Abstract
Non-coding RNAs and epigenetics are remarkable mechanisms of cellular control. In this review we underline the processes by which non-coding RNAs (ncRNAs), shown to be involved in various diseases, are capable of modifying and being modified by the epigenetic machinery, emphasizing the clinical importance of this network in cancer. Many ncRNAs have been described that play important roles in the establishment and maintenance of the epigenome. However, only a few studies deeply take into account the role of ncRNAs from a clinicopathological standpoint. The wide range of interactions between the non-coding RNome and the epigenome, and the roles of these networks in the pathogenesis, prognosis and early diagnosis of many diseases, present new challenges and opportunities for future studies regarding therapeutic strategies in oncology.
Introduction
For many decades, since molecular biology arose as a shedding light in the oncology field, cancer was believed to be a purely genetic disease. However, with the emergence of epigenetics, the unquestionable intricacy of this disease became even more evident. Derived from Latin (the prefix epi means upon, over), epigenetics is the study of alterations that are beyond those at the genetics level—Watson and Crick DNA pairing properties; the term is used to describe those events that cannot be solely explained by genetics but can be inherited by mitosis or meiosis.Citation1,Citation2 These events are of outstanding importance, as they are known to contribute to deregulation of gene expression. Among the most important epigenetic mechanisms are DNA methylation, histone modifications and non-coding RNAs (ncRNAs) effects.
Non-coding RNAs are an extensive class of evolutionarily conserved RNAs that are not translated into proteins.Citation4 Their discovery was a breakthrough in science, responsible for promoting a new vision of the former biology “central dogma” (DNA→RNA→Protein) and for the “junk RNA” theory, in which the term “junk RNA” comprised all those RNAs that were not translated—about 70% of the genome is transcribed but approximately only 2% is translated—and, therefore, were believed to have no function.Citation5-Citation7 By now, these molecules have been extensively shown to play roles in the regulation of the translation and transcription of both coding and non-coding genes. As a result, the cancer transcriptome is currently viewed as more complex than previously imagined.Citation4,Citation8
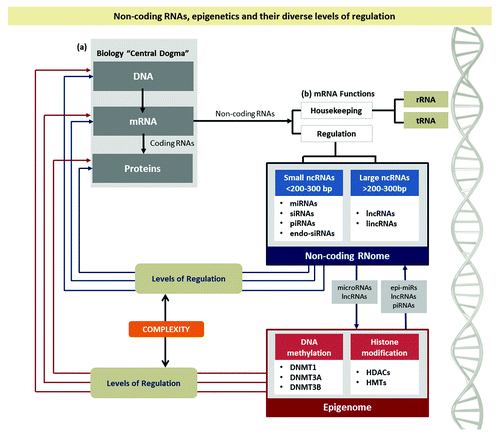
Many important functions are encompassed by ncRNAs, including, (1) architectural functions, such as structuring ribosomal subunits (rRNAs); (2) transportation functions during the translation of proteins, in which tRNA (tRNA) act together with RNA ligands and mRNACitation9 and; (3) regulatory functions through modulation of RNA, DNA, and proteins. Depending on their sizes, regulatory ncRNAs can be classified into small ncRNAs (< 200–300 bp)—including microRNAs (miRNAs), short interfering RNAs (siRNAs), endo-siRNAs and PIWI-interacting RNAs (piRNAs)— and large ncRNAs (lncRNAs; >200–300 bp).Citation8 provides a broad overview of ncRNAs and their epigenetic levels of regulation.
Figure 1. Non-coding RNAs, epigenetics and their diverse levels of regulation. (A) The gray box illustrates the classical Biology “Central Dogma” and the emergence of a new class of RNAs, the ncRNAs, and their main functions (B). The non-coding RNome is shown in the blue box, comprised by short and long ncRNAs and the epigenome is shown as a red box. Their levels of regulation are shown by arrows. The complexity of the interactions between the miRNome and the epigenome can be easily visualized.
Since their discovery,Citation10,Citation11 much has been said about miRNAs. These small ncRNAs are approximately 22 nucleotides long, single-stranded RNAs that are involved in the pathogenesis of all tumor types studied so far, acting in a very complex manner. Their regulatory networks involve, among others, the translational repression and/or cleavage of target complementary mRNA, the targeting of mRNA-binding proteins, and the binding of promoters and regions other than 3′ UTRs, such as genic regions.Citation12
Piwi-interacting RNAs (piRNAs) represent a set of small ncRNAs that are 26–31 nucleotides long and were discovered by their association Piwi, a protein from the Argonaute subfamily restricted to germline. piRNAs possess a 5′ uridine and target antisense transposon sequences.Citation4,Citation8 They are produced from the cleavage of selfish and repetitive genomic element transcripts.Citation13
Long ncRNAs are RNAs ranging from 200 bp to multiple kilobases in size. In several pathological states, including cancer, they have been verified as major players in gene expression regulation, representing the largest class of ncRNAs and accounting for a great part of the transcribed genome.Citation5 However, in spite of their magnitude, they are the least comprehended entity generated by the eukaryotic genome.Citation4 Similarly to RNAs that encode for proteins, lncRNAs are 5′ capped, spliced, and polyadenylated; the absence of a protein-coding open reading frame is what makes them different from mRNAs. Long ncRNAs can be divided into several major subtypes, the long intergenic ncRNAs (lincRNAs) and the natural antisense transcripts (NATs) being the most well-known so far. LincRNAs are located between protein-coding loci or in introns; NATs are RNAs with sequence complementarity to another RNA transcript.Citation5,Citation14
Non-coding RNAs and epigenetics are remarkable mechanisms of cellular control. In this review, we attempted to provide a better understanding of how the epigenome regulates and is regulated by ncRNAs and what clinical importance these interactions may have in cancer. A summary of the most relevant findings in regard to epigenetics and ncRNAs is provided in .
Table 1. Summary of the most relevant findings in regard to epigenetics and non-coding RNAs
Epigenetics and Non-Coding RNAs: Clinical Aspects
Epigenetic regulation of miRNAs
Throughout the last decade, many studies have contributed to a better understanding concerning the role of epigenetics in miRNA expression.Citation15 Furthermore, ever since the detection of DNA methylation evolved as a promising biomarker in cancer, many studies have anticipated that epigenetically regulated miRNAs could have new roles in prognosis, early detection, and predictive values in cancer.Citation16,Citation17
A recent study by Lu et al.Citation18 employing a whole-genome approach demonstrated that miR-129-2 was frequently methylated in hepatocellular carcinoma cells (HCC) but not in normal liver cells and tissues. miRNAs are well known for their ability to posttranscriptionally regulate the expression of target mRNAs. Deregulation of these small molecules contributes to carcinogenesis through affecting major hallmarks of cancer.Citation12,Citation19 Similarly, in samples from patients, methylation levels of miR-129-2 were increased in tumors when compared with normal samples. Moreover, miR-129-2 methylation was detected in plasma samples from patients with tumors but not in healthy individuals or patients with cirrhosis, suggesting miR-129-2 methylation as a specific, potential marker for early diagnosis in HCC. Moreover, miR-129-2 methylation was found in lymphoid malignancies as adversely impacting survival in chronic lymphocytic leukemia (CLL). In myeloid malignancies—acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL)—miR-129-2 was not methylated but, in multiple myeloma (MM), this miRNA was associated with the progression from monoclonal gammopathy of undetermined significance (MGUS) to symptomatic MM.Citation20 For a review of epigenetics and miRNAs in acute leukemia see ref.Citation21
Increased methylation of miR-124-3 CpG islands in renal carcinomas (clear cell histology) was associated with unfavorable clinicopathological aspects, such as tumor grade and diameter, metastasis, and advanced disease. This hypermethylation was tumor-specific and related to poor recurrence-free survival, implying that it could be involved in cancer progression, making miR-124-3 a putative biomarker for patients that can benefit from targeted therapy by means of small molecule effectors.Citation22 Similarly, in ALL, hypermethylation of this microRNA locus was found as an independent prognostic factor for overall and disease-free survival and associated with higher relapse and mortality rates. Epigenetic regulation of miR-124a was shown to mediate increased expression of CDK6, which contributed to abnormal proliferation profiles of ALL cells in vitro and in vivo.Citation23
Another miRNA described as epigenetically regulated is miR-34a, which, when hypermethylated, was associated with metastasis to liver and lymph nodes in colon cancer. Repressed expression of miR-34a due to methylation was probably the trigger to the metastatic phenotype obtained by subsequent upregulation of c-Met, Snail, and b-catenin.Citation24 By treating lymph node metastatic cells with a DNA demethylating agent (5-aza-2'-deoxycytidine), Lujambio et al.Citation25 have associated particular DNA methylation signatures of miRNAs with the metastatic behavior of tumors. This was valid for lymph node metastasis cancer cells derived from colon, melanoma and head and neck. Epigenetic silencing of miR-148a and miR-34b/c was shown to mediate the activation of oncogenic and metastasis target genes as c-MYC, E2F3, CDK6, and TGIF2.
Histone modifications, rather than DNA methylation, can also drive miRNAs suppression. A study by Incoronato et al.Citation26 showed silencing of miR-212 in lung cancer and correlation to later stages of the disease (T3/T4). The authors found that the promoter element of this miRNA is inserted in a CpG island, but its inactivation in lung cancer is linked to alterations in the methylation status of histone tails and not to DNA methylation status.
The wide range of interactions between the miRNome and the epigenome provide new challenges and opportunities for future studies regarding therapeutic strategies in oncology.
Regulation of lncRNAs by epigenetic mechanisms
Long ncRNAs have been shown to exhibit patterns of expression much more specific to each tissue than those exhibited by protein-coding genes,Citation27 offering a variety of benefits in clinical applications,Citation28 both for early diagnosis and disease prognosis. In hepatocellular carcinomas, the lncRNA H19 has been found to be involved in disease progression, behaving as an oncogene.Citation29 Zhang and colleaguesCitation30 showed that epigenetic activation of miR-200 family promotes H19 downregulation, driving metastasis through EMT induction. Also, H19 was found to be underexpressed in intratumoral tissues (T) when compared with peritumoral tissues (R), and low T/R ratio was associated with shorter disease-free survival, more aggressive tumors with metastatic properties and, therefore, worse prognosis. Moreover, in the same study, restoration of H19 levels was capable of inhibiting the invasion capacity of hepatocellular carcinoma cells.
A lncRNA signature was obtained in glioblastoma multiforme (GBM) and a prognostic value, independently of O6-methylguanine methyltransferase (MGMT) promoter methylation status, was found. The lncRNAs KIAA0495, PART1, MGC21881, MIAT, GAS5, and PAR5 expression—named the six-lncRNA signature—were linked with overall survival, alluding that this signature may supply new markers for prognosis treatment and outcome prediction in GBM.Citation31
Epigenetic regulation has effects on lincRNAs as well. ANRIL, also known as CDKN2b antisense or CDKN2BAS, is a lincRNA mapped in the INK4b-ARF-INK4a locus, which was found to be regulated by the Polycomb repressive complex (PRC). This locus has been shown to be one of the most recurrently altered in tumorsCitation32,Citation33 and, in a subset of tumors, epigenetic modifications contribute to its silencing, which may bring both diagnostic and prognostic perspectives in the future. For a deeper review, please check ref.Citation34
Regulation of the Epigenome by Non-Coding Rnas
Epigenome regulation by the miRNome: The turn of epi-miRs
The idea that miRNAs could act as regulators of the epigenome first came with the work published by Fabbri et al.,Citation35 in which members of the miR-29 family (29a, 29b, and 29c) were shown to negatively modulate DNMT3A and DNMT3B enzymes in lung cancer, causing lower survival rates in patients with higher levels of DNMT3A. These miRNAs were then named epi-miRs, referring to a group of miRNAs capable of targeting effectors of the epigenetic system—directly or indirectly—via a regulatory loop, by affecting its regulating enzymes.Citation12 Also, DNMT3B was described as a target for miR-29b in AMLCitation36; however, no clinical significance was assessed for this tumor.
In hepatocellular carcinomas, downregulation of miR-22, known to target HDAC4, was correlated with decreased disease-free survival and worse prognosis, suggesting that it may be involved in the carcinogenesis and progression of this tumor.Citation37
Other miRNAs have also been implicated as regulators of the epigenetic machinery, through the target of crucial enzymes. Some of the examples are miR-449a targeting HDAC in prostatic cancer,Citation38 miR-144, targeting EZH2 in bladder cancer,Citation39 and miR-101, also targeting EZH2 in bladder,Citation40 prostate,Citation41,Citation42 and breast cancer.Citation42 DNMTs have been demonstrated as targets of miR-152 in hepatobiliary cancerCitation43 and in ALL,Citation44 and as target of miR-148 in various cell lines.Citation45 Although the field of epi-miRs is very promising, the literature is still scarce in regard to clinical aspects in cancer detection, prognosis and predictive values of miRNAs.
lncRNAs as regulators of the epigenome
XIST (X-inactive-specific transcript), one of the first discovered lncRNAs in mammals,Citation46 is 17 kb long and possesses a very important and well known function: it is responsible for the epigenetic regulation in the inactivation of X chromosome. Through coating one of the X chromosomes, XIST is capable of inactivating it,Citation47 driven by its binding to PRC2.Citation48 Unmethylated XIST is a frequent finding in testicular germ cell tumors, and the methylation status of this gene is being considered for evaluation in male-derived plasma as a biomarker for early tumor detection and prognosis of these types of tumors.Citation49
HOTAIR (homeobox transcript antisense RNA), a lincRNA, is overexpressed in laryngeal squamous cell carcinomas when compared with adjacent tumors, and associated with poorly differentiated tumors, lymph node metastasis and advanced clinical stages, suggesting this lincRNA as a marker of poor prognosis.Citation50 HOTAIR seems to bring this oncogenic phenotype through promotion of PTEN methylationCitation50 and was also described as capable of altering histone H3 methylation in a PRC2 regulation-dependent fashion. HOTAIR expression was increased in both primary breast and metastasis tumors, posing as a predictor of metastasis and shorter overall survival.Citation51
piRNAs regulation of the epigenome
piRNAs have been shown to be capable of maintaining the integrity of the genome by epigenetically silencing transposons through DNA methylation. piRNAs interact only with members from the Piwi family, but its human ortholog, Hiwi, has been described as aberrantly expressed in many cancer types.Citation47 In human esophageal squamous cell carcinoma, cytoplasmic expression of Hiwi was associated with evidences of poor prognosis.Citation52
Concluding Remarks: Challenges and Opportunities
The accurate comprehension of the molecular machineries involved in cancer development and progression is critical for diagnosis, prognosis, and treatment. Much data have been generated in the last years regarding ncRNAs, their regulation by the epigenetic machinery and, in turn, their role in the regulation of epigenetic processes. The knowledge of how these two mechanisms of regulation are integrated in the cellular context is opening up new avenues for the identification of precise predictive and prognostic factors for new therapeutic strategies in oncology. Still, much has to be done. Cancer is part of an intricate network; clarifying the complex relationship between epigenetics and other mechanisms, such as ncRNAs, and translating these into the clinical practice, remains a big challenge.
The wide range of interactions between the non-coding RNome and the epigenome present new challenges and opportunities for the development of new therapeutic strategies in oncology. Even though ncRNAs are promising candidates for their role in the regulation of the epigenetic machinery, few studies have explored their function in a clinicopathological standpoint. The study of their capacity to regulate one of the most important machineries in cancer remains not only a big challenge, but also a big opportunity in oncology.
| Abbreviations: | ||
| ncRNAs | = | non-coding RNAs |
| DNMTs | = | DNA methyltransferases |
| HDAC | = | histone deacetylases |
| HMTs | = | methyltransferases |
| rRNAs | = | ribosomal RNAs |
| tRNAs | = | transfer RNAs |
| siRNAs | = | short interfering RNAs |
| endo-siRNAs | = | endogenous small interfering RNAs |
| microRNAs | = | miRNAs |
| piRNAs | = | PIWI-interacting RNAs |
| lncRNAs | = | long non-coding RNAs |
| lincRNAs | = | long intergenic non-coding RNAs |
| NATs | = | natural antisense transcripts |
| HCC | = | hepatocellular carcinoma |
| CLL | = | chronic lymphocytic leukemia |
| AML | = | acute myeloid leukemia |
| ALL | = | acute lymphoblastic leukemia |
| MM | = | multiple myeloma |
| MGUS | = | monoclonal gammopathy of undetermined significance |
| c-MYC | = | v- myc avian myelocytomatosis viral oncogene homolog |
| E2F3 | = | E2F transcription factor 3 |
| CDK6 | = | cyclin-dependent kinase 6 |
| TGIF2 | = | TGFB-induced factor homeobox 2 |
| GBM | = | glioblastoma multiforme |
| MGMT | = | O6-methylguanine methyltransferase |
| PRC | = | Polycomb repressive complex |
| EZH2 | = | enhancer of zeste homolog 2 |
| HOTAIR | = | Homeobox transcript antisense RNA |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Calin G is The Alan M. Gewirtz Leukemia and Lymphoma Society Scholar. He is supported also as a Fellow at The University of Texas MD Anderson Research Trust, as a University of Texas System Regents Research Scholar and by the CLL Global Research Foundation. Work in Calin’s laboratory is supported in part by the NIH/NCI (CA135444), a Department of Defense Breast Cancer Idea Award, Developmental Research Awards in Breast Cancer, Ovarian Cancer, Brain Cancer, Prostate Cancer, Multiple Myeloma, Leukemia (P50 CA100632), and Head and Neck (P50 CA097007) SPOREs, a SINF MDACC_DKFZ grant in CLL, a SINF grant in colon cancer, the Laura and John Arnold Foundation, the RGK Foundation, and the Estate of CG Johnson, Jr. Maia B receives scholarship from São Paulo Research Foundation (FAPESP), grant # 2013/04075-5.
References
- Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem 2002; 3:274 - 93; http://dx.doi.org/10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S; PMID: 11933228
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell 2007; 128:635 - 8; http://dx.doi.org/10.1016/j.cell.2007.02.006; PMID: 17320500
- Costa FF. Non-coding RNAs: Meet thy masters. Bioessays 2010; 32:599 - 608; http://dx.doi.org/10.1002/bies.200900112; PMID: 20544733
- Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 2013; 12:433 - 46; http://dx.doi.org/10.1038/nrd4018; PMID: 23722346
- Carninci P, Yasuda J, Hayashizaki Y. Multifaceted mammalian transcriptome. Curr Opin Cell Biol 2008; 20:274 - 80; http://dx.doi.org/10.1016/j.ceb.2008.03.008; PMID: 18468878
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 2005; 308:1149 - 54; http://dx.doi.org/10.1126/science.1108625; PMID: 15790807
- Röther S, Meister G. Small RNAs derived from longer non-coding RNAs. Biochimie 2011; 93:1905 - 15; http://dx.doi.org/10.1016/j.biochi.2011.07.032; PMID: 21843590
- Graifer D, Karpova G. Structural and functional topography of the human ribosome. Acta Biochim Biophys Sin (Shanghai) 2012; 44:281 - 99; http://dx.doi.org/10.1093/abbs/gmr118; PMID: 22257731
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843 - 54; http://dx.doi.org/10.1016/0092-8674(93)90529-Y; PMID: 8252621
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993; 75:855 - 62; http://dx.doi.org/10.1016/0092-8674(93)90530-4; PMID: 8252622
- Kunej T, Godnic I, Horvat S, Zorc M, Calin GA. Cross talk between microRNA and coding cancer genes. Cancer J 2012; 18:223 - 31; http://dx.doi.org/10.1097/PPO.0b013e318258b771; PMID: 22647358
- Berry B, Deddouche S, Kirschner D, Imler JL, Antoniewski C. Viral suppressors of RNA silencing hinder exogenous and endogenous small RNA pathways in Drosophila. PLoS One 2009; 4:e5866; http://dx.doi.org/10.1371/journal.pone.0005866; PMID: 19516905
- Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 2011; 12:136 - 49; http://dx.doi.org/10.1038/nrg2904; PMID: 21245830
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857 - 66; http://dx.doi.org/10.1038/nrc1997; PMID: 17060945
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005; 353:1793 - 801; http://dx.doi.org/10.1056/NEJMoa050995; PMID: 16251535
- Calin GA, Trapasso F, Shimizu M, Dumitru CD, Yendamuri S, Godwin AK, Ferracin M, Bernardi G, Chatterjee D, Baldassarre G, et al. Familial cancer associated with a polymorphism in ARLTS1. N Engl J Med 2005; 352:1667 - 76; http://dx.doi.org/10.1056/NEJMoa042280; PMID: 15843669
- Lu CY, Lin KY, Tien MT, Wu CT, Uen YH, Tseng TL. Frequent DNA methylation of MiR-129-2 and its potential clinical implication in hepatocellular carcinoma. Genes Chromosomes Cancer 2013; 52:636 - 43; PMID: 23580407
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100:57 - 70; http://dx.doi.org/10.1016/S0092-8674(00)81683-9; PMID: 10647931
- Wong KY, Yim RL, Kwong YL, Leung CY, Hui PK, Cheung F, Liang R, Jin DY, Chim CS. Epigenetic inactivation of the MIR129-2 in hematological malignancies. J Hematol Oncol 2013; 6:16; http://dx.doi.org/10.1186/1756-8722-6-16; PMID: 23406679
- Agirre X, Martínez-Climent JA, Odero MD, Prósper F. Epigenetic regulation of miRNA genes in acute leukemia. Leukemia 2012; 26:395 - 403; http://dx.doi.org/10.1038/leu.2011.344; PMID: 22143672
- Gebauer K, Peters I, Dubrowinskaja N, Hennenlotter J, Abbas M, Scherer R, Tezval H, Merseburger AS, Stenzl A, Kuczyk MA, et al. Hsa-mir-124-3 CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br J Cancer 2013; 108:131 - 8; http://dx.doi.org/10.1038/bjc.2012.537; PMID: 23321515
- Agirre X, Vilas-Zornoza A, Jiménez-Velasco A, Martin-Subero JI, Cordeu L, Gárate L, San José-Eneriz E, Abizanda G, Rodríguez-Otero P, Fortes P, et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res 2009; 69:4443 - 53; http://dx.doi.org/10.1158/0008-5472.CAN-08-4025; PMID: 19435910
- Siemens H, Neumann J, Jackstadt R, Mansmann U, Horst D, Kirchner T, Hermeking H. Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and β-catenin predicts distant metastasis of colon cancer. Clin Cancer Res 2013; 19:710 - 20; http://dx.doi.org/10.1158/1078-0432.CCR-12-1703; PMID: 23243217
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A 2008; 105:13556 - 61; http://dx.doi.org/10.1073/pnas.0803055105; PMID: 18768788
- Incoronato M, Urso L, Portela A, Laukkanen MO, Soini Y, Quintavalle C, Keller S, Esteller M, Condorelli G. Epigenetic regulation of miR-212 expression in lung cancer. PLoS One 2011; 6:e27722; http://dx.doi.org/10.1371/journal.pone.0027722; PMID: 22110741
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature 2012; 489:101 - 8; http://dx.doi.org/10.1038/nature11233; PMID: 22955620
- Beckedorff FC, Amaral MS, Deocesano-Pereira C, Verjovski-Almeida S. Long non-coding RNAs and their implications in cancer epigenetics. Biosci Rep 2013; 33:e00061; http://dx.doi.org/10.1042/BSR20130054; PMID: 23875687
- Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One 2007; 2:e845; http://dx.doi.org/10.1371/journal.pone.0000845; PMID: 17786216
- Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS, Xu D, Bi HS, Wang F, Sun SH. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 2013; 34:577 - 86; http://dx.doi.org/10.1093/carcin/bgs381; PMID: 23222811
- Zhang XQSS, Sun S, Lam KF, Kiang KM, Pu JK, Ho AS, Lui WM, Fung CF, Wong TS, Leung GK. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol Dis 2013; 58:123 - 31; http://dx.doi.org/10.1016/j.nbd.2013.05.011; PMID: 23726844
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010; 463:899 - 905; http://dx.doi.org/10.1038/nature08822; PMID: 20164920
- Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, et al. Signatures of mutation and selection in the cancer genome. Nature 2010; 463:893 - 8; http://dx.doi.org/10.1038/nature08768; PMID: 20164919
- Popov N, Gil J. Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics 2010; 5:685 - 90; http://dx.doi.org/10.4161/epi.5.8.12996; PMID: 20716961
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A 2007; 104:15805 - 10; http://dx.doi.org/10.1073/pnas.0707628104; PMID: 17890317
- Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood 2009; 114:5331 - 41; http://dx.doi.org/10.1182/blood-2009-03-211938; PMID: 19850741
- Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu S, Wu M, Pan Z, Zhou W. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer 2010; 103:1215 - 20; http://dx.doi.org/10.1038/sj.bjc.6605895; PMID: 20842113
- Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 2009; 28:1714 - 24; http://dx.doi.org/10.1038/onc.2009.19; PMID: 19252524
- Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang Z, Qiu F, Lin J. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J 2013; 280:4531 - 8; http://dx.doi.org/10.1111/febs.12417; PMID: 23815091
- Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res 2009; 69:2623 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-08-3114; PMID: 19258506
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008; 322:1695 - 9; http://dx.doi.org/10.1126/science.1165395; PMID: 19008416
- Ren G, Baritaki S, Marathe H, Feng J, Park S, Beach S, Bazeley PS, Beshir AB, Fenteany G, Mehra R, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res 2012; 72:3091 - 104; http://dx.doi.org/10.1158/0008-5472.CAN-11-3546; PMID: 22505648
- Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 2010; 51:881 - 90; PMID: 20146264
- Stumpel DJ, Schotte D, Lange-Turenhout EA, Schneider P, Seslija L, de Menezes RX, Marquez VE, Pieters R, den Boer ML, Stam RW. Hypermethylation of specific microRNA genes in MLL-rearranged infant acute lymphoblastic leukemia: major matters at a micro scale. Leukemia 2011; 25:429 - 39; http://dx.doi.org/10.1038/leu.2010.282; PMID: 21116279
- Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA 2008; 14:872 - 7; http://dx.doi.org/10.1261/rna.972008; PMID: 18367714
- Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992; 71:527 - 42; http://dx.doi.org/10.1016/0092-8674(92)90520-M; PMID: 1423611
- Siddiqi S, Matushansky I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem 2012; 113:373 - 80; http://dx.doi.org/10.1002/jcb.23363; PMID: 21928326
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008; 322:750 - 6; http://dx.doi.org/10.1126/science.1163045; PMID: 18974356
- Lind GE, Skotheim RI, Lothe RA. The epigenome of testicular germ cell tumors. APMIS 2007; 115:1147 - 60; http://dx.doi.org/10.1111/j.1600-0463.2007.apm_660.xml.x; PMID: 18042148
- Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J, Liu M. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol 2013; 182:64 - 70; http://dx.doi.org/10.1016/j.ajpath.2012.08.042; PMID: 23141928
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464:1071 - 6; http://dx.doi.org/10.1038/nature08975; PMID: 20393566
- He W, Wang Z, Wang Q, Fan Q, Shou C, Wang J, Giercksky KE, Nesland JM, Suo Z. Expression of HIWI in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. BMC Cancer 2009; 9:426; http://dx.doi.org/10.1186/1471-2407-9-426; PMID: 19995427
