Abstract
DNA methylation changes are known to occur in gastric cancers and in premalignant lesions of the gastric mucosae. In order to examine variables associated with methylation levels, we quantitatively evaluated DNA methylation in tumors, non-tumor gastric mucosae, and in gastric biopsies at promoters of 5 genes with methylation alterations that discriminate gastric cancers from non-tumor epithelia (EN1, PCDH10, RSPO2, ZIC1, and ZNF610). Among Colombian subjects at high and low risk for gastric cancer, biopsies from subjects from the high-risk region had significantly higher levels of methylation at these 5 genes than samples from subjects in the low risk region (p ≤ 0.003). When results were stratified by Helicobacter pylori infection status, infection with a cagA positive, vacA s1m1 strain was significantly associated with highest methylation levels, compared with other strains (p = 0.024 to 0.001). More severe gastric inflammation and more advanced precancerous lesions were also associated with higher levels of DNA methylation (p ≤ 0.001). In a multivariate model, location of residence of the subject and the presence of cagA and vacA s1m1 in the H. pylori strain were independent variables associated with higher methylation in all 5 genes. High levels of mononuclear cell infiltration were significantly related to methylation in PCDH10, RSPO2, and ZIC1 genes. These results indicate that for these genes, levels of methylation in precancerous lesions are related to H. pylori virulence, geographic region and measures of chronic inflammation. These genes seem predisposed to sustain significant quantitative changes in DNA methylation at early stages of the gastric precancerous process.
Introduction
Gastric cancer was responsible for approximately 738 000 deaths worldwide in 2008, and is the 3rd most common lethal cancer in men and 5th in women.Citation1 This disease is distinctive in its association with two infectious agents, the bacterium Helicobacter pylori (H. pylori) and Epstein-Barr virus (EBV). While EBV is associated with approximately 10% of gastric cancer cases, H. pylori is a risk factor for at least 80–90% of cases, especially for non-cardiac tumors.Citation2 Furthermore, H. pylori has been classified as a human carcinogenCitation3 and, as a bacterium, can be eradicated with antibiotics. Yet, half the world’s population harbors this infection. Even if cost were not an issue, treatment of large populations with antibiotics poses the risk of development of antibiotic-resistant strains of H. pylori and other pathogenic bacteria, as well as side effects from therapy. Identification of better biomarkers for gastric cancer risk would allow more efficient targeting of prevention efforts.
H. pylori isolates are genetically diverse, and virulence differs from strain to strain. A well-established virulence factor is encoded by the cagA gene, a marker for a pathogenicity island present in some strains. The CagA protein is injected into the gastric epithelial cell by a Type IV secretion system and becomes tyrosine phosphorylated by Abl and Src family kinases. Phosphorylated CagA protein may then bind to Src Homology2-containing tyrosine phosphatase (SHP-2) and disrupt normal cell signaling.Citation4 In addition, CagA proteins are polymorphic, having three or more EPIYA (Glu-Pro-Ile-Tyr-Ala) motifs in their C-termini. Within Western H. pylori strains, CagA proteins with more than three EPIYA motifs bind more strongly to SHP-2.Citation5 Infection with such strains is associated with an increased risk for gastric cancer. Another known H. pylori virulence gene is vacA, which encodes the vacuolating cytotoxin. This protein has a polymorphic signal peptide, present as either s1 (with s1a, s1b, and s1c subtypes), or s2. The middle portion of vacA is also polymorphic, present in either m1 or m2 forms. Strains bearing vacA s1m1 alleles are associated with higher toxin production and more severe gastric pathology than strains with vacA s2m2 alleles.
H. pylori infection is associated with alterations of DNA methylation in cells of the gastric epithelium, in both humans and in the Mongolian gerbil model of gastric cancer. Chan et al. reported methylation of the CDH1 promoter to be associated with H. pylori infection in dyspeptic patients.Citation6 Using quantitative methods, Maekita et al. noted H. pylori-associated promoter methylation in LOX, HAND1, THBD, HRASLS, FLNC, ARC, and CDKN2A.Citation7 Additional hypermethylated gene promoters associated with H. pylori infection have been identified.Citation8,Citation9 Such hypermethylation may at least partially regress after H. pylori eradication.Citation10 Increased methylation is associated with higher risk for gastric cancer.Citation7
Hypermethylated promoter DNA is of additional interest as representing potential biomarkers for gastric cancer. Hypermethylated DNA for ReprimoCitation11 (RPRM, a tumor suppressor gene in gastric cancer), CDKN2A,Citation12 and other markersCitation13 have been detected in serum or plasma of gastric cancer patients at levels significantly different from those found in control patients. These findings raise hope that biomarkers may eventually be developed, that will facilitate early detection and thus reduce mortality of this disease.
Gastric cancer of the intestinal subtype develops over decades, through a series of well-defined premalignant lesions.Citation14 Infection with H. pylori induces gastritis, which may progress to multifocal atrophic gastritis (MAG), intestinal metaplasia (IM), dysplasia, and carcinoma. Although, in theory, the many years typically required for these stages to advance provide an opportunity for intervention, in practice, gastric cancers are typically diagnosed at late stages, especially in developing countries where screening is not common. To investigate DNA methylation with respect to this process of progression, we examined several variables, including severity of lesions and H. pylori virulence, to determine how these influenced levels of DNA methylation in 5 genes that become hypermethylated in gastric cancers.
Results
DNA methylation in tumors
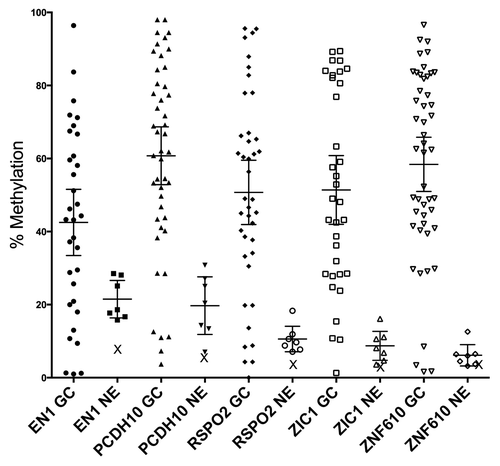
When gastric cancer samples were analyzed by pyrosequencing for methylation at EN1, PCDH10, RSPO2, ZIC1, and ZNF610, results for the set of diffuse tumors were not significantly different from that of the set of intestinal tumors, so results for both subtypes were combined. Mean methylation was 48% (38.1–57.8, 95% CI) for EN1, 67.9% (61–74.7, 95% CI) for PCDH10, 53.2% (43.4–63, 95% CI) for RSPO2, 51.5% (40.5–62.4, 95% CI) for ZIC1, and 61.8% (53.4–70.2, 95% CI) for ZNF610 (). Corresponding values for histologically normal mucosae were 22.1% (16.1–28.2, 95% CI), 21.8% (14.4–29.2, 95% CI), 11.2% (7.2–15.1, 95% CI), 9.6% (5.4–13.8, 95% CI), and 7.2% (3.3–11.1, 95% CI), respectively, all significantly different from the tumors (p < 0.0001 for all). As discriminators of tumor from non-tumor mucosae, the area under the curve (AUC) for ZIC1 was 0.95 (95% CI, 0.85–0.99); for ZNF610, 0.94 (95% CI, 0.85–0.99); for RSPO2, 0.89 (95% CI, 0.77–0.97); for PCDH10, 0.90 (95% CI, 0.77–0.96); and for EN1, 0.77 (95% CI, 0.62–0.89).
Figure 1. Methylation measurements from gastric cancers, control epithelia, and normal blood. Pyrosequencing measurements of gastric cancers reveal a large increase compared with control epithelia (p < 0.0001 for each gene). Bars indicate means, with 95% CI. X = value for pooled normal blood DNA. GaCa, gastric cancer; NE, non-tumor gastric epithelia.
Univariate analysis of biopsy results
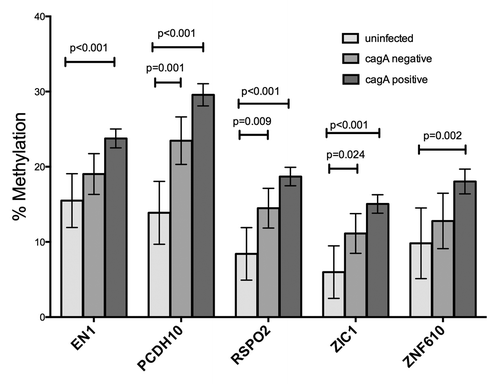
For the biopsy analysis, provides information regarding the study population and infecting H. pylori strains. Subjects residing in the high-risk region showed more advanced mucosal lesions (p = 0.024). For each of the five genes (EN1, PCDH10, RSPO2, ZIC1, and ZNF610), we stratified DNA methylation results by geographic risk region, by histopathological diagnosis, and by characteristics of the infecting H. pylori strains, including cagA, vacA s and m alleles, and the number of EPIYA motifs (3 vs. more than 3) within the CagA of the infecting strains. For all five genes, methylation was significantly increased in DNA from subjects from the high-risk region compared with those from the low risk region (P values ranging from 0.003 to 0.0003; ). When results from all subjects were stratified into categories of uninfected, infected (cagA negative) and infected (cagA positive), samples associated with cagA positive strains showed significantly higher methylation than did those from uninfected persons. Results associated with persons harboring cagA negative H. pylori strains were intermediate in DNA methylation, although for EN1 and ZNF610, the difference in methylation between DNA from uninfected persons and those with cagA negative infections was not significant (). The vacA s1m1 genotype is highly associated with cagA positivity and the vacA s2m2 genotype with cagA negativity, so the relationship of vacA genotypes to methylation produced an almost identical pattern to that of the cagA analysis (data not shown). The level of methylation was not related to the number of EPIYA motifs or to strain ancestry (European vs. African) as characterized by MLST (data not shown).
Table 1. Study population and characteristics of infecting H. pylori strains
Figure 2. Area of residence and methylation. Geographic location of residence of the subjects was significantly associated with levels of methylation in the 5 genes, both in the univariate and multivariate models. Bars indicate means, with 95% CI. LR, low risk; HR, high risk.
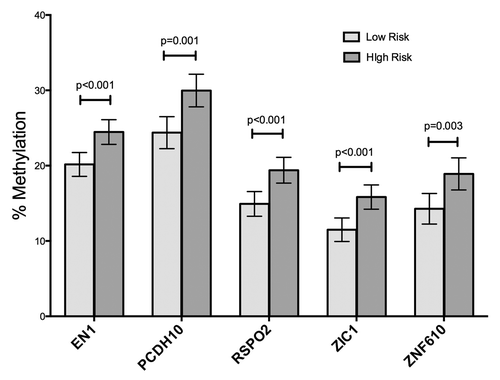
Figure 3. Virulence of the infecting H. pylori strain and methylation. The highly virulent cagA positive, vacA s1m1 strains were associated with highest levels of methylation in the 5 genes. The presence of the cagA gene is strongly associated with the presence of vacA s1 and m1 alleles. Bars indicate means, with 95% CI. Uninf, subjects having no detectable H. pylori infection; cagA negative, methylation in biopsies from subjects with H. pylori strains lacking the cagA gene; cagA positive, methylation in biopsies from subjects with H. pylori infections bearing the cagA gene.
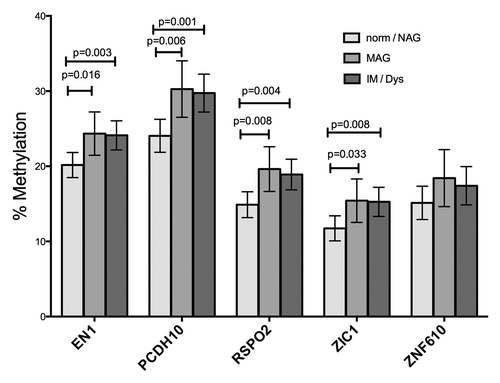
When the relationship of diagnoses to methylation was evaluated, MAG and IM/dysplasia categories had higher methylation in all genes except ZNF610, compared with samples from uninfected persons or persons with non-atrophic gastritis (NAG) (). Methylation levels measured in samples from subjects diagnosed with MAG vs IM / dysplasia were not different from each other. Results were similar, for evaluation by both categories, or by histopathology score in a multivariate model. Biopsies diagnosed with dysplasia were too few to analyze separately, so they were grouped with those diagnosed with IM. In the age range of our subjects (39 to 60 y), methylation was not significantly related to age for any of the five genes. Methylation in all 5 genes was highly correlated with methylation in the other genes, with correlation coefficients for each comparison ranging from 0.7882 for EN1 with ZNF610 to 0.9609 for ZIC1 with RSPO2. In univariate analysis, the mononuclear cell (MN) score was significantly related to methylation in all 5 genes (p < 0.001 for all); however, because the MN score is closely associated with H. pylori virulence, region and histology score, a multivariate approach was necessary. In univariate analysis, the PMN score was significantly related to methylation for PCDH10 only (p = 0.007).
Figure 4. Histological diagnosis and methylation. Pyrosequencing measurements from gastric biopsy DNA samples were stratified by the most advanced gastric lesion diagnosed in the subjects. Histological diagnosis was significantly associated in the univariate model with methylation in 4 of the 5 genes, but lost significance in the multivariate model, when cagA status was included.
Multivariate analysis of biopsy results
shows multivariate analysis of methylation differences in gastric biopsies. Location of residence (HR vs. LR) and H. pylori virulence factors cagA and vacA s and m remained significant variables in the model. Histological diagnosis lost significance in the model, once the H. pylori virulence factors were included. In a multivariate model for average methylation across the 5 genes, region and H. pylori virulence remained as significant variables. Chronic inflammation, as measured by the level of infiltration of MN cells, remained significant for methylation at PCDH10, RSPO2, and ZIC1 after adjusting for other variables. The PMN score, significantly related to methylation only for PCDH10 in univariate analysis, lost significance in the multivariate analysis.
Table 2. Multivariate generalized linear model* analysis
Discussion
For analyses of DNA methylation, we selected genes with possible functionality in promoting tumor development, and which showed marked discrimination of tumor from non-tumor gastric mucosae in their levels of promoter methylation. Notably, these genes also showed variation in methylation in premalignant lesions, unlike some others we previously reported, for example, APC.Citation15 EN1 (engrailed homeobox 1) encodes a homeodomain-containing protein involved in development of the central nervous system. EN1 lies within the middle portion of a region of long-range epigenetic silencing in colorectal cancer, located on chromosome 2q14.2. EN1 is hypermethylated in 70% of colorectal cancers and has been suggested as a potential biomarker for this tumor type.Citation16 In addition, EN1 is hypermethylated in 65% of prostate cancersCitation17 and in 80% of serous ovarian cancers.Citation18
PCDH10, a member of the protocadherin family, encodes a calcium-dependent cell-cell adhesion molecule. Although expressed at highest levels in the central nervous system, PCDH10 has tumor suppressor activity in gastric cancer,Citation19,Citation20 as well as in nasopharyngeal, esophageal, and colorectal cancers.Citation21PCDH10 is hypermethylated in 82% of nasopharyngeal carcinomas, 51% of esophageal carcinomas, 45% of breast cancers, 42% of hepatocellular carcinomas, and in 42%Citation21 to over 80% of gastric cancers.Citation19
RSPO2 (R spondin family member 2) encodes a secreted protein that regulates β-catenin signaling and can modulate invasiveness of cultured mammary epithelial cells. RSPO2 is an important regulator of lung, limb, and tracheal development. Although reports mainly indicate a growth-promoting effect of RSPO2 expression,Citation22 stable expression of transfected RSPO2 in mammary epithelial cells was associated with lowered rates of cell proliferation in monolayer culture.Citation23 The RSPO2 protein may maintain, rather than increase Wnt pathway activation.Citation24 In a genetic screen following insertion of a transposon, RSPO2 was identified as a gene that when dysregulated, may lead to colorectal cancer in a mouse model.Citation25 Of interest, cagA-positive H. pylori strains can activate β-catenin.Citation26
ZIC1 (zinc finger protein of the cerebellum 1) encodes a zinc finger protein important in organogenesis in the central nervous system, especially for development of the dorsal spinal cord and cerebellum. The ZIC1 protein regulates Nodal and Sonic Hedgehog signaling, as well as retinoic acid signaling,Citation27 and ZIC1 may serve as a tumor suppressor gene in colorectal cancers.Citation28 In earlier studies, ZIC1 was reported to be hypermethylated in 95% of gastric cancersCitation29 and in 85% of colorectal cancers.Citation27 When the ZIC1 protein was ectopically expressed in gastric cancer cell lines, colony formation was reduced.Citation29 Zhong et al. showed that when the ZIC1 protein was ectopically expressed in gastric cancer cell lines, cell cycle distributions were altered (more cells were found in G1 phase), expression of the cyclin-dependent kinase inhibitors p21 and p27 was increased, and expression of sonic hedgehog was reduced.Citation30 While methylation of the ZIC1 promoter had a numerically higher AUC value in discriminating tumors from non-tumor mucosae, the difference from the other genes was not significant.
ZNF610 encodes zinc finger protein 610, which belongs to the Krüppel C2H2-type zinc finger protein family. ZNF610 encodes 9 C2H2-type zinc fingers and 1 Krüppel-associated box (KRAB) domain. KRAB domains may serve as transcriptional repressors by recruiting histone-modifying proteins and interacting with the co-repressor KAP1.Citation31 Other members of the KRAB family of proteins act to inhibit transformation by MycCitation32 and affect p53-dependent apoptosis.Citation33
Our study detected quantitative alterations in methylation occurring in biopsies of subjects diagnosed with MAG, compared with biopsies from uninfected subjects or those with NAG. This finding is interesting, in light of the fact that MAG (or chronic atrophic gastritis) is the gastric lesion recognized as the earliest in the precancerous cascade.Citation14 This finding is also consistent with the Epigenetic Progenitor model of cancer development,Citation34 which proposes that the earliest disruptions in the development of a malignancy are epigenetic ones. Notably, we found no significant differences between MAG and IM lesions in the biopsies, but in the gastric cancers, levels of methylation were highly variable, with some approaching 100% and some having levels similar to those of the precancerous lesions.
Subjects infected with the more virulent strains of H. pylori had higher levels of methylation. Levels of hypermethylation increased progressively in DNA from uninfected subjects, to subjects with cagA negative infections, and to subjects with cagA positive infections. This effect of the H. pylori virulence had a stronger association than the histology variable in the multivariate model. We were unable to separate the effect of cagA from that of vacA s1m1, due to the strong association of these virulence factors with each other. Although the association of H. pylori infection with DNA methylation has been known for some years,Citation6,Citation7 the effect of virulence determinants has been less explored. Examination of gastric washings from gastric cancer patients and controls indicated no association with cagA positivity in the strains.Citation35 However, Sepulveda et al. found a higher proportion of their set of gastric biopsies designated as positive for MGMT methylation to be associated with cagA positivity vs cagA negativity in the infecting strain.Citation36 In contrast to these prior studies, our study measured average levels of methylation in individual tissues, which may reflect the cell composition and expansion of cell compartments.
The finding of the association of mononuclear inflammatory cell infiltration with high levels of methylation in three of the genes is consistent with the idea that chronic inflammation promotes aberrant DNA methylation. In the gerbil model of H. pylori-induced gastric cancer, treatment of infected animals with cyclosporin A to repress inflammation suppressed methylation while not altering the presence of the H. pylori, suggesting inflammation, not infection in itself, is responsible for the hypermethylation effect.Citation37 The importance of inflammation was underscored by Yoshida et al. in a study that associated levels of DNA methylation in gastric mucosae with titers of antibodies against H. pylori and with pepsinogen I/pepsinogen II ratios.Citation38 In a study of methylation assayed by qualitative means, examining 11 genes in gastric biopsies, Kang et al. found that the number of the 11 genes marked with methylation was greater in biopsies with high monocyte infiltration.Citation39 Our study examines the effect of chronic inflammation within individual tissues, noting quantitative differences in methylation of individual genes.
We observed a wide range of methylation levels among the set of gastric cancers tested. Yet, given this heterogeneity, for the biopsy material, it is notable that patterns of univariate analysis, stratifying by geographic region, H. pylori virulence, and histological diagnosis, are similar for all 5 genes examined in detail. In a quantitative study of promoter methylation in 4 candidate genes (APC, MGMT, RPRM, and TWIST1), only RPRM showed an association of promoter hypermethylation with more virulent infecting H. pylori strains, more severe gastric lesions, and geographic location.Citation15 The genes EN1, PCDH10, RSPO2, ZIC1, and ZNF610, with RPRM, form a list of genes that seem prone to sustain significant measurable changes in DNA methylation at early stages of disease.
In conclusion, we have identified a panel of genes that sustain DNA hypermethylation in gastric cancers, and that also show significant methylation differences in gastric mucosae in early stages of the precancerous process. These differences are associated with H. pylori virulence, geographic area and (for PCDH10, RSPO2, and ZIC1) differences in chronic inflammation, in a multivariate model. All these variables are known to be related to gastric cancer risk. The practical utilities of these genes as markers for disease progression in high-risk populations are under investigation.
Methods
Human gastric cancer tissues
A total of 53 gastric adenocarcinomas (9 frozen, 44 formalin-fixed, paraffin-embedded, or FFPE) were used in the study. Of these, 43 were of the intestinal subtype and 10 were diffuse. De-identified frozen gastric adenocarcinomas (n = 9) plus matched non-tumor gastric mucosae from the same patients were obtained from Vanderbilt’s Translational Pathology Shared Resource and from the Cooperative Human Tissue Network. These tissues were collected with approval of Vanderbilt’s Institutional Review Board, in association with the Cooperative Human Tissue Network. Sections (5 microns) were harvested on slides, fixed in 70% ethanol and dehydrated in graded ethanol solutions and xylene. FFPE gastric adenocarcinomas were obtained from Temuco, Chile; Pasto, Colombia; New Orleans, LA; San Antonio, TX; and Nashville, TN. Chilean tumors (n = 22 FFPE) came from the Hospital Regional Temuco. Colombian tumors (n = 14 FFPE) were provided from the Hospital Fundacion San Pedro. Gastric adenocarcinoma tissues from North American patients (n = 8 FFPE) were obtained from University Hospital, Touro Infirmary, and Baptist Medical Center. The use of these tissues was approved by the Ethics Committee of the Universidad de la Frontera, the Ethics Committee of the Hospital Fundacion San Pedro, the Institutional Review Boards of the Louisiana State University Health Sciences Center, the Vanderbilt University Medical Center, and the University of Texas Health Sciences Center. Cancer cells or normal epithelia were harvested either by laser capture microdissection (LCM) with an Arcturus Pixcell IIe instrument (Life Technologies), or by manual microdissection when the desired cell type was at least 50% of total cells. Following LCM, harvested cells were digested overnight from the Capsure® using proteinase K, heated to denature the proteinase K, and subjected to bisulfite modification without additional purification.
Human gastric mucosal biopsies with premalignant lesions
Subjects providing gastric biopsies were 88 men with dyspeptic symptoms, aged 39 to 60 y old, who underwent upper endoscopy in gastroenterology clinics in two public hospitals in Colombia between June and September 2006. Subjects were residents either of the Andean community of Tuquerres, or of Tumaco on the Pacific coast. The mountain community has incidence rates of gastric cancer approximately 25 times that of the coastal community.Citation40 All subjects provided informed consent and ethics committees of the participating hospitals and of the Universidad del Valle approved the study protocol. Exclusion criteria for the study were chronic conditions such as diabetes or heart disease, or prior gastrectomy, or use of proton pump inhibitors, H2-receptor antagonists, or antimicrobials in the month before the endoscopy. Biopsy samples were taken from the antrum (greater curvature, within 5 cm of the pylorus), incisura angularis (lesser curvature), and corpus (middle anterior wall) by a single experienced gastroenterologist. One biopsy from each site was fixed in buffered formalin and embedded in paraffin for histopathology. One antral biopsy was frozen in glycerol/thioglycolate for culture of H. pylori organisms, and another for analysis of DNA methylation. Frozen biopsies were shipped on dry ice to Vanderbilt University and stored at −80 °C.
Histopathology
From paraffin blocks, 4-micron sections were stained with hematoxylin and eosin for independent diagnosis by two experienced pathologists (MBP and PC). Discordant diagnoses were reviewed until a consensus developed. Diagnosis was performed by established guidelinesCitation41,Citation42 discriminating categories of normal, non-atrophic gastritis (NAG), multifocal atrophic gastritis without intestinal metaplasia (MAG), intestinal metaplasia (IM), or dysplasia. Infiltration by polymorphonuclear (PMN) and mononuclear (MN) cells was evaluated in biopsies from antrum and incisura angularis and classified with a 0–3 scale (normal, mild, moderate, marked) according to published criteria.Citation41 Augmented histopathology scores were characterized as described previously.Citation43 Average inflammation scores were calculated for all biopsies from antrum and incisura angularis for each subject. An individual was designated as uninfected with H. pylori only if both these conditions were met: no H. pylori isolates could be cultured, and no H. pylori organisms were visible in Steiner stained preparations of any of 3 biopsies. This protocol identified 8 subjects who were designated as currently uninfected; of these, 4 had normal mucosae, 3 had NAG, and one had IM. Pathologists evaluated diagnoses and inflammation without knowledge of residence area of the subjects.
Quantitative analysis of DNA methylation by pyrosequencing
Up to 2 μg of DNA per sample was bisulfite modified using a Zymo EZ Methylation Direct kit (Zymo Research Corp.) or an Epitect Bisulfite kit (Qiagen). Laser captured templates were used undiluted from the 20 μl sample eluted from the column supplied with the bisulfite modification kit. Modified DNA (20 ng per reaction for DNA from frozen tissue) was amplified by PCR, using Pyromark assays for EN1 (Hs_EN1_02_PM, PM00101752), PCDH10 (Hs_PCDH10_06_PM, PM00111783), RSPO2 (Hs_RSPO2_01_PM, PM00036267), ZIC1 (Hs_ZIC1_03_PM, PM00015099), or ZNF610 (HS_ZNF610_01_PM, PM00191331), using Amplitaq Gold DNA polymerase (Applied Biosystems, Life Technologies). The amplification program was 95°C for 15 min., then 45 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s, followed by a 10 min. incubation at 72 °C. PCR products were analyzed by gel electrophoresis in 2% agarose, to determine that a single band was obtained. The biotinylated strand of each PCR product was isolated from 10 μl of each PCR product according to the manufacturer’s protocol. Pyrosequencing® reactions were performed using a PyroMark MD Pyrosequencing instrument (Qiagen), following the manufacturer’s recommendations. Bisulfite-modified Methylated HeLa DNA (New England Biolabs) was employed as a positive control with each assay; bisulfite-modified pooled normal human blood DNA (Promega Corp) and no-template buffer were used as negative controls, at least one of which was used with each assay. Methylation values used for analysis were the mean of the percent methylation of the first 2 to 4 potential methylation sites quantitated in the assay following the sequencing primer.
H. pylori culture and genotyping
Culture of H. pylori organisms (one colony each, from one antral biopsy), isolation of DNA, and genotyping for cagA and vacA s and m regions for this set of subjects was previously reported.Citation15 Sanger sequencing of the polymorphic 3′ end of the cagA gene was used to characterize EPIYA motifs.Citation44 Previously, H. pylori strains from these subjects were characterized for ancestry by Multilocus Sequence Typing (MLST).Citation43
Statistical analysis
Histological diagnoses, age, H. pylori virulence genes, numbers of EPIYA motifs, and levels of methylation at the five gene promoters were compared by risk area using χ2, Spearman, and the Student t tests, as appropriate. To assess the contribution of the different variables to levels of methylation and account for confounders, multivariate generalized linear models were employed. For binary and continuous variables, the coefficients represent the average difference in percentage of methylation for a one unit change in the predictor (i.e., for every year of age, or low vs. high risk area). For H. pylori genotypes, the coefficient represents the average difference in percentage of methylation associated with membership in the specified category (cagA and vacA s1m1 or other genotypes), compared with the reference group (uninfected). Nonparametric estimation of receiver operating characteristic (ROC) curves was used to compute AUC values for percent methylation of all genes. Data were analyzed using Stata MP v13 (Stata Corporation).
To assess reproducibility of the Pyrosequencing results for the biopsies, Lin’s concordance correlation coefficient was calculated on 152 replicates. The coefficient’s value was 0.939 (95% CI 0.920–0.958). The average difference between the replicates was 0.031 with a standard deviation of 2.6 (95% limit of agreement, −5.201–5.263).
| Abbreviations: | ||
| EBV | = | Epstein Barr virus |
| LCM | = | laser capture microdissection |
| FFPE | = | formalin-fixed, paraffin-embedded |
| MLST | = | multi-locus sequence typing |
| HR | = | high risk |
| LR | = | low risk |
| H. pylori | = | Helicobacter pylori |
| NAG | = | non-atrophic gastritis |
| IM | = | intestinal metaplasia |
| MAG | = | multi-focal atrophic gastritis |
| DYS | = | dysplasia |
| PMN | = | polymorphonuclear cells |
| MN | = | mononuclear cells |
| CI | = | confidence interval |
| IRB | = | institutional review board |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was supported by the National Center for Research Resources, grant UL1 RR024975-01, which is now at the National Center for Advancing Translational Sciences, Grant 2 UL2 TR000445-06; grants P01 CA28842, R01 CA095103, R01 CA77955, R01 CA93999, P01 CA116387 from the National Cancer Institute, P30 DK058404, R01 DK053620, and R01 DK58587. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute, NIH, Department of Veterans Affairs or Vanderbilt University. We wish to acknowledge assistance of Anthony Frazier in Vanderbilt’s Translational Pathology Shared Resource. Frozen gastric cancers and matched non-tumor epithelia were provided by the Vanderbilt Translational Pathology Shared Resource and the Cooperative Human Tissue Network.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69 - 90; http://dx.doi.org/10.3322/caac.20107; PMID: 21296855
- Webb PM, Law M, Varghese C, Forman D, Yuan JM, Yu M, et al, Helicobacter and Cancer Collaborative Group. Gastric cancer and Helicobacter pylori: a combined analysis of 12 case control studies nested within prospective cohorts. Gut 2001; 49:347 - 53; http://dx.doi.org/10.1136/gut.49.3.347; PMID: 11511555
- IARC Working Group. IARC monograph on the evaluation of carcinogenic risks to humans: Schistosomes, liver flukes and Helicobacter pyloried., vol. 61. Lyon: International Agency for Research on Cancer, 1994.
- Hatakeyama M. SagA of CagA in Helicobacter pylori pathogenesis. Curr Opin Microbiol 2008; 11:30 - 7; http://dx.doi.org/10.1016/j.mib.2007.12.003; PMID: 18243773
- Naito M, Yamazaki T, Tsutsumi R, Higashi H, Onoe K, Yamazaki S, Azuma T, Hatakeyama M. Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. Gastroenterology 2006; 130:1181 - 90; http://dx.doi.org/10.1053/j.gastro.2005.12.038; PMID: 16618412
- Chan AO, Lam SK, Wong BC, Wong WM, Yuen MF, Yeung YH, Hui WM, Rashid A, Kwong YL. Promoter methylation of E-cadherin gene in gastric mucosa associated with Helicobacter pylori infection and in gastric cancer. Gut 2003; 52:502 - 6; http://dx.doi.org/10.1136/gut.52.4.502; PMID: 12631658
- Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res 2006; 12:989 - 95; http://dx.doi.org/10.1158/1078-0432.CCR-05-2096; PMID: 16467114
- Kang GH, Lee S, Cho NY, Gandamihardja T, Long TI, Weisenberger DJ, Campan M, Laird PW. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab Invest 2008; 88:161 - 70; http://dx.doi.org/10.1038/labinvest.3700707; PMID: 18158559
- Nakajima T, Yamashita S, Maekita T, Niwa T, Nakazawa K, Ushijima T. The presence of a methylation fingerprint of Helicobacter pylori infection in human gastric mucosae. Int J Cancer 2009; 124:905 - 10; http://dx.doi.org/10.1002/ijc.24018; PMID: 19035455
- Leung WK, Man EP, Yu J, Go MY, To KF, Yamaoka Y, Cheng VY, Ng EK, Sung JJ. Effects of Helicobacter pylori eradication on methylation status of E-cadherin gene in noncancerous stomach. Clin Cancer Res 2006; 12:3216 - 21; http://dx.doi.org/10.1158/1078-0432.CCR-05-2442; PMID: 16707623
- Bernal C, Aguayo F, Villarroel C, Vargas M, Díaz I, Ossandon FJ, Santibáñez E, Palma M, Aravena E, Barrientos C, et al. Reprimo as a potential biomarker for early detection in gastric cancer. Clin Cancer Res 2008; 14:6264 - 9; http://dx.doi.org/10.1158/1078-0432.CCR-07-4522; PMID: 18829507
- Abbaszadegan MR, Moaven O, Sima HR, Ghafarzadegan K, A’rabi A, Forghani MN, Raziee HR, Mashhadinejad A, Jafarzadeh M, Esmaili-Shandiz E, et al. p16 promoter hypermethylation: a useful serum marker for early detection of gastric cancer. World J Gastroenterol 2008; 14:2055 - 60; http://dx.doi.org/10.3748/wjg.14.2055; PMID: 18395906
- Zheng Y, Chen L, Li J, Yu B, Su L, Chen X, Yu Y, Yan M, Liu B, Zhu Z. Hypermethylated DNA as potential biomarkers for gastric cancer diagnosis. Clin Biochem 2011; 44:1405 - 11; http://dx.doi.org/10.1016/j.clinbiochem.2011.09.006; PMID: 21945024
- Correa P. Human Gastric Carcinogenesis: A multistep and multifactorial process–First American Cancer Society award lecture on cancer epidemiology and prevention. Cancer Res 1992; 52:6735 - 40; PMID: 1458460
- Schneider BG, Peng DF, Camargo MC, Piazuelo MB, Sicinschi LA, Mera R, Romero-Gallo J, Delgado AG, Bravo LE, Wilson KT, et al. Promoter DNA hypermethylation in gastric biopsies from subjects at high and low risk for gastric cancer. Int J Cancer 2010; 127:2588 - 97; http://dx.doi.org/10.1002/ijc.25274; PMID: 20178103
- Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet 2006; 38:540 - 9; http://dx.doi.org/10.1038/ng1781; PMID: 16642018
- Devaney J, Stirzaker C, Qu W, Song JZ, Statham AL, Patterson KI, Horvath LG, Tabor B, Coolen MW, Hulf T, et al. Epigenetic deregulation across chromosome 2q14.2 differentiates normal from prostate cancer and provides a regional panel of novel DNA methylation cancer biomarkers. Cancer Epidemiol Biomarkers Prev 2011; 20:148 - 59; http://dx.doi.org/10.1158/1055-9965.EPI-10-0719; PMID: 21098650
- Montavon C, Gloss BS, Warton K, Barton CA, Statham AL, Scurry JP, Tabor B, Nguyen TV, Qu W, Samimi G, et al. Prognostic and diagnostic significance of DNA methylation patterns in high grade serous ovarian cancer. Gynecol Oncol 2012; 124:582 - 8; http://dx.doi.org/10.1016/j.ygyno.2011.11.026; PMID: 22115852
- Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN, Geng H, Tian LW, Wong YP, Tong JH, Ying JM, et al. Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology 2009; 136:640 - 51, e1; http://dx.doi.org/10.1053/j.gastro.2008.10.050; PMID: 19084528
- Li Z, Chim JC, Yang M, Ye J, Wong BC, Qiao L. Role of PCDH10 and its hypermethylation in human gastric cancer. Biochim Biophys Acta 2012; 1823:298 - 305; http://dx.doi.org/10.1016/j.bbamcr.2011.11.011; PMID: 22206871
- Ying J, Li H, Seng TJ, Langford C, Srivastava G, Tsao SW, Putti T, Murray P, Chan AT, Tao Q. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene 2006; 25:1070 - 80; http://dx.doi.org/10.1038/sj.onc.1209154; PMID: 16247458
- Kim KA, Zhao J, Andarmani S, Kakitani M, Oshima T, Binnerts ME, Abo A, Tomizuka K, Funk WD. R-Spondin proteins: a novel link to beta-catenin activation. Cell Cycle 2006; 5:23 - 6; http://dx.doi.org/10.4161/cc.5.1.2305; PMID: 16357527
- Klauzinska M, Baljinnyam B, Raafat A, Rodriguez-Canales J, Strizzi L, Greer YE, Rubin JS, Callahan R. Rspo2/Int7 regulates invasiveness and tumorigenic properties of mammary epithelial cells. J Cell Physiol 2012; 227:1960 - 71; http://dx.doi.org/10.1002/jcp.22924; PMID: 21732367
- Baljinnyam B, Klauzinska M, Saffo S, Callahan R, Rubin JS. Recombinant R-spondin2 and Wnt3a up- and down-regulate novel target genes in C57MG mouse mammary epithelial cells. PLoS One 2012; 7:e29455; http://dx.doi.org/10.1371/journal.pone.0029455; PMID: 22238613
- Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O’Sullivan MG, Matise I, Dupuy AJ, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 2009; 323:1747 - 50; http://dx.doi.org/10.1126/science.1163040; PMID: 19251594
- Franco AT, Israel DA, Washington MK, Krishna U, Fox JG, Rogers AB, Neish AS, Collier-Hyams L, Perez-Perez GI, Hatakeyama M, et al. Activation of beta-catenin by carcinogenic Helicobacter pylori.. Proc Natl Acad Sci U S A 2005; 102:10646 - 51; http://dx.doi.org/10.1073/pnas.0504927102; PMID: 16027366
- Maurus D, Harris WA. Zic-associated holoprosencephaly: zebrafish Zic1 controls midline formation and forebrain patterning by regulating Nodal, Hedgehog, and retinoic acid signaling. Genes Dev 2009; 23:1461 - 73; http://dx.doi.org/10.1101/gad.517009; PMID: 19528322
- Gan L, Chen S, Zhong J, Wang X, Lam EK, Liu X, Zhang J, Zhou T, Yu J, Si J, et al. ZIC1 is downregulated through promoter hypermethylation, and functions as a tumor suppressor gene in colorectal cancer. PLoS One 2011; 6:e16916; http://dx.doi.org/10.1371/journal.pone.0016916; PMID: 21347233
- Wang LJ, Jin HC, Wang X, Lam EK, Zhang JB, Liu X, Chan FK, Si JM, Sung JJ. ZIC1 is downregulated through promoter hypermethylation in gastric cancer. Biochem Biophys Res Commun 2009; 379:959 - 63; http://dx.doi.org/10.1016/j.bbrc.2008.12.180; PMID: 19135984
- Zhong J, Chen S, Xue M, Du Q, Cai J, Jin H, Si J, Wang L. ZIC1 modulates cell-cycle distributions and cell migration through regulation of sonic hedgehog, PI(3)K and MAPK signaling pathways in gastric cancer. BMC Cancer 2012; 12:290; http://dx.doi.org/10.1186/1471-2407-12-290; PMID: 22799764
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol 2003; 4:231; http://dx.doi.org/10.1186/gb-2003-4-10-231; PMID: 14519192
- Hennemann H, Vassen L, Geisen C, Eilers M, Möröy T. Identification of a novel Krüppel-associated box domain protein, Krim-1, that interacts with c-Myc and inhibits its oncogenic activity. J Biol Chem 2003; 278:28799 - 811; http://dx.doi.org/10.1074/jbc.M207196200; PMID: 12748187
- Tian C, Xing G, Xie P, Lu K, Nie J, Wang J, Li L, Gao M, Zhang L, He F. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol 2009; 11:580 - 91; http://dx.doi.org/10.1038/ncb1864; PMID: 19377469
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet 2006; 7:21 - 33; http://dx.doi.org/10.1038/nrg1748; PMID: 16369569
- Watanabe Y, Kim HS, Castoro RJ, Chung W, Estecio MR, Kondo K, Guo Y, Ahmed SS, Toyota M, Itoh F, et al. Sensitive and specific detection of early gastric cancer with DNA methylation analysis of gastric washes. Gastroenterology 2009; 136:2149 - 58; http://dx.doi.org/10.1053/j.gastro.2009.02.085; PMID: 19375421
- Sepulveda AR, Yao Y, Yan W, Park DI, Kim JJ, Gooding W, Abudayyeh S, Graham DY. CpG methylation and reduced expression of O6-methylguanine DNA methyltransferase is associated with Helicobacter pylori infection. Gastroenterology 2010; 138:1836 - 44; http://dx.doi.org/10.1053/j.gastro.2009.12.042; PMID: 20044995
- Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, Ichinose M, Tatematsu M, Ushijima T. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res 2010; 70:1430 - 40; http://dx.doi.org/10.1158/0008-5472.CAN-09-2755; PMID: 20124475
- Yoshida T, Kato J, Maekita T, Yamashita S, Enomoto S, Ando T, Niwa T, Deguchi H, Ueda K, Inoue I, et al. Altered mucosal DNA methylation in parallel with highly active Helicobacter pylori-related gastritis. Gastric Cancer 2013; http://dx.doi.org/10.1007/s10120-012-0230-x; PMID: 23292007
- Kang GH, Lee HJ, Hwang KS, Lee S, Kim JH, Kim JS. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol 2003; 163:1551 - 6; http://dx.doi.org/10.1016/S0002-9440(10)63511-0; PMID: 14507661
- Correa P, Cuello C, Duque E, Burbano LC, Garcia FT, Bolanos O, Brown C, Haenszel W. Gastric cancer in Colombia. III. Natural history of precursor lesions. J Natl Cancer Inst 1976; 57:1027 - 35; PMID: 1003539
- Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol 1996; 20:1161 - 81; http://dx.doi.org/10.1097/00000478-199610000-00001; PMID: 8827022
- Rugge M, Correa P, Dixon MF, Hattori T, Leandro G, Lewin K, Riddell RH, Sipponen P, Watanabe H. Gastric dysplasia: the Padova international classification. Am J Surg Pathol 2000; 24:167 - 76; http://dx.doi.org/10.1097/00000478-200002000-00001; PMID: 10680883
- de Sablet T, Piazuelo MB, Shaffer CL, Schneider BG, Asim M, Chaturvedi R, Bravo LE, Sicinschi LA, Delgado AG, Mera RM, et al. Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut 2011; 60:1189 - 95; http://dx.doi.org/10.1136/gut.2010.234468; PMID: 21357593
- Sicinschi LA, Correa P, Peek RM Jr., Camargo MC, Piazuelo MB, Romero-Gallo J, Hobbs SS, Krishna U, Delgado A, Mera R, et al. CagA C-terminal variations in Helicobacter pylori strains from Colombian patients with gastric precancerous lesions. Clin Microbiol Infect 2010; 16:369 - 78; http://dx.doi.org/10.1111/j.1469-0691.2009.02811.x; PMID: 19456839