and C/EBPa as well as the cistromic function of PPARγ.Citation44 This can directly inhibit cell-fate commitment, terminal differentiation and maintain a pluripotent stateCitation44 and could explain why lithium is not associated with an increase in fat mass.Citation53
Receptors, protein kinases, and histone methylation
The limited literature in peripheral or brain tissue indicates that antipsychotics most frequently implicated in obesity have replicable, but variable effects on epigenetic modifications. Dopamine antagonists, including haloperidol, clozapine, and sulpiride can induce DNA demethylation.Citation49,Citation54 Dopamine antagonists can also increase global histone acetylation.Citation55 These effects appear to be amplified when the antipsychotic is administered with an HDAC inhibitor, such as valproic acid.Citation54
All known antipsychotics, and many antidepressants, regulate the levels of cAMP in the cytosol through their interaction with G-protein coupled receptors and the adenylate cyclase enzyme responsible for the cyclization of ATP to cAMP. cAMP is a major secondary messenger capable of activating protein kinase A (PKA), with downstream effects on a variety of nuclear proteins. Antipsychotics antagonize dopamine receptors, increase adenylate cyclase activity and increase cAMP. Antidepressants are also known to regulate levels of cAMP often by their actions on phosphidiesterase enzymes responsible for cAMP breakdown.Citation56 The cAMP/PKA signaling system is now demonstrated to directly regulate the activity of chromatin modifying enzymes, namely the lysine demethylase PHF2, which acts on H3K9me2,Citation57 and the shuttling capabilities of the HDAC5 histone deacetylase.Citation58 Consequently, the histone modifications described in this review are now understood to be in the direct path of the actions of most psychotropic drugs.
Administration of the antidepressant fluoxetine for a ten day period can increase HDAC2 immunoreactivity, and reduce global levels of histone acetylation.Citation59 Further, fluoxetine working through the 5HT1A serotonin receptor, can enhance plasticity in the visual cortex that is coordinated with increases in histone acetylation.Citation60
Conclusion
The development of obesity in psychiatric patients can clearly have many antecedents. The bench model of pre-adipocyte differentiation is illustrative, whereby the 3T3-L1 pre-adipocyte cell line is commonly differentiated into fat containing adipocytes by the application of a cocktail containing a phosphodiesterase inhibitor (IDMX), a steroid hormone (cortisol), and a trophic factor (insulin). This “cocktail” is conceptually analogous to the biochemical makeup of psychiatric patients who are being treated with antipsychotics (acting similarly to IDMX as noted above), are psychologically stressed (increased cortisol), and are obese (insulin resistance with increased insulin levels).
This review clearly focuses on peripheral epigenetic targets of adipose tissue accumulation. There are also many mechanisms in the central nervous system that psychotropic drugs could act though to promote positive energy balance and body weight gain. An increasing number of single nucleotide polymorphism (SNP) studies have shown compelling evidence of association with obesity.Citation61 Interestingly, these genes are also known to be expressed in both the central nervous system and adipose tissue, including the RNA demethylase FTO and neuropeptide Y.Citation62,Citation63 While beyond the scope of this review, other processes of fat accumulation could prove to be as important.
Psychiatric illness thus evolves into a complex of multiple incoming pathological signals; some emerging from the chronicity and dysfunction associated with a lifelong illness, but others from the clearly adverse obesogenic effects of psychotropic drugs. As we present in this review, multiple incoming signals are organized at the level of chromatin by histone modifications, which have known capacity to facilitate developmental routines. Histone modifying enzymes make attractive targets for developing small molecule pharmacology that can target the primary histone modification. This approach is very practical with the demonstration that the clinically prescribed antidepressant, tranylcypromine, has attractive properties in basic models.
Acknowledgments
The research was supported by funding awarded to Rajiv P Sharma from the National Institutes of health (NIH) R01 MH094358. This work was supported in part by PHS grants MH069839 and MH094358 (RPS)
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Dent R, Blackmore A, Peterson J, Habib R, Kay GP, Gervais A, Taylor V, Wells G. Changes in body weight and psychotropic drugs: a systematic synthesis of the literature. PLoS One 2012; 7:e36889; http://dx.doi.org/10.1371/journal.pone.0036889; PMID: 22719834
- Patten SB, Williams JV, Lavorato DH, Brown L, McLaren L, Eliasziw M. Major depression, antidepressant medication and the risk of obesity. Psychother Psychosom 2009; 78:182 - 6; http://dx.doi.org/10.1159/000209349; PMID: 19321971
- Smits JA, Rosenfield D, Mather AA, Tart CD, Henriksen C, Sareen J. Psychotropic medication use mediates the relationship between mood and anxiety disorders and obesity: findings from a nationally representative sample. J Psychiatr Res 2010; 44:1010 - 6; http://dx.doi.org/10.1016/j.jpsychires.2010.04.007; PMID: 20434171
- Klemp M, Tvete IF, Skomedal T, Gaasemyr J, Natvig B, Aursnes I. A review and Bayesian meta-analysis of clinical efficacy and adverse effects of 4 atypical neuroleptic drugs compared with haloperidol and placebo. J Clin Psychopharmacol 2011; 31:698 - 704; http://dx.doi.org/10.1097/JCP.0b013e31823657d9; PMID: 22020356
- Jin H, Shih PA, Golshan S, Mudaliar S, Henry R, Glorioso DK, Arndt S, Kraemer HC, Jeste DV. Comparison of longer-term safety and effectiveness of 4 atypical antipsychotics in patients over age 40: a trial using equipoise-stratified randomization. J Clin Psychiatry 2013; 74:10 - 8; http://dx.doi.org/10.4088/JCP.12m08001; PMID: 23218100
- Heitmann BL, Garby L. Patterns of long-term weight changes in overweight developing Danish men and women aged between 30 and 60 years. Int J Obes Relat Metab Disord 1999; 23:1074 - 8; http://dx.doi.org/10.1038/sj.ijo.0801035; PMID: 10557028
- Leslie WS, Hankey CR, Lean ME. Weight gain as an adverse effect of some commonly prescribed drugs: a systematic review. QJM 2007; 100:395 - 404; http://dx.doi.org/10.1093/qjmed/hcm044; PMID: 17566010
- Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, Bergmann O, Blomqvist L, Hoffstedt J, Näslund E, Britton T, et al. Dynamics of fat cell turnover in humans. Nature 2008; 453:783 - 7; http://dx.doi.org/10.1038/nature06902; PMID: 18454136
- Peng L, Jia Z, Yin X, Zhang X, Liu Y, Chen P, Ma K, Zhou C. Comparative analysis of mesenchymal stem cells from bone marrow, cartilage, and adipose tissue. Stem Cells Dev 2008; 17:761 - 73; http://dx.doi.org/10.1089/scd.2007.0217; PMID: 18393634
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science 2000; 289:950 - 3; http://dx.doi.org/10.1126/science.289.5481.950; PMID: 10937998
- Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 2009; 458:757 - 61; http://dx.doi.org/10.1038/nature07777; PMID: 19194461
- Pavan C, Vindigni V, Michelotto L, Rimessi A, Abatangelo G, Cortivo R, Pinton P, Zavan B. Weight gain related to treatment with atypical antipsychotics is due to activation of PKC-β. Pharmacogenomics J 2010; 10:408 - 17; http://dx.doi.org/10.1038/tpj.2009.67; PMID: 20029385
- Hemmrich K, Gummersbach C, Pallua N, Luckhaus C, Fehsel K. Clozapine enhances differentiation of adipocyte progenitor cells. Mol Psychiatry 2006; 11:980 - 1; http://dx.doi.org/10.1038/sj.mp.4001892; PMID: 17063181
- Fajas L, Schoonjans K, Gelman L, Kim JB, Najib J, Martin G, Fruchart JC, Briggs M, Spiegelman BM, Auwerx J. Regulation of peroxisome proliferator-activated receptor gamma expression by adipocyte differentiation and determination factor 1/sterol regulatory element binding protein 1: implications for adipocyte differentiation and metabolism. Mol Cell Biol 1999; 19:5495 - 503; PMID: 10409739
- Kim JB, Wright HM, Wright M, Spiegelman BM. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc Natl Acad Sci U S A 1998; 95:4333 - 7; http://dx.doi.org/10.1073/pnas.95.8.4333; PMID: 9539737
- Yang LH, Chen TM, Yu ST, Chen YH. Olanzapine induces SREBP-1-related adipogenesis in 3T3-L1 cells. Pharmacol Res 2007; 56:202 - 8; http://dx.doi.org/10.1016/j.phrs.2007.05.007; PMID: 17651982
- Yang Z, Yin JY, Gong ZC, Huang Q, Chen H, Zhang W, Zhou HH, Liu ZQ. Evidence for an effect of clozapine on the regulation of fat-cell derived factors. Clin Chim Acta 2009; 408:98 - 104; http://dx.doi.org/10.1016/j.cca.2009.07.021; PMID: 19682446
- Fernø J, Vik-Mo AO, Jassim G, Håvik B, Berge K, Skrede S, Gudbrandsen OA, Waage J, Lunder N, Mørk S, et al. Acute clozapine exposure in vivo induces lipid accumulation and marked sequential changes in the expression of SREBP, PPAR, and LXR target genes in rat liver. Psychopharmacology (Berl) 2009; 203:73 - 84; http://dx.doi.org/10.1007/s00213-008-1370-x; PMID: 18989661
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006; 7:885 - 96; http://dx.doi.org/10.1038/nrm2066; PMID: 17139329
- Sugii S, Evans RM. Epigenetic codes of PPARγ in metabolic disease. FEBS Lett 2011; 585:2121 - 8; http://dx.doi.org/10.1016/j.febslet.2011.05.007; PMID: 21605560
- Tamori Y, Masugi J, Nishino N, Kasuga M. Role of peroxisome proliferator-activated receptor-gamma in maintenance of the characteristics of mature 3T3-L1 adipocytes. Diabetes 2002; 51:2045 - 55; http://dx.doi.org/10.2337/diabetes.51.7.2045; PMID: 12086932
- Imai T, Takakuwa R, Marchand S, Dentz E, Bornert JM, Messaddeq N, Wendling O, Mark M, Desvergne B, Wahli W, et al. Peroxisome proliferator-activated receptor gamma is required in mature white and brown adipocytes for their survival in the mouse. Proc Natl Acad Sci U S A 2004; 101:4543 - 7; http://dx.doi.org/10.1073/pnas.0400356101; PMID: 15070754
- Couturier J, Patel SG, Iyer D, Balasubramanyam A, Lewis DE. Human monocytes accelerate proliferation and blunt differentiation of preadipocytes in association with suppression of C/EBPΑ mRNA. Obesity (Silver Spring) 2012; 20:253 - 62; http://dx.doi.org/10.1038/oby.2011.275; PMID: 21869759
- Musri MM, Gomis R, Párrizas M. A chromatin perspective of adipogenesis. Organogenesis 2010; 6:15 - 23; http://dx.doi.org/10.4161/org.6.1.10226; PMID: 20592861
- Fajas L, Egler V, Reiter R, Hansen J, Kristiansen K, Debril MB, Miard S, Auwerx J. The retinoblastoma-histone deacetylase 3 complex inhibits PPARgamma and adipocyte differentiation. Dev Cell 2002; 3:903 - 10; http://dx.doi.org/10.1016/S1534-5807(02)00360-X; PMID: 12479814
- Sharma RP. Schizophrenia, epigenetics and ligand-activated nuclear receptors: a framework for chromatin therapeutics. Schizophr Res 2005; 72:79 - 90; http://dx.doi.org/10.1016/j.schres.2004.03.001; PMID: 15560954
- Sharma RP, Gavin DP, Grayson DR. CpG methylation in neurons: message, memory, or mask?. Neuropsychopharmacology 2010; 35:2009 - 20; http://dx.doi.org/10.1038/npp.2010.85; PMID: 20631690
- Sharma RP, Gavin DP, Chase KA. Heterochromatin as an incubator for pathology and treatment non-response: implication for neuropsychiatric illness. Pharmacogenomics J 2012; 12:361 - 7; http://dx.doi.org/10.1038/tpj.2011.64; PMID: 22249356
- Sharma RP, Chase KA. Increasing neuronal ‘stemness’: chromatin relaxation and the expression of reprogramming genes in post-mitotic neurons. Med Hypotheses 2012; 78:553 - 4; http://dx.doi.org/10.1016/j.mehy.2011.12.018; PMID: 22326495
- Musri MM, Corominola H, Casamitjana R, Gomis R, Párrizas M. Histone H3 lysine 4 dimethylation signals the transcriptional competence of the adiponectin promoter in preadipocytes. J Biol Chem 2006; 281:17180 - 8; http://dx.doi.org/10.1074/jbc.M601295200; PMID: 16613853
- Yoo EJ, Chung JJ, Choe SS, Kim KH, Kim JB. Down-regulation of histone deacetylases stimulates adipocyte differentiation. J Biol Chem 2006; 281:6608 - 15; http://dx.doi.org/10.1074/jbc.M508982200; PMID: 16407282
- Couturier J, Patel SG, Iyer D, Balasubramanyam A, Lewis DE. Human monocytes accelerate proliferation and blunt differentiation of preadipocytes in association with suppression of C/EBPΑ mRNA. Obesity (Silver Spring) 2012; 20:253 - 62; http://dx.doi.org/10.1038/oby.2011.275; PMID: 21869759
- Fujiki K, Kano F, Shiota K, Murata M.. Expression of the peroxisome proliferator activated receptor gamma gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol 2009; 7:38 - 7007
- Wang L, Xu S, Lee JE, Baldridge A, Grullon S, Peng W, Ge K. Histone H3K9 methyltransferase G9a represses PPARγ expression and adipogenesis. EMBO J 2013; 32:45 - 59; http://dx.doi.org/10.1038/emboj.2012.306; PMID: 23178591
- Mann J, Chu DC, Maxwell A, Oakley F, Zhu NL, Tsukamoto H, Mann DA. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology 2010; 138:705 - 14
- Takada I, Mihara M, Suzawa M, Ohtake F, Kobayashi S, Igarashi M, Youn MY, Takeyama K, Nakamura T, Mezaki Y, et al. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol 2007; 9:1273 - 85; http://dx.doi.org/10.1038/ncb1647; PMID: 17952062
- Wiper-Bergeron N, Wu D, Pope L, Schild-Poulter C, Haché RJ. Stimulation of preadipocyte differentiation by steroid through targeting of an HDAC1 complex. EMBO J 2003; 22:2135 - 45; http://dx.doi.org/10.1093/emboj/cdg218; PMID: 12727880
- Inagaki T, Tachibana M, Magoori K, Kudo H, Tanaka T, Okamura M, Naito M, Kodama T, Shinkai Y, Sakai J. Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells 2009; 14:991 - 1001; http://dx.doi.org/10.1111/j.1365-2443.2009.01326.x; PMID: 19624751
- Lee J, Saha PK, Yang QH, Lee S, Park JY, Suh Y, Lee SK, Chan L, Roeder RG, Lee JW. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci U S A 2008; 105:19229 - 34; http://dx.doi.org/10.1073/pnas.0810100105; PMID: 19047629
- Cho YW, Hong S, Jin Q, Wang L, Lee JE, Gavrilova O, Ge K. Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell Metab 2009; 10:27 - 39; http://dx.doi.org/10.1016/j.cmet.2009.05.010; PMID: 19583951
- Hino S, Sakamoto A, Nagaoka K, Anan K, Wang Y, Mimasu S, Umehara T, Yokoyama S, Kosai K, Nakao M. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat Commun 2012; 3:758; http://dx.doi.org/10.1038/ncomms1755; PMID: 22453831
- Ge K.. Epigenetic regulation of adipogenesis by histone methylation. Biochim Biophys Acta 2012; 1819:727 - 732
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science 2000; 289:950 - 3; http://dx.doi.org/10.1126/science.289.5481.950; PMID: 10937998
- Wang L, Jin Q, Lee JE, Su IH, Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc Natl Acad Sci U S A 2010; 107:7317 - 22; http://dx.doi.org/10.1073/pnas.1000031107; PMID: 20368440
- Noer A, Sørensen AL, Boquest AC, Collas P. Stable CpG hypomethylation of adipogenic promoters in freshly isolated, cultured, and differentiated mesenchymal stem cells from adipose tissue. Mol Biol Cell 2006; 17:3543 - 56; http://dx.doi.org/10.1091/mbc.E06-04-0322; PMID: 16760426
- Sharma RP. Schizophrenia, epigenetics and ligand-activated nuclear receptors: a framework for chromatin therapeutics. Schizophr Res 2005; 72:79 - 90; http://dx.doi.org/10.1016/j.schres.2004.03.001; PMID: 15560954
- Dong E, Grayson DR, Guidotti A, Costa E. Antipsychotic subtypes can be characterized by differences in their ability to modify GABAergic promoter methylation. Epigenomics 2009; 1:201 - 11; http://dx.doi.org/10.2217/epi.09.2; PMID: 22122643
- Kim SH, Lee HY, Yi H, Ahn YM, Kim YS. Haloperidol induces demethylation and expression of the dual specificity phosphatase 6 gene in MIA PaCa-2 human pancreatic cancer cells. Life Sci 2012; 91:1317 - 22; http://dx.doi.org/10.1016/j.lfs.2012.10.002; PMID: 23063941
- Shimabukuro M, Jinno Y, Fuke C, Okazaki Y. Haloperidol treatment induces tissue- and sex-specific changes in DNA methylation: a control study using rats. Behav Brain Funct 2006; 2:37; http://dx.doi.org/10.1186/1744-9081-2-37; PMID: 17132176
- Sani G, Napoletano F, Forte AM, Kotzalidis GD, Panaccione I, Porfiri GM, Simonetti A, Caloro M, Girardi N, Telesforo CL, et al. The wnt pathway in mood disorders. Curr Neuropharmacol 2012; 10:239 - 53; http://dx.doi.org/10.2174/157015912803217279; PMID: 23449817
- Sutton LP, Honardoust D, Mouyal J, Rajakumar N, Rushlow WJ. Activation of the canonical Wnt pathway by the antipsychotics haloperidol and clozapine involves dishevelled-3. J Neurochem 2007; 102:153 - 69; http://dx.doi.org/10.1111/j.1471-4159.2007.04527.x; PMID: 17472703
- Borcherding DC, Hugo ER, Idelman G, De Silva A, Richtand NW, Loftus J, Ben-Jonathan N. Dopamine receptors in human adipocytes: expression and functions. PLoS One 2011; 6:e25537; http://dx.doi.org/10.1371/journal.pone.0025537; PMID: 21966540
- Sachs GS, Guille C. Weight gain associated with use of psychotropic medications. J Clin Psychiatry 1999; 60:Suppl 21 16 - 9; PMID: 10548137
- Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci U S A 2008; 105:13614 - 9; http://dx.doi.org/10.1073/pnas.0805493105; PMID: 18757738
- Li J, Guo Y, Schroeder FA, Youngs RM, Schmidt TW, Ferris C, Konradi C, Akbarian S. Dopamine D2-like antagonists induce chromatin remodeling in striatal neurons through cyclic AMP-protein kinase A and NMDA receptor signaling. J Neurochem 2004; 90:1117 - 31; http://dx.doi.org/10.1111/j.1471-4159.2004.02569.x; PMID: 15312167
- Halene TB, Siegel SJ. PDE inhibitors in psychiatry--future options for dementia, depression and schizophrenia?. Drug Discov Today 2007; 12:870 - 8; http://dx.doi.org/10.1016/j.drudis.2007.07.023; PMID: 17933689
- Baba A, Ohtake F, Okuno Y, Yokota K, Okada M, Imai Y, Ni M, Meyer CA, Igarashi K, Kanno J, et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat Cell Biol 2011; 13:668 - 75; http://dx.doi.org/10.1038/ncb2228; PMID: 21532585
- Ha CH, Kim JY, Zhao J, Wang W, Jhun BS, Wong C, Jin ZG. PKA phosphorylates histone deacetylase 5 and prevents its nuclear export, leading to the inhibition of gene transcription and cardiomyocyte hypertrophy. Proc Natl Acad Sci U S A 2010; 107:15467 - 72; http://dx.doi.org/10.1073/pnas.1000462107; PMID: 20716686
- Cassel S, Carouge D, Gensburger C, Anglard P, Burgun C, Dietrich JB, Aunis D, Zwiller J. Fluoxetine and cocaine induce the epigenetic factors MeCP2 and MBD1 in adult rat brain. Mol Pharmacol 2006; 70:487 - 92; http://dx.doi.org/10.1124/mol.106.022301; PMID: 16670375
- Maya Vetencourt JF, Tiraboschi E, Spolidoro M, Castrén E, Maffei L. Serotonin triggers a transient epigenetic mechanism that reinstates adult visual cortex plasticity in rats. Eur J Neurosci 2011; 33:49 - 57; http://dx.doi.org/10.1111/j.1460-9568.2010.07488.x; PMID: 21156002
- O’Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature 2009; 462:307 - 14; http://dx.doi.org/10.1038/nature08532; PMID: 19924209
- Willer CJ, Speliotes EK, Loos RJ, Li S, et al., Genetic Investigation of Anthropometric Traits Consortium. Six new loci associated.
- Obuchowicz E, Krysiak R, Herman ZS. Does neuropeptide Y (NPY) mediate the effects of psychotropic drugs?. Neurosci Biobehav Rev 2004; 28:595 - 610; http://dx.doi.org/10.1016/j.neubiorev.2004.08.006; PMID: 15527865