Abstract
The initiation of angiogenesis can mark the transition from tumor dormancy to active growth and recurrence. Mechanisms that regulate recurrence in human cancers are poorly understood, in part because of the absence of relevant models. The induction of ARHI (DIRAS3) induces dormancy and autophagy in human ovarian cancer xenografts but produces autophagic cell death in culture. The addition of VEGF to cultures maintains the viability of dormant autophagic cancer cells, thereby permitting active growth when ARHI is downregulated, which mimics the “recurrence” of growth in xenografts. Two inducible ovarian cancer cell lines, SKOv3-ARHI and Hey-ARHI, were used. The expression level of angiogenesis factors was evaluated by real-time PCR, immunohistochemistry, immunocytochemistry and western blot; their epigenetic regulation was measured by bisulfite sequencing and chromatin immunoprecipitation. Six of the 15 angiogenesis factors were upregulated in dormant cancer cells (tissue inhibitor of metalloproteinases-3, TIMP3; thrombospondin-1, TSP1; angiopoietin-1; angiopoietin-2; angiopoietin-4; E-cadherin, CDH1). We found that TIMP3 and CDH1 expression was regulated epigenetically and was related inversely to the DNA methylation of their promoters in cell cultures and in xenografts. Increased H3K9 acetylation was associated with higher TIMP3 expression in dormant SKOv3-ARHI cells, while decreased H3K27me3 resulted in the upregulation of TIMP3 in dormant Hey-ARHI cells. Elevated CDH1 expression during dormancy was associated with an increase in both H3K4me3 and H3K9Ac in two cell lines. CpG demethylating agents and/or histone deacetylase inhibitors inhibited the re-growth of dormant cancer cells, which was associated with the re-expression of anti-angiogenic genes. The expression of the anti-angiogenic genes TIMP3 and CDH1 is elevated during dormancy and is reduced during the transition to active growth by changes in DNA methylation and histone modification.
Introduction
Ovarian cancer is the third most common gynecologic cancer, but it is clearly the most lethal because it has a 5-y mortality greater than 60%.Citation1 While more than 70% of ovarian cancers will respond to primary cytoreductive surgery and combination chemotherapy, the majority of cancers recur, accounting for the high mortality rate. Recurrence can occur after many months or years, which suggests that ovarian cancer, similar to other epithelial neoplasms, can remain dormant. At “second-look” operations after primary therapy, small, poorly vascularized deposits of ovarian cancer cells are found within collagen-rich nodules on the surface of the parietal or visceral peritoneum. Several mechanisms have been proposed for the dormancy of individual cancer cells, including G0-G1 arrest, immunosurveillance and failure to initiate angiogenesis.Citation2 The outgrowth of dormant cancer cells has been associated with the initiation of angiogenesis, although other mechanisms could be important.Citation3 Prolonged dormancy has been associated with impaired angiogenesis in several models. The transition from a microenvironment with predominantly anti-angiogenic factors and dormant cancer cells to an environment with pro-angiogenic factors and rapid tumor growth has been termed the “angiogenic switch.”Citation4 In a model for ovarian cancer dormancy, the outgrowth of human ovarian cancer cell spheroids after subcutaneous injection in nu/nu mice coincided with the ingrowth and maturation of blood vessels and the induction of vascular endothelial growth factor (VEGF) by hypoxia in cancer cells.Citation5
Recent studies have produced an inducible model for studying the dormancy of human ovarian epithelial cancer, which depends on the inducible expression of an aplasia ras homolog member (ARHI), which is also called DIRAS3. ARHI is a maternally imprinted tumor suppressor gene that is widely expressed by normal tissues and is downregulated in cancers of breast, lung, prostate and ovary.Citation6-Citation10 While ARHI is strongly expressed in normal ovarian epithelial cells,Citation11 it is downregulated in 60% of ovarian cancers and is associated with decreased progression-free survival.Citation9 ARHI is downregulated by several mechanisms, including loss of heterozygosity, promoter DNA methylation, transcriptional regulation, and shortened RNA half-life.Citation12-Citation18 ARHI encodes a 26 kD GTPase with a 50–60% homology to Ras and Rap. The function of ARHI depends critically on a 33 amino acid N-terminal extension.Citation19 Re-expression of ARHI at physiologic levels inhibits the proliferation,Citation16 decreases the motility,Citation20 and blocks the growth of xenografts, and it initiates autophagy and induces tumor dormancy.Citation21
The development of stable sublines of SKOv3 and Hey ovarian cancer cells with tet-on inducible expression of ARHI (SKOv3-ARHI and Hey-ARHI) has permitted studies of the effect of ARHI re-expression in cell cultures and in xenografts. Feeding doxycycline (DOX) to mice bearing SKOv3-ARHI xenografts blocks tumor growth and induces autophagy. When DOX is withdrawn after 6 weeks of suppression, xenografts grow promptly to kill the mice. If chloroquine is administered while xenografts are dormant, the outgrowth is significantly delayed, which is consistent with the importance of autophagy in sustaining dormant cancer cells.Citation21 In cell cultures, treatment with DOX induces autophagic cell death. Autophagic ovarian cancer cells can, however, be rescued by treatment with growth factors found in xenografts, including VEGF, interleukin-8(IL-8) and insulin growth factor (IGF-1). In this report, we have found that cancer cells that are treated with both DOX and VEGF remain dormant in cell cultures and can regrow when DOX is removed. This finding has provided the opportunity to examine the expression of pro-angiogenic and anti-angiogenic genes while cells are dormant in cultures or growing actively after the removal of DOX. Tissue inhibitor of metalloproteinases-3 (TIMP3), thrombospondin-1 (TSP1), angiopoietin-1 (Ang1), angiopoietin-2 (Ang2), angiopoietin-4 (Ang4), and E-cadherin (CDH1) are upregulated in dormancy and downregulated in recurrence, both in cell cultures and in xenografts. The expression of TIMP3 and CDH1 is regulated epigenetically by DNA methylation and histone modification. Thus, TIMP3 and CDH1 could play a key role in the angiogenic switch that occurs when dormancy gives way to tumor outgrowth. In this setting, epigenetic therapy might significantly delay ovarian cancer recurrence.
Results
In the presence of VEGF, the re-expression of ARHI induces dormancy and autophagy in cultures of ovarian cancer cells
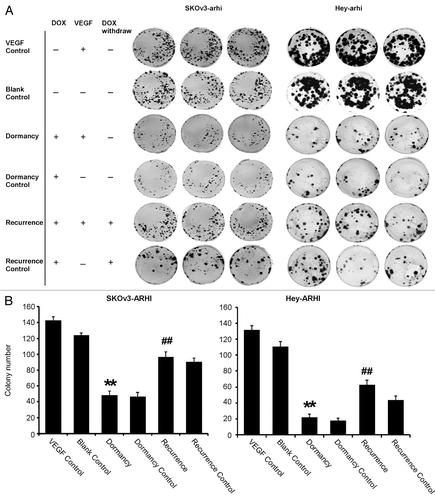
In previous studies, the re-expression of ARHI with the addition of DOX induced autophagic cell death within 2–3 d in the absence of additional growth factors.Citation21 To determine whether incubation with VEGF would induce dormancy rather than cell death in culture, SKOv3-ARHI and Hey-ARHI cells were treated with VEGF without the induction of ARHI, with a transient induction of ARHI for 6 d and with a persistent induction of ARHI for the duration of the 14-d experiment. The results of clonogenic assays of SKOv3-ARHI cells are presented in (left). Treatment with VEGF alone in the absence of DOX increased the number of colonies when compared with blank controls at 14 d (colony number: 143 ± 5 vs. 124 ± 3, P < 0.05). The induction of ARHI by treatment with DOX inhibited clonogenic growth (colony number: 47 ± 4 vs. 124 ± 3, P < 0.05). The induction of ARHI for 14 d in the presence of VEGF in the “dormancy” group significantly reduced the number of colonies observed after treatment with VEGF alone (colony number: 49 ± 5 vs. 143 ± 5, P < 0.05). When the re-expression of ARHI was terminated after 6 d in the “recurrence” group, the number of colonies increased significantly (91 ± 4 vs. 49 ± 5, P < 0.05), which indicates that dormant cells were still capable of proliferating. The number of colonies observed was, however, lower than that observed in the VEGF control group (91 ± 4 vs. 143 ± 5, P < 0.05). However, without the presence of VEGF, the cells in the recurrence control group reduced the ability of proliferating after DOX withdrawal (56 ± 7 vs. 91 ± 4, P < 0.05). To confirm these results, we repeated the clonogenic assays in Hey-ARHI cells. Similar results were obtained, as shown in (right), which indicates that inducing ARHI expression with VEGF can induce dormancy without losing the ability of proliferating in ovarian cancer cells.
Figure 1. Re-expression of ARHI induces dormancy, and the downregulation of ARHI permits recurrent growth in cultures of ovarian cancer cells that are stimulated with VEGF. (A) Clonogenic growth of SKOv3-ARHI and Hey-ARHI cells after different treatments. The cells were divided into six groups: the blank control group (medium only), dormancy control group (cells treated with 1 μg/ml DOX for 14 d), VEGF control group (cells treated with 20 ng/ml VEGF for 14 d), dormancy group (cells treated with both 20 ng/ml VEGF and 1 μg/ml DOX for 14 d), recurrence group (cells treated with VEGF for 14 d and DOX for only 6 d followed by DOX removal), and recurrence control group (cells treated with 1 μg/ml DOX for 6 d followed by DOX removal). (B) Colony numbers of the SKOv3-ARHI (left) and Hey-ARHI (right) cell lines in the six groups were counted. The experiments were performed in triplicate, and the data are presented as the mean ± SD of the three separate experiments. **Compared with the VEGF control group, P < 0.01. ##Compared with the dormancy group, P < 0.01
Studies with real-time PCR indicated that ARHI had been upregulated in the dormancy group with continuous DOX and VEGF treatment and downregulated after the withdrawal of DOX in the recurrence group (), in both SKOv3-ARHI and Hey-ARHI cells. The ARHI protein expression changes in SKOv3-ARHI cells were similar to the mRNA changes, as shown in .
Figure 2. ARHI induces autophagy in ovarian cancer cells. (A) Expression of ARHI in the ovarian cancer dormancy-recurrence in-vitro model as measured by real-time PCR. Top: SKOv3-ARHI; bottom: Hey-ARHI. (B) Increased cleavage of LC3-I to LC3-II and higher ARHI expression were shown in the dormancy group, whereas the withdrawal of DOX resulted in decreased LC3-II and ARHI expression in SKOv3-ARHI (C) The percentage of autophagic cells was measured by acridine orange staining and flow cytometry. FL3H detects red fluorescence (650 nm), and FL1H detects blue fluorescence (488 nm). Dot plots that show increased autophage formation in the dormancy (VEGF+DOX treatment) group and decreased autophage formation after the transition of dormancy to recurrence (VEGF + DOX withdrawn). *Compared with the VEGF control group, P < 0.05, **Compared with the VEGF control group, P < 0.01; #Compared with the dormancy group, P < 0.01, ##Compared with the dormancy group, P < 0.01.
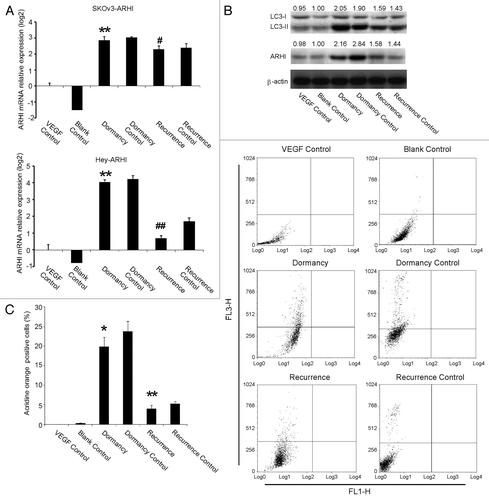
Because ARHI induces autophagy and maintains tumor dormancy in human ovarian cancer xenografts, we observed whether ARHI would induce autophagy in the presence of VEGF in cell culture. Flow cytometry was used to measure acidic vesicular organelles () in SKOv3-ARHI cells. Compared with the VEGF control group, the number of autophagic cells increased significantly in the dormancy group when the cells were treated with VEGF and DOX for 14 d (16% vs. 0.21%, P < 0.05). The number of autophagic cells decreased significantly in the recurrence group compared with the dormancy group (2.58% vs. 16%, P < 0.05). Furthermore, using western blot, we confirmed an increased cleavage of LC3-I to LC3-II, a well-characterized marker for autophagy ().
Anti-angiogenic genes are activated in dormant ovarian cancer cells
Using real-time PCR, we investigated the expression changes of 15 angiogenesis-related genes: TSP1, angiopoietin-like 2 (ANGPTL2), Ang1, Ang2, Ang4, insulin-like growth factor binding protein-3 (IGFBP3), tyrosine kinase with immunoglobulin-like and EGF-like domains 1 (TIE-1), tyrosine kinase with immunoglobulin-like and EGF-like domains 2 (TIE-2), TIMP3, H-cadherin (CDH13), CDH1, secreted acidic cysteine rich glycoprotein (SPARC), reversion-inducing-cysteine-rich protein with kazal motifs (RECK), sulfatase 1 (SULF-1) and EGF containing fibulin-like extracellular matrix protein 1 (EFEMP1).
First, we tested gene expression in six groups of SKOv3-ARHI cells that were treated for 14 d. We compared their expression in two settings. The first setting contained three groups (VEGF, dormancy and recurrence), which were all treated with VEGF and/or DOX. Among these pro-angiogenic and anti-angiogenic factors, mRNAs of 6 genes (TIMP3, TSP1, Ang1, Ang2, Ang4, and CDH1) were upregulated in dormant tumor cells compared with the VEGF control group cells and downregulated in progressively growing ovarian cancer cells compared with dormant cells, which indicates that those factors are involved in the transition from dormancy to growth (). The second setting contained three control groups that were all treated without VEGF. The results showed that these six genes were upregulated when SKOv3-ARHI cells were treated with DOX and downregulated after DOX was withdrawn (Fig. S1A).
Figure 3. Anti-angiogenic genes are activated in dormant and repressed in recurrent ovarian cancer cells. (A) The expression of angiogenesis-related factors in dormancy and the recurrence transition measured by real-time PCR in SKOv3-ARHI cells. Of the 15 genes, the six genes (Ang1, Ang2, Ang4, TSP1, CDH1, and TIMP3) that were upregulated during dormancy and decreased again in recurrence were presented. (B) The expression of the six genes in the Hey-ARHI cells (C) Dynamics of TIMP3 and CDH1 expression in SKOv-3 ARHI cells. This expression was measured by real-time RT-PCR at four time points (0, 6, 10, and 14 d). (D) Dynamics of TIMP3 and CDH1 expression in Hey-3 ARHI cells. **Compared with the control group, P < 0.01. ##Compared with the dormancy group, P < 0.01
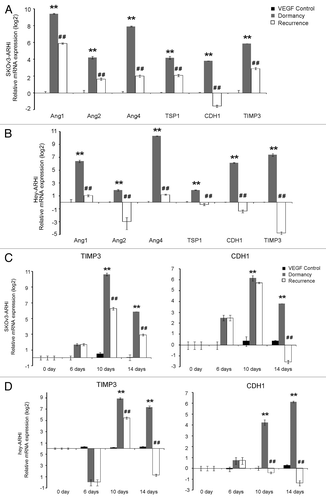
Because our main intention was to observe the angiogenesis factors during the dormancy-to-recurrence transition in this VEGF-added, in-vitro model that mimics the in-vivo environment, we further investigated the dynamics of the expression of the six genes in the VEGF control, dormancy, and recurrence groups. shows that TIMP3 and CDH1 expression elevated on day 6, reached a peak on day 10 and slightly decreased on day 14 in the dormancy group, when treated with DOX+VEGF. Compared with the expression level in the dormancy group, four days after DOX withdrawal (day 10), the expression of TIMP3 and CDH1 was obviously decreased and downregulated greatly on day 14.
We evaluated the expression of 6 genes in Hey-ARHI cells. shows that all 6 genes were elevated in the dormancy group and decreased in the recurrence group. The dynamics of TIMP3 and CDH1 expression were similar to that of SKOv3-ARHI cells (). The dynamics of expression of the other four genes are shown in Fig. S1B.
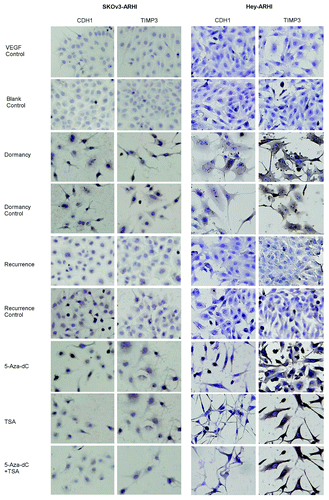
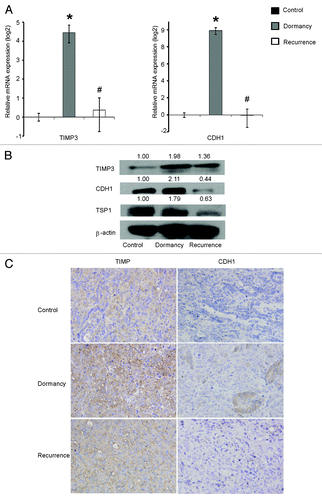
Immunocytochemistry confirmed that the expression of CDH1 and TIMP3 protein increased in dormancy compared with the VEGF control and decreased in the recurrent growth compared with the dormant state (). The quantitative scores are shown in Fig. S2.
Figure 4. TIMP3 and CDH1 protein expression increases in dormancy, decreases in recurrence and are upregulated by 5-Aza-dC and TSA. The cells were divided into nine groups: the Blank control, VEGF control, Dormancy, Dormancy control, Recurrence and Recurrence control, which were mentioned in ; the 5-Aza-dC group: recurrence group treated with 5-Aza-dC for the last 72 h; the TSA group: recurrence group treated for TSA for the last 24 h; the 5-Aza-dC+TSA group: recurrence group treated with 5-Aza-dC for the last 72 h and with TSA for the last 24 h. The cells were stained with CDH1 and TIMP3 antibodies with the indicated concentrations. Brown staining means positive.
DNA methylation of TIMP3 and CDH1 decreased during dormancy and increased during recurrent growth
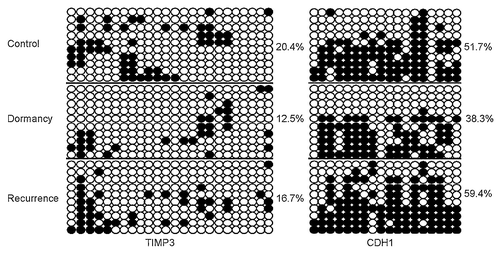
To study the DNA methylation status of 15 angiogenesis-related factors, bisulfite-treated DNA was sequenced. Overall, in the basal state of the ovarian carcinoma cell line SKOv3-ARHI, the promoters of Ang2, IGFBP3, RECK, EFEMP1, TIE1, TIE2, and SPARC were hypermethylated (>80%); the promoters of Ang1, ANGPTL2, TSP1 were hypomethylated (<10%); and the promoters of Ang4, TIMP3, CDH13, CDH1 and SULF1 were moderately methylated (10%~40%).
Because suppression of some genes is commonly associated with CpG island hypermethylation within a promoter, we asked whether the gene expression changes during dormancy-to-recurrence transitions were due to de novo DNA methylation. We first examined DNA methylation of these 15 angiogenesis-related genes in the VEGF control, dormancy and recurrence groups of SKOV3-ARHI cells that were treated for 14 d. TIMP3 promoter showed decreased methylation in dormancy and increased methylation in recurrence (VEGF control: 22.5%, dormancy: 9.2%, recurrence: 30%), which was in accordance to their elevated expression in dormancy and decreased expression in recurrence. CDH1 showed dramatically hypermethylation in recurrence (VEGF control: 44.4%, dormancy: 60.6%, recurrence: 87.8%). In addition, sequencing results showed that both TIMP3 and CDH1 were hypomethylated in dormancy and regained methylation in recurrence in Hey-ARHI cells. Furthermore, these two genes represented varying changes of specific CpG sites (). However, TSP1 did not show a dramatic change in DNA methylation (TSP1, control: 1%, dormancy: 0, recurrence: 6.7%), and the methylation changes of the other 12 genes did not show a significant difference (data not shown).
Figure 5. DNA methylation of TIMP3 and CDH1 decreases during dormancy and increases during recurrent growth. (A) Methylation status of TIMP3 and CDH1 was measured by bisulfate sequencing in the in-vitro VEGF control, dormancy, recurrence and 5-Aza-dC groups. Ten clones were sequenced and are shown here. Unmethylated CpG sites are shown as ○, whereas methylated CpG sites are indicated as ●. The proportion of methylated CpG sites among the total number of CpG sites in the 10 clones analyzed is given on the right. Note the low degree of methylation in the dormancy and 5-Aza-dC treatment groups. (B) Dynamics of TIMP3 and CDH1 methylation in SKOv-3 ARHI and Hey-ARHI cells measured by bisulfite sequencing (BSP) at four time points (0, 6, 10, and 14 d).
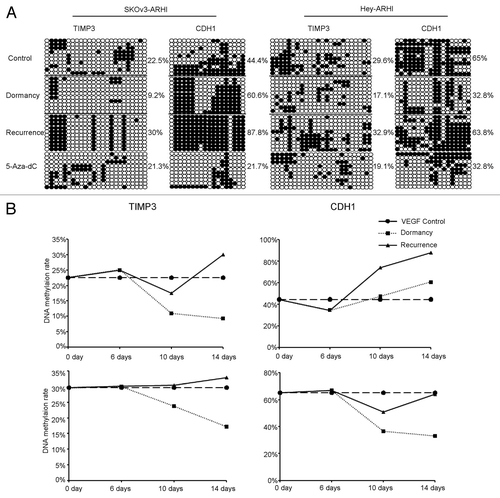
Because the tumor dormancy-to-recurrence transition is a dynamic process that can be induced by DOX withdrawal in our in vitro model, we further investigated the dynamics of epigenetic changes of TIMP3 and CDH1 at four sequential time points (0, 6, 10, and 14 d) in both SKOv3-ARHI and Hey-ARHI cells. We noticed that the timing of TIMP3 and CDH1 methylation fell slightly behind the upregulation of TIMP3 and CDH1 (). For example, TIMP3 and CDH1 expression were obviously increased on day 6 in the dormancy group of Hey-ARHI cells, whereas DNA methylation was significantly decreased on day 10 and was further decreased on day 14. In accordance with that finding, in the recurrence group, on day 10 (4 d after DOX withdrawal), TIMP3 and CDH1 hypomethylation was reversed. On day 14 (8 d after DOX withdrawal), the methylation level of TIMP3 and CDH1 reached or even exceeded the level observed in the control group, which was also consistent with the expression changes.
H3K9Ac, H3K4me3, and H3K27me3 regulate the expression of TIMP3 and CDH1
Because chromatin modifications play a critical role in controlling gene expression and because H3K9me2 and H3K27me3 are generally linked to gene repression in euchromatin while H3K4me3 and H3K9Ac are linked to gene activation, we examined histone modifications of the CDH1 and TIMP3 promoter using ChIP assays. We designed 7 pairs of primers for TIMP3 and 6 pairs of primers for CDH1, which covered from the promoter region near the transcription start site to the CpG island in exon 1 (Fig. S3A) . After testing, we found that histone markers were modulated in the DNA fragment between T7 (TIMP3) and C2 (CDH1).
The TIMP3 CpG island that was associated with H3K9Ac and H3K9Ac was significantly increased in dormancy compared with the recurrent growth in SKOv3-ARHI (Fig.S3B; P < 0.05), which is consistent with the increased expression of TIMP3 in dormancy and its decreased expression in recurrence. However, in Hey-ARHI cells, decreased H3K27me3 (a repressive marker) at TIMP3 is associated with increased TIMP3 expression (Fig.S3C). In contrast, the level of H3K9me2 and H3K4me3 associated with the TIMP3 CpG island did not change during dormancy or recurrence.
CDH1 regulation was associated with different histone markers. Two active markers, H3K4me3 and H3K9Ac, were significantly enriched at the CDH1 promoter during dormancy and were reduced during recurrence in both cell lines (Figs. S3B and C). These results suggest that the accumulation of H3K4me3 and H3K9Ac at the CDH1 promoter plays a critical role in upregulating the expression of CDH1 in dormant cells.
Inhibition of DNMT and/or HDAC prevents recurrent growth of ovarian cancer cells after downregulation of ARHI and upregulates angiogenesis-related factors
SKOv3-ARHI cells were treated with VEGF for 14 d and DOX for 6 d to observe the recurrent growth. Further treatment with the DNMT inhibitor 5-Aza-deoxycytidine (5-Aza-dC), the HDAC inhibitor Trichostatin (TSA) or a combination of both agents significantly inhibited colony formation when compared with the recurrent growth (recurrence group: 151 ± 4, 5-Aza-dC group: 32 ± 6, TSA group: 50 ± 3, TSA and 5-Aza-dC: 36 ± 5, P < 0.05) (). The DNMT inhibitor and the HDAC inhibitor altered the expression of angiogenesis-related factors after the downregulation of ARHI. The expression of Ang1, Ang2, ANGPTL2, IGFBP3, EFEMP1, TIMP3, TIE1, TIE2, SPARC, and CDH1 was reactivated by 5-Aza-dC treatment, whereas the expression of Ang2, IGFBP3, EFEMP1, TIMP3, TIE1, and CDH1 was elevated after TSA treatment in the in-vitro recurrence model ().
Figure 6. The 5-Aza-dC and /or TSA treatment regulates ovarian cancer cell colony formation and the expression of angiogenesis-related genes. (AandC) Colony number of four groups (recurrence, 5-Aza-dC, TSA, and 5-Aza-dC + TSA) in SKOv3-ARHI (A) and Hey-ARHI (C) cells. *Compared with the recurrence group, P < 0.05. (BandD) Results of 10 genes that showed significant changes after 5-Aza-dC and/or TSA treatment in the recurrence groups measured by real-time RT-PCR were presented for SKOv3-ARHI cells (B) and in Hey-ARHI cells (D). *Compared with the recurrence group, P < 0.05. Independent experiments were repeated at least three times to confirm the reproducibility of the results.
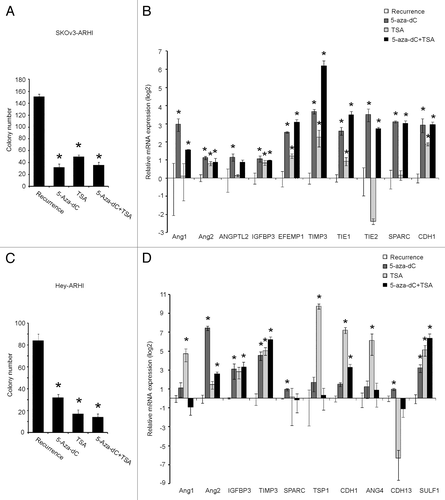
Similar colony formation inhibition effects of 5-Aza-dC and TSA on Hey-ARHI recurrent cells were observed (). However, the changed pattern on these genes was slightly different. The expression of Ang2, IGFBP3, TIMP3, SPARC, CDH1, CDH13, and SULF1 was reactivated by 5-Aza-dC treatment, whereas the expression of Ang1, TIMP3, TSP1, CDH1, Ang4, and SULF1 was elevated after TSA treatment in the in-vitro Hey-ARHI recurrence model ().
DNMT inhibitor reduces DNA promoter methylation of TIMP3 and CDH1 in ovarian cancer cells after the downregulation of ARHI
To determine whether treatment with 5-Aza-dC could reverse DNA hypermethylation of angiogenesis-related factors during recurrent growth after the withdrawal of DOX and the downregulation of ARHI, bisulfite-treated DNA was sequenced. As expected, the methylation rates of TIMP3 and CDH1 of the recurrence group were all reduced by 5-Aza-dC treatment (SKOv3-ARHI cells: from 30% to 21.3% in TIMP3 and from 87.8% to 21.7% in CDH1; Hey-ARHI cells: from 32.9% to 19.1% in TIMP3 and from 63.8% to 32.8% in CDH1) ().
Induction of ARHI induces tumor dormancy in human ovarian cancer xenografts associated with decreased staining for PCNA and CD31
When SKOv3-ARHI cells were grown as xenografts in nu/nu mice, the induction of ARHI by feeding DOX to the tumor-bearing animals significantly inhibited xenograft growth. Control SKOv3-ARHI tumors without ARHI induction grew to more than 1.4 cm in diameter by 75 d. When ARHI induction was withdrawn after 32 d, xenografts grew rapidly (); this finding suggests that cancer cells remained viable during the 32 d of ARHI induction, which is consistent with a dormant state.
Figure 7. Induction of ARHI induces tumor dormancy in human ovarian cancer xenografts. (A) The control and dormancy groups were provided without DOX or with DOX in their drinking water. In the recurrence group, DOX was withdrawn after 32 d of treatment. Arrowheads indicate the day of DOX withdrawal. The results shown were from three independent experiments. *Compared with the control group, P < 0.01. ##Compared with the recurrence group, P < 0.01. (B) LC3-I, LC3-II and ARHI expression in the three groups were detected by Western Blot. *Compared with the control group, P < 0.05. #Compared with the recurrence group, P < 0.05 (C) PCNA (a cell proliferation marker) and CD31 (an angiogenesis marker) expression detected by immunohistochemistry.
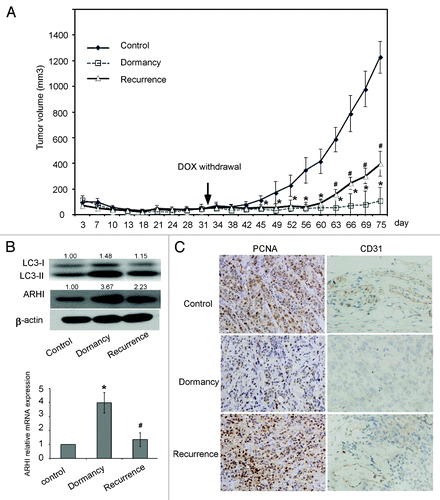
Real-time PCR and western blot results all showed that ARHI was induced in dormant xenografts and was decreased in the group that had progressive growth after the withdrawal of DOX (). Moreover, western blot analysis demonstrated that the expression of ARHI correlated with an increased cleavage of LC3-I to LC3-II, a well-characterized marker for autophagy (). Our data confirm that dormant xenografts undergo ARHI-mediated autophagy, although our matrigel-based model is slightly different from the previous study by Lu et al.Citation21
To assess the changes in cell proliferation and angiogenesis in the in vivo model, we performed immunohistochemical staining of PCNA and CD31. PCNA stained nuclei of cancer cells in the xenografts. PCNA staining was reduced in dormant xenografts that expressed ARHI relative to control xenografts that did not, whereas PCNA staining increased once again in xenografts that had progressive growth after the withdrawal of DOX and that reduced the expression of ARHI. In addition, the expression of CD31, an angiogenesis marker, followed the same trend as PCNA (). Thus, cell proliferation and angiogenesis were decreased in dormant xenografts.
TIMP3 and CDH1 are upregulated in dormant SKOv3-ARHI xenografts
We examined the expression of the two angiogenesis-related genes (TIMP3 and CDH1) that had been measured in the cell culture model. Real-time PCR showed that the two genes were upregulated more than a hundred fold in dormant xenografts and were downregulated during recurrent growth (). The increased expression of TIMP3 and CDH1 protein was confirmed by immunohistochemistry and western blot analysis (). To clarify whether these changes were caused by DNA methylation and histone modification, we also measured the promoter methylation of the three genes and the general level of histone modification sites H3K4me3, H3K9ac and H3K9me2 in xenograft tissues. The results showed that the DNA methylation level of TIMP3 and CDH1 decreased in dormancy and increased during recurrent growth (). No significant changes in the general histone modification were found during the transition from dormancy to recurrent growth in vivo (Fig. S5).
Figure 8. TIMP3 and CDH1 are upregulated in dormancy and downregulated in recurrent SKOv3-ARHI xenografts. (A) The expression of CDH1 and TIMP3 in xenografts of three groups was measured by real-time PCR. *Compared with the control group, P < 0.05. #Compared with the dormancy group, P < 0.05. (BandC): TIMP3 and CDH3 expression in three groups were detected by western blot (B) and by immunohistochemistry (C).
Discussion
This report documents that multiple anti-angiogenic factors are upregulated in dormant human ovarian cancer cells in culture and in xenografts. Our findings support studies at other tumor sites that indicate that angiogenesis-related genes are the most frequently affected genes within the consensus dormancy signature.Citation23 Among the 15 angiogenesis factors, six (TIMP3, TSP1, Ang1, Ang2, Ang4, and CDH1) were upregulated in dormancy and downregulated in recurrence. During the transition from dormancy to progressive growth, TIMP3 and CDH1 are downregulated by hypermethylation and histone modification. DNMT inhibitors and HDAC inhibitors can reverse these changes, upregulating those two factors.
Both genetic and epigenetic changes contribute to the process of metastasis. Epigenetic alterations generally consist of DNA methylation and histone modification. Abnormal methylation patterns with malignancies are generally governed by widespread DNA hypomethylation of tumor-promoting genes along with site-specific DNA hypermethylation of tumor-suppressor genes.Citation24 Of the angiogenesis factors that we studied, TIMP3 and CDH1 appeared to be regulated by epigenetic mechanisms.
TIMP3, TSP1, and CDH1 all inhibit angiogenesis. TIMP3 belongs to a family of MMPsCitation25 and regulates VEGF-mediated angiogenesis by binding directly to VEGF-2 and inhibiting the downstream signaling that is required for the stimulation of endothelial and cancer cells.Citation26 CDH1 is a member of the transmembrane glycoprotein family, which is expressed in epithelial tissue and is responsible for calcium-dependent, cell-to-cell adhesionCitation27 and angiogenesis.Citation28 Studies of various human cancers have shown a close relationship between CDH1 dysfunction and invasion and metastasis.Citation29 TSP1, a matrix-bound adhesive glycoprotein, exerts its anti-angiogenic activity via binding to the CD36 receptor by triggering an apoptotic signaling pathway.Citation30 CDH1 has been found to be upregulated in a dormancy model for breast cancer.Citation31 Several studies have shown that the 3 genes can be downregulated in different cancers by DNA methylation. In our model, TIMP3 and CDH1 were upregulated in dormancy and downregulated in recurrence. The methylation of TIMP3 and CDH1 was significantly lower in dormant cancer cells than in cells that were undergoing recurrent growth. DNA hypermethylation inhibited the expression of the two genes, decreasing their anti-angiogenic activity and contributing to tumor angiogenesis. However, the change in TSP1 expression during the dormancy-to-recurrence transition did not appear to be mainly regulated by DNA methylation.
Unlike genetic changes in cancer, epigenetic changes are potentially reversible, which raises the possibility that epigenetic therapy might prolong progression-free survival and eliminate dormant cancer cells. Many phase I clinical trials have indicated that HDAC and DNMT inhibitors are well tolerated. The US FDA has approved the use of two DNA-demethylating agents, 5-azacytidine and 5-aza-2'-deoxycytidine, for the treatment of myelodysplastic syndrome.Citation32 The HDAC inhibitor SAHA has been approved by the FDA for the treatment of cutaneous T-cell lymphoma.Citation33 In addition, a combination of decitabine and SAHA inhibited ovarian cancer xenograft growth.Citation34 The anti-angiogenic activity of DNMT inhibitors and HDAC inhibitors has been documented in cell culture and xenografts.Citation35,Citation36
In this report, 5-Aza-dC and TSA significantly inhibited colony formation that occurred after the withdrawal of doxycycline and the downregulation of ARHI. The expression of angiogenesis-related genes reacted to both types of agents. 5-Aza-dC elevated the expression levels of Ang1, Ang2, ANGPTL2, IGFBP3, EFEMP1, TIMP3, TIE1, TIE2, SPARC, and CDH1 in SKOv3-ARHI cells and upregulated the expression of Ang2, IGFBP3, TIMP3, CDH1, SPARC, CDH13, and SULF1 in Hey-ARHI cells, which indicates that methylation plays a crucial role in their regulation during tumor recurrence. All of the results presented above further proved that ovarian cancer recurrence can be reversed by epigenetic agents. In addition, the sequencing results of the 5-Aza-dC-treated recurrence model further discovered that the hypermethylated promoters of TIMP3 and CDH1 had all been distinguishingly reversed. Among these genes, the most significant change was observed for CDH1, whose methylation level was reduced from 87.8% in recurrence to 21.7% after 5-Aza-dC treatment, which is even lower than the level in dormancy. In Hey-ARHI, CDH1 methylation also experienced a large decrease after 5-Aza-dC treatment, from 63.8% in recurrence to 32.8%. Taking the dramatic upregulation of 5-Aza-dC on CDH1 expression in both cell lines together, DNA methylation might be vitally important in regulating CDH1 during this process.
An interesting finding was that TIMP3 and CDH1 DNA methylation fell slightly behind the changes of expression in the dormancy group, which suggests that gene expression could also be regulated by other mechanisms. Histone modifications and other epigenetic mechanisms work together in maintaining gene activity states. Epigenetic information in chromatin includes covalent modifications, such as acetylation, methylation, phosphorylation and ubiquitination of histones.Citation37 We found a subset of genes (Ang2, IGFBP3, EFEMP1, TIMP3, TIE1, and CDH1 in SKOv3-ARHI cells; Ang1, TIMP3, TSP1, CDH1, Ang4, and SULF1 in Hey-ARHI) could be re-activated by TSA treatment, which suggests that those genes are potentially controlled by histone acetylation. Among these genes, TIMP3 expression was also dramatically upregulated by a combination of 5-Aza-dC and TSA in both cell lines. With our finding that elevated TIMP3 expression was associated with an increased H3K9Ac marker at the TIMP3_CpG island in dormant cells and a loss in progressively growing SKOv3-ARHI cells, our results indicate that both DNA methylation and histone acetylation can regulate TIMP3 expression. In addition, decreased H3K27me3 that is associated with the upregulation of TIMP3 during dormancy in Hey-ARHI suggests that histone methylation might also regulate TIMP3 expression. Our results are consistent, in part, with studies of prostate cancer in which the reversal of TIMP3 repression by TSA was associated with decreased H3K27me3 and increased H3K9Ac histone markers at the TIMP3 promoter.Citation38 CDH1 expression was regulated by both DNA and histone modification during the transition from dormancy to recurrence. H3K4me3 and H3K9Ac, two markers of transcriptional activation, were enriched at the CDH1 promoter in dormant cells and were reduced in progressively growing cells. Thus, our results suggest that bivalent histone methylation and acetylation markers potentially influence CDH1 expression and the transition from dormancy to regrowth.
This report confirms and extends an earlier study that documented that re-expression of the imprinted tumor suppressor gene ARHI inhibits clonogenic growth, induces autophagy and produces tumor dormancy in xenografts.Citation21 For our studies, a cell culture model was established, in which concurrent treatment with VEGF prevents autophagic death after the upregulation of ARHI and permits additional clonogenic growth after the downregulation of ARHI. This model provided a convenient method for studying the expression of anti-angiogenic factors that predict expression in the xenograft model. Matrigel was found to facilitate the growth of xenograft.
Taken together, our studies are consistent with the importance of anti-angiogenic factors in maintaining the dormant state in models of ovarian cancer. DNA methylation and histone modification were essential in regulating TIMP3 and CDH1. Other anti-angiogenic and pro-angiogenic factors are likely to be regulated by other mechanisms. Epigenetic therapy is, however, worthy of further study as consolidative therapy to inhibit or eliminate dormant ovarian cancer cells.
Materials and Methods
Antibodies and reagents
McCoy’s 5A (GNM16600) and RPMI 1640 (GNM31800) medium was purchased from Jinuo Co, Ltd. Matrigel was purchased from BD Bioscience (356234). FBS and DOX were purchased from Clontech (631106, 631311). G418 was supplied by Gibco-BRL (11811). VEGF was purchased from R&D Systems (293-ve). TSA, 5-Aza-dC and Blasticidin were purchased from Sigma-Aldrich (T8552, A3656, 15205). Antibodies against human TSP1, TIMP3, H3K4me3, H3K27me3, and H3K9Ac were purchased from Abcam (ab1823, ab58804, Ab8580, Ab6002, and Ab4441, respectively). Antibodies against CDH1 and β-actin were purchased from Cell Signaling Technology (3195, 4970). Anti-H3K9me2 was purchased from Millipore (17-648). Anti-PCNA and anti-LC3 were purchased from Epitomics (2577-1, 2057-1). Anti-ARHI was provided by the Bast Laboratory at the University of Texas MD Anderson Cancer Center. All of the other chemicals were purchased from Takara Biotechnology and Tiangen Biotech.
Cell lines and culture conditions
The human ovarian epithelial cancer cell lines SKOv3-ARHI and Hey-ARHI were obtained from the Bast Laboratory at the University of Texas MD Anderson Cancer Center.Citation21 SKOv3-ARHI was routinely maintained in McCoy’s medium, which contained 10% FBS, 500 µg/ml G418, 50 ng/ml puromycin, 100 U/ml penicillin and 100 µg/ml streptomycin. Hey-ARHI was maintained in RPMI 1640 medium, which contained 10% FBS, 1% Glutamine, 25 μg/ml blasticidin and 1 μg/ml puromycin. ARHI expression was induced by adding 1 μg/ml DOX to the culture medium. Cells were incubated at 37 °C in 5% CO2. All of the experiments were performed by using cells in the exponential phase of growth in culture.
Establishment of an ovarian carcinoma dormancy and recurrence model in cell culture
Cells (1 × 103) were seeded in six-well culture dishes and incubated overnight in growth medium. The cells were divided into six groups: the blank control group (medium only), dormancy control group (cells treated with 1 µg/ml DOX for 14 d), VEGF control group (cells treated with 20 ng/ml VEGF for 14 d), dormancy group (cells treated with both 20 ng/ml VEGF and 1 µg/ml DOX for 14 d), recurrence group (cells treated with VEGF for 14 d and DOX for 6 d followed by DOX removal) and recurrence control group (cells treated with 1 µg/ml DOX for 6 d followed by DOX removal). All of the media were replenished daily.
Treatment of ovarian cancer cells with DNA methyltransferase inhibitor and histone deacetylase inhibitors
Cells (1 × 103) were seeded in six-well culture dishes and incubated overnight in growth medium. The cells were divided into six groups: the VEGF control group, dormancy group, recurrence group, 5-Aza-dC group (recurrence group treated with 5 µM 5-Aza-dC in the last 72 h), TSA group (recurrence group treated with 0.4 µM TSA in the last 24 h) and 5-Aza-dC + TSA group (recurrence group treated with 5 µM 5-Aza-dC in the last 72 h and with 0.4 µM TSA in the last 24 h). All of the media were replenished daily, and all of the cells were harvested after 14 d of treatment.
Clonogenic assays
Cells (1 × 103) were cultured in control medium or medium containing VEGF, DOX, TSA or 5-Aza-dC for 14 d. The colonies in each well were stained with Coomassie blue and were counted in a phase-contrast microscope.
cDNA synthesis and SYBR green real-time PCR
RNA was isolated from cells in culture and from xenografts using the RNAprep Micro kit (Tiangen Biotech, DP420). Reverse transcription was performed using a RevertAid First Strand cDNA Synthesis kit (Fermentas, K1622). Semi-quantitative RT-PCR was performed using a SYBR Green Master Mix (Takara Bio Inc., DRR820A) and was monitored on the Illumina Eco Real-time PCR System. The levels of angiogenesis-related factor mRNAs were calculated using the equation 2−ΔΔCt and were normalized to human β-actin mRNA levels. The sequences of the specific primers are listed in Table S1.
DNA isolation, bisulfite modification and bisulfite sequencing
Genomic DNA was extracted from cultured cells and tumor xenografts. Genomic DNA was isolated using the TIANamp Micro DNA kit (Tiangen Biotech, DP316). Bisulfite reactions were performed using the EZ DNA Methylation-Gold kit (ZYMO Research, D5006) according to the manufacturer’s recommendations. For bisulfate sequencing PCR reactions, specific primers were designed using “Methprimer” (http://www.urogene.org/methprimer/index1.html) or as previously described.Citation39,Citation40 Primer sequences and annealing temperatures are listed in Table S2. The amplified fragments were subcloned using the pBLUE-T vector kit (Aidlab Biotech, CV04). After cloning, 10 clones from each sample were randomly selected for DNA sequencing. Inserts were sequenced using M13 primers.
Flow cytometry of AVO
The control and treated cells were stained with acridine orange (Songon, A742007) at a final concentration of 1 μg/ml for a period of 15 min at 37°C while avoiding light. After washing twice with PBS, the cells were trypsinized, suspended in PBS containing 1% FBS and analyzed immediately. The fluorescence emission was measured by flow cytometry (FACS Calibur, BD) through the FL1/ FL3 channels using CellQuest 7.0 software (Beckman Coulter Co).
Western blotting
Antibodies against TSP1 (1:200), TIMP3 (1:200), H3K4me3 (1:500), H3K9Ac (1:200), CDH1 (1:1000), β-actin (1:2000), H3K9me2 (1:200), LC3 (1:1000) and ARHI (1:1000) were used in western blotting. Cells and tissues were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris-base, 5 mM EDTA, 1% NP-40, 0.25% deoxycholate, pH 7.4). Protein concentrations were measured using the BCA Protein Assay. Equal amounts of protein were resolved by SDS-PAGE, transferred to PVDF membranes, and incubated with appropriate primary antibodies with the indicated concentration. Immune complexes were detected with HRP-conjugated second antibodies (Santa Cruz, sc-2004 and sc2005) and ECL chemiluminescence reagent (Thermo, 32109).
Establishment of an ovarian carcinoma dormancy model in vivo
We used matrigel to facilitate the establishment of SKOv3-ARHI xenografts. Eight-week-old BALB/c nu/nu mice were purchased from Sino-British SIPPR/BK Lab Animal Ltd., Co (Shanghai, PRC). Matrigel was used as described.Citation22 Briefly, SKOv3-ARHI cells (1 × 107 in 150 µl) were added to liquid matrigel at 4°C. The mixture (final volume 300 µl) was slowly injected SC into the flanks of each mouse, where the matrigel immediately polymerized to form a solid gel. Twenty-four mice were divided into three groups (8 mice per group). The control and dormancy groups were given drinking water without DOX or with DOX (2 mg/ml). In the recurrence group, DOX was withdrawn after 32 d of treatment. The tumor size was measured every third day using a caliper. The tumor volume was calculated with the use of the following formula: tumor volume (mm3) = L × I2 × 0.5, where L is the longest diameter, I is the shortest diameter, and 0.5 is a constant to calculate the volume of the ellipsoid. At the end of the experiment (day 75), the mice were killed by cervical dislocation. Tumors were harvested by dissection and were either snap-frozen or fixed in formalin.
Immunohistochemistry
Formalin-fixed specimens were embedded in paraffin, and 4-µM-thick sections were prepared. Endogenous peroxidase activity was blocked using 0.3% hydrogen peroxide. Antigen retrieval was performed by heating the sections for 10 min in a microwave oven with 10 mM sodium citrate (pH 6.0). The slides were then incubated with monoclonal antibodies against CDH1 (1:200), TIMP3 (1:50), PCNA (1:100), and CD31 (1:100) at 4 °C overnight. The slides were washed and incubated with biotinylated secondary antibody for 30 min at room temperature and were then washed with PBS. The slides were incubated with avidin-biotin-peroxidase for 10 min at room temperature and were incubated with DAB for 2 min. Finally, the slides were counterstained with hematoxylin and evaluated at a 200 × magnification using light microscopy. Semi-quantitative estimates were made using a composite score that included the staining intensity values and the relative abundance of the positive cells. The intensities were graded as 0 (negative), 1 (weakly positive), 2 (moderately positive), and 3 (strongly positive). The abundance of positive cells was graded from 0 to 4 (0, <5% positive cells; 1, 5–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%). The scores were determined independently by two observers, and the average of their scores was used for the evaluation.
Immunocytochemistry
Cultured cells were washed with PBS and fixed with 4% paraformaldehyde. The slides were washed again, treated with 1% Triton for 15 min and blocked with 3% H2O2 for 20 min. After washing, the slides were blocked with normal goat serum for 10 min at room temperature and incubated with anti-CDH1 (1:200), anti-TSP-1(1:25), or anti-TIMP3 (1:200) antibody at 4°C overnight. The slides were then incubated with biotinylated anti-rabbit secondary antibody for 30 min at room temperature. The slides were then incubated with avidin-biotin-peroxidase for 10 min at room temperature and incubated with DAB for 2 min. Finally, they were counterstained with hematoxylin and evaluated at 400× magnification using light microscopy. Semi-quantitative estimates of staining intensity were performed as described above in immunohistochemistry.
Chromatin immunoprecipitation (ChIP) assays
Cells were subjected to ChIP based on the EZ-ChIP protocol (Millipore, 17-371). Briefly, chromatin and DNA were cross-linked by treatment with 1% formaldehyde. Cell lysates were collected and sonicated to shear DNA. Soluble chromatin was incubated for 2 h at 4 °C with 60 μl of protein A-agarose-salmon sperm DNA. Pre-cleared lysate was incubated overnight at 4 °C with 2 μg of anti-H3K9Ac, anti-H3K4me3, anti-H3K27me3 or anti-H3K9me2. The antibody-protein-DNA complexes were precipitated with 60 μl of protein A-agarose beads at 4 °C for 2 h. The complexes were eluted in elution buffer (0.1 mM NaHCO3 and 1% SDS) before the reversal of cross-links overnight at 65°C under high salt conditions (0.5 M NaCl). After proteinase K digestion, DNA was extracted in 25:24:1 phenol/chloroform/isoamyl alcohol and precipitated overnight in ethanol at −20 °C, and DNA was then eluted in Tris/EDTA buffer. The presence of TIMP3 and CDH1 gene promoter sequences in immunoprecipitated DNA was identified by PCR using the primer sequences listed in Table S3. The number of PCR cycle for CDH1 is 34 cycles, and for TIMP3 is 35 cycles. In control samples, the primary antibody was replaced with non-immune IgG. PCR products were semi-quantified by ImageJ software. All of the experiments were repeated at least three times.
Statistical analysis
Statistical analysis was conducted using SPSS version 15.0 (SPSS). The level of significance for all of the tests was defined as α = 0.05. Student t-tests were used for the analysis of categorical data, and all of the statistical tests were 2-sided. All of the data were expressed as the mean ± SD for the experiments that were performed at least 3 times. A P value of less than 0.05 was considered to be significant.
| Abbreviations: | ||
| ARHI | = | aplasia ras homolog member I |
| VEGF | = | vascular endothelial growth factor |
| TIMP3 | = | tissue inhibitor of metalloproteinases-3 |
| TSP1 | = | thrombospondin-1 |
| Ang1 | = | angiopoietin-1 |
| Ang2 | = | angiopoietin-2 |
| Ang4 | = | angiopoietin-4 |
| CDH1 | = | E-cadherin |
| DOX | = | doxycycline |
| ANGPTL2 | = | angiopoietin-like 2 |
| IGFBP3 | = | insulin-like growth factor binding protein-3 |
| TIE-1 | = | tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| TIE-2 | = | tyrosine kinase with immunoglobulin-like and EGF-like domains 2 |
| CDH13 | = | H-cadherin |
| SPARC | = | secreted acidic cysteine rich glycoprotein |
| RECK | = | reversion-inducing-cysteine-rich protein with kazal motifs |
| SULF-1 | = | sulfatase 1 |
| EFEMP1 | = | EGF containing fibulin-like extracellular matrix protein 1 |
| DNMT | = | DNA methyltransferase |
| HDAC | = | histone deacetylase |
| MMPs | = | endogenous matrix metalloproteinases |
| FDA | = | food and drug administration |
| SAHA | = | suberoylanilide hydroxamic acid |
| TSA | = | trichostatin |
| 5-Aza-dC | = | 5-Aza-deoxycytidine |
| PCNA | = | proliferating cell nuclear antigen |
| AVO | = | acidic vesicular organelles |
| HRP | = | horseradish peroxidase |
| DAB | = | diaminobenzidine |
| ChIP | = | chromatin immunoprecipitation |
| BSP | = | bisulfite sequencing |
Additional material
Download Zip (661.8 KB)Acknowledgments
This work is supported by the National Natural Science Foundation of China (grant number: 30973185) to WF; RO1 CA135354-01 from the National Institutes of Health; MD Anderson SPORE in Ovarian Cancer NCI P50 CA83639, the National Foundation for Cancer Research to RCB and kind gifts from Stuart and Gaye Lynn Zarrow.
Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012; 62:10 - 29; http://dx.doi.org/10.3322/caac.20138; PMID: 22237781
- Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer 2007; 7:834 - 46; http://dx.doi.org/10.1038/nrc2256; PMID: 17957189
- Almog N. Molecular mechanisms underlying tumor dormancy. Cancer Lett 2010; 294:139 - 46; http://dx.doi.org/10.1016/j.canlet.2010.03.004; PMID: 20363069
- Páez D, Labonte MJ, Bohanes P, Zhang W, Benhanim L, Ning Y, Wakatsuki T, Loupakis F, Lenz HJ. Cancer dormancy: a model of early dissemination and late cancer recurrence. Clin Cancer Res 2012; 18:645 - 53; http://dx.doi.org/10.1158/1078-0432.CCR-11-2186; PMID: 22156560
- Gilead A, Neeman M. Dynamic remodeling of the vascular bed precedes tumor growth: MLS ovarian carcinoma spheroids implanted in nude mice. Neoplasia 1999; 1:226 - 30; http://dx.doi.org/10.1038/sj.neo.7900032; PMID: 10935477
- Wu X, Liang L, Dong L, Yu Z, Fu X. Effect of ARHI on lung cancer cell proliferation, apoptosis and invasion in vitro. Mol Biol Rep 2013; 40:2671 - 8; http://dx.doi.org/10.1007/s11033-012-2353-x; PMID: 23247805
- Wang Y, Yu Q, Cho AH, Rondeau G, Welsh J, Adamson E, Mercola D, McClelland M. Survey of differentially methylated promoters in prostate cancer cell lines. Neoplasia 2005; 7:748 - 60; http://dx.doi.org/10.1593/neo.05289; PMID: 16207477
- Dalai I, Missiaglia E, Barbi S, Butturini G, Doglioni C, Falconi M, Scarpa A. Low expression of ARHI is associated with shorter progression-free survival in pancreatic endocrine tumors. Neoplasia 2007; 9:181 - 3; http://dx.doi.org/10.1593/neo.06838; PMID: 17401457
- Rosen DG, Wang L, Jain AN, Lu KH, Luo RZ, Yu Y, Liu J, Bast RC Jr.. Expression of the tumor suppressor gene ARHI in epithelial ovarian cancer is associated with increased expression of p21WAF1/CIP1 and prolonged progression-free survival. Clin Cancer Res 2004; 10:6559 - 66; http://dx.doi.org/10.1158/1078-0432.CCR-04-0698; PMID: 15475444
- Wang L, Hoque A, Luo RZ, Yuan J, Lu Z, Nishimoto A, Liu J, Sahin AA, Lippman SM, Bast RC Jr., et al. Loss of the expression of the tumor suppressor gene ARHI is associated with progression of breast cancer. Clin Cancer Res 2003; 9:3660 - 6; PMID: 14506155
- Yu Y, Xu F, Peng H, Fang X, Zhao S, Li Y, Cuevas B, Kuo WL, Gray JW, Siciliano M, et al. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc Natl Acad Sci U S A 1999; 96:214 - 9; http://dx.doi.org/10.1073/pnas.96.1.214; PMID: 9874798
- Feng W, Marquez RT, Lu Z, Liu J, Lu KH, Issa JP, Fishman DM, Yu Y, Bast RC Jr.. Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer 2008; 112:1489 - 502; http://dx.doi.org/10.1002/cncr.23323; PMID: 18286529
- Feng W, Shen L, Wen S, Rosen DG, Jelinek J, Hu X, Huan S, Huang M, Liu J, Sahin AA, et al. Correlation between CpG methylation profiles and hormone receptor status in breast cancers. Breast Cancer Res 2007; 9:R57; http://dx.doi.org/10.1186/bcr1762; PMID: 17764565
- Fujii S, Luo RZ, Yuan J, Kadota M, Oshimura M, Dent SR, Kondo Y, Issa JP, Bast RC Jr., Yu Y. Reactivation of the silenced and imprinted alleles of ARHI is associated with increased histone H3 acetylation and decreased histone H3 lysine 9 methylation. Hum Mol Genet 2003; 12:1791 - 800; http://dx.doi.org/10.1093/hmg/ddg204; PMID: 12874100
- Hisatomi H, Nagao K, Wakita K, Kohno N. ARHI/NOEY2 inactivation may be important in breast tumor pathogenesis. Oncology 2002; 62:136 - 40; http://dx.doi.org/10.1159/000048259; PMID: 11914599
- Yu Y, Fujii S, Yuan J, Luo RZ, Wang L, Bao J, Kadota M, Oshimura M, Dent SR, Issa JP, et al. Epigenetic regulation of ARHI in breast and ovarian cancer cells. Ann N Y Acad Sci 2003; 983:268 - 77; http://dx.doi.org/10.1111/j.1749-6632.2003.tb05981.x; PMID: 12724231
- Yuan J, Luo RZ, Fujii S, Wang L, Hu W, Andreeff M, Pan Y, Kadota M, Oshimura M, Sahin AA, et al. Aberrant methylation and silencing of ARHI, an imprinted tumor suppressor gene in which the function is lost in breast cancers. Cancer Res 2003; 63:4174 - 80; PMID: 12874023
- Lu Z, Luo RZ, Peng H, Rosen DG, Atkinson EN, Warneke C, Huang M, Nishmoto A, Liu J, Liao WS, et al. Transcriptional and posttranscriptional down-regulation of the imprinted tumor suppressor gene ARHI (DRAS3) in ovarian cancer. Clin Cancer Res 2006; 12:2404 - 13; http://dx.doi.org/10.1158/1078-0432.CCR-05-1036; PMID: 16638845
- Luo RZ, Fang X, Marquez R, Liu SY, Mills GB, Liao WS, Yu Y, Bast RC. ARHI is a Ras-related small G-protein with a novel N-terminal extension that inhibits growth of ovarian and breast cancers. Oncogene 2003; 22:2897 - 909; http://dx.doi.org/10.1038/sj.onc.1206380; PMID: 12771940
- Badgwell DB, Lu Z, Le K, Gao F, Yang M, Suh GK, Bao JJ, Das P, Andreeff M, Chen W, et al. The tumor-suppressor gene ARHI (DIRAS3) suppresses ovarian cancer cell migration through inhibition of the Stat3 and FAK/Rho signaling pathways. Oncogene 2012; 31:68 - 79; http://dx.doi.org/10.1038/onc.2011.213; PMID: 21643014
- Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare S, Kondo S, Kondo Y, Yu Y, Mills GB, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest 2008; 118:3917 - 29; PMID: 19033662
- Albini A, Fontanini G, Masiello L, Tacchetti C, Bigini D, Luzzi P, Noonan DM, Stetler-Stevenson WG. Angiogenic potential in vivo by Kaposi’s sarcoma cell-free supernatants and HIV-1 tat product: inhibition of KS-like lesions by tissue inhibitor of metalloproteinase-2. AIDS 1994; 8:1237 - 44; http://dx.doi.org/10.1097/00002030-199409000-00004; PMID: 7528513
- Almog N, Ma L, Raychowdhury R, Schwager C, Erber R, Short S, Hlatky L, Vajkoczy P, Huber PE, Folkman J, et al. Transcriptional switch of dormant tumors to fast-growing angiogenic phenotype. Cancer Res 2009; 69:836 - 44; http://dx.doi.org/10.1158/0008-5472.CAN-08-2590; PMID: 19176381
- Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene 2002; 21:5400 - 13; http://dx.doi.org/10.1038/sj.onc.1205651; PMID: 12154403
- Anand-Apte B, Bao L, Smith R, Iwata K, Olsen BR, Zetter B, Apte SS. A review of tissue inhibitor of metalloproteinases-3 (TIMP-3) and experimental analysis of its effect on primary tumor growth. Biochem Cell Biol 1996; 74:853 - 62; http://dx.doi.org/10.1139/o96-090; PMID: 9164653
- Qi JH, Ebrahem Q, Moore N, Murphy G, Claesson-Welsh L, Bond M, Baker A, Anand-Apte B. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med 2003; 9:407 - 15; http://dx.doi.org/10.1038/nm846; PMID: 12652295
- Pedrazzani C, Corso G, Marrelli D, Roviello F. E-cadherin and hereditary diffuse gastric cancer. Surgery 2007; 142:645 - 57; http://dx.doi.org/10.1016/j.surg.2007.06.006; PMID: 17981184
- Pinheiro H, Carvalho J, Oliveira P, Ferreira D, Pinto MT, Osório H, Licastro D, Bordeira-Carriço R, Jordan P, Lazarevic D, et al. Transcription initiation arising from E-cadherin/CDH1 intron2: a novel protein isoform that increases gastric cancer cell invasion and angiogenesis. Hum Mol Genet 2012; 21:4253 - 69; http://dx.doi.org/10.1093/hmg/dds248; PMID: 22752307
- Yi Kim D, Kyoon Joo J, Kyu Park Y, Yeob Ryu S, Soo Kim H, Kyun Noh B, Hwa Lee K, Hyuk Lee J. E-cadherin expression in early gastric carcinoma and correlation with lymph node metastasis. J Surg Oncol 2007; 96:429 - 35; http://dx.doi.org/10.1002/jso.20732; PMID: 17786966
- Jiménez B, Volpert OV, Crawford SE, Febbraio M, Silverstein RL, Bouck N. Signals leading to apoptosis-dependent inhibition of neovascularization by thrombospondin-1. Nat Med 2000; 6:41 - 8; http://dx.doi.org/10.1038/71517; PMID: 10613822
- Wendt MK, Taylor MA, Schiemann BJ, Schiemann WP. Down-regulation of epithelial cadherin is required to initiate metastatic outgrowth of breast cancer. Mol Biol Cell 2011; 22:2423 - 35; http://dx.doi.org/10.1091/mbc.E11-04-0306; PMID: 21613543
- Mack GS. Epigenetic cancer therapy makes headway. J Natl Cancer Inst 2006; 98:1443 - 4; http://dx.doi.org/10.1093/jnci/djj447; PMID: 17047192
- Thompson CA. Vorinostat approved for rare lymphoma. Am J Health Syst Pharm 2006; 63:2168; http://dx.doi.org/10.2146/news060020; PMID: 17090730
- Chen MY, Liao WS, Lu Z, Bornmann WG, Hennessey V, Washington MN, Rosner GL, Yu Y, Ahmed AA, Bast RC Jr.. Decitabine and suberoylanilide hydroxamic acid (SAHA) inhibit growth of ovarian cancer cell lines and xenografts while inducing expression of imprinted tumor suppressor genes, apoptosis, G2/M arrest, and autophagy. Cancer 2011; 117:4424 - 38; http://dx.doi.org/10.1002/cncr.26073; PMID: 21491416
- Hellebrekers DM, Jair KW, Viré E, Eguchi S, Hoebers NT, Fraga MF, Esteller M, Fuks F, Baylin SB, van Engeland M, et al. Angiostatic activity of DNA methyltransferase inhibitors. Mol Cancer Ther 2006; 5:467 - 75; http://dx.doi.org/10.1158/1535-7163.MCT-05-0417; PMID: 16505122
- Kim MS, Kwon HJ, Lee YM, Baek JH, Jang JE, Lee SW, Moon EJ, Kim HS, Lee SK, Chung HY, et al. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nat Med 2001; 7:437 - 43; http://dx.doi.org/10.1038/86507; PMID: 11283670
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012; 150:12 - 27; http://dx.doi.org/10.1016/j.cell.2012.06.013; PMID: 22770212
- Shinojima T, Yu Q, Huang SK, Li M, Mizuno R, Liu ET, Hoon DS, Lessard L. Heterogeneous epigenetic regulation of TIMP3 in prostate cancer. Epigenetics 2012; 7:1279 - 89; http://dx.doi.org/10.4161/epi.22333; PMID: 23023649
- Pesta M, Kulda V, Topolcan O, Safranek J, Vrzalova J, Cerny R, Holubec L. Significance of methylation status and the expression of RECK mRNA in lung tissue of patients with NSCLC. Anticancer Res 2009; 29:4535 - 9; PMID: 20032402
- Barski D, Wolter M, Reifenberger G, Riemenschneider MJ. Hypermethylation and transcriptional downregulation of the TIMP3 gene is associated with allelic loss on 22q12.3 and malignancy in meningiomas. Brain Pathol 2010; 20:623 - 31; http://dx.doi.org/10.1111/j.1750-3639.2009.00340.x; PMID: 19922547