Abstract
Although most cancer research has focused in mRNA, non-coding RNAs are also an essential player in tumorigenesis. In addition to the well-recognized microRNAs, recent studies have also shown that epigenetic silencing by CpG island hypermethylation of other classes of non-coding RNAs, such as transcribed ultraconserved regions (T-UCRs) or small nucleolar RNAs (snoRNAs), also occur in human neoplasia. Herein we have studied the putative existence of epigenetic aberrations in the activity of PIWI proteins, an Argonaute family protein subclass, and the small regulatory PIWI-interacting RNAs (piRNAs) in testicular cancer, as the PIWI/piRNA pathway plays a critical role in male germline development. We have observed the existence of promoter CpG island hypermethylation-associated silencing of PIWIL1, PIWIL2, PIWIL4, and TDRD1 in primary seminoma and non-seminoma testicular tumors, in addition to testicular germ cell tumor cell lines. Most importantly, these epigenetic lesions occur in a context of piRNA downregulation and loss of DNA methylation of the LINE-1 repetitive sequences, one of the target genomic loci where the PIWI/piRNA machinery acts as a caretaker in non-transformed cells.
Introduction
piRNAs (PIWI-interacting RNAs) are a peculiar class of small non-coding RNAs of 24–30 nt in length that are mainly expressed in the germline.Citation1,Citation2 They are transcribed from regions in the genome that contain transcribed transposable elements and other repetitive elements.Citation1,Citation2 piRNAs are Dicer-independent and bind the PIWI subfamily of Argonaute family proteins.Citation1,Citation3 The PIWI/piRNA pathway seem to be involved in the maintenance of genomic stability and germ cell function by two different mechanisms that have been described in Drosophila melanogaster: cleavage of transposable element transcripts by PIWI proteins, a process that is mediated through piRNA base-pairing recognition, and heterochromatin-mediated transcriptional silencing associated with a gain of DNA methylation.Citation1-Citation3 PIWI-class proteins, such as PIWIL2 and PIWIL4, are also involved in a so-denominated “ping-pong” amplification cycle, creating antisense piRNAs that are capable of repressing the transcript of origin.Citation1-Citation3 In fact, knockout mice models for the proteins involved in the piRNA biogenesis (such as, MIWI, MILI, and MIWI2) revealed a recovery of transposon activity, which is thought to be the cause of the observed sterility due to meiotic arrest.Citation4,Citation5 In this regard, DNA methylation regulated expression of piRNAs occurs in human spermatozoaCitation6 and non-genetic infertility syndromes in males could be associated with epigenetic disruption of PIWI proteins.Citation7
Interestingly, piRNAs have recently been detected also in human cancer cells and somatic cells, and it has been suggested that piRNAs control gene expression more broadly than previously.Citation2 Herein, we have wondered if in their natural functional context (the normal human testis), the PIWI/piRNA pathway undergoes aberrant DNA methylation events that compromise their activity in the corresponding transformed cells (human testicular germ cell tumors). To this aim, we have analyzed the 5′end promoter CpG islands of the main PIWI-class protein genes (PIWIL1, PIWIL2, and PIWIL4) and its associated protein TDRD1 (Tudor domain containing protein 1). PIWIL1 is expressed after birth in the pachytene stage and acts in translational control in the latest stages of spermatogenesis.Citation8 PIWIL2 has essential roles in the initial phases of spermatogenesis: transposon silencing in fetal gonocytes, germline stem cell self-renewal and early prophase of mammalian testis.Citation9 Furthermore, PIWIL2 has been implicated in translational regulation of many genes during early spermatogenesis since it binds piRNAs and mRNAs.Citation9 TDRD1 binds directly to PIWIL2 and PIWIL1.Citation10 Although it does not affect the ability of PIWI proteins to associate with piRNAs in embryonic testes, it controls the entry of correct transcripts into the normal pool of piRNAs.Citation11 Our data indicate that epigenetic disruption of the entire PIWI/piRNA pathway is indeed a hallmark for the development of testicular tumors.
Results and Discussion
Gain of 5′end promoter CpG island methylation for the PIWIL1, PIWIL2, PIWIL4 and TDRD1 genes occurs in primary testicular tumors in association with their transcriptional silencing
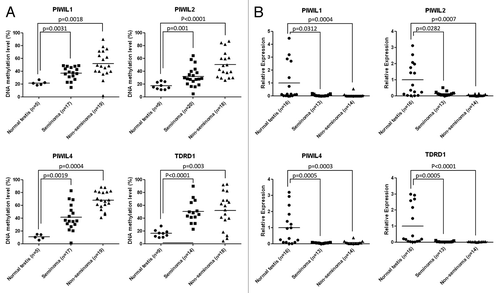
The PIWIL1, PIWIL2, PIWIL4, and TDRD1 genes have 5′end CpG islands surrounding the corresponding transcription start sites and, thus, they are candidate genes to be epigenetically inactivated by promoter CpG island hypermethylation in human cancer.Citation12,Citation13 Using bisulfite modification of DNA coupled with pyrosequencing, we observed that the four genes undergo hypermethylation events in primary testicular cancer in comparison to normal testicular tissues that show unmethylated CpG islands. The gain of 5′end CpG island methylation of PIWIL1, PIWIL2, PIWIL4, and TDRD1 occurred both in seminoma and non-seminoma tumors (). To demonstrate the transcriptional silencing of these PIWI-class protein genes in cancer cells in association with the presence of CpG island hypermethylation, we measured PIWIL1, PIWIL2, PIWIL4, and TDRD1 levels by quantitative RT-PCR. The expression of the four studied PIWI/piRNA pathway genes was significantly downregulated in testicular tumors in comparison to normal testis. The diminished expression of PIWIL1, PIWIL2, PIWIL4, and TDRD1 occurred both in seminoma and non-seminoma tumors ().
Figure 1. Epigenetic inactivation of genes encoding piRNA-related proteins in primary testicular germ cell tumors. (A) DNA methylation levels at the 5′end CpG islands of the PIWIL1, PIWIL2, PIWIL4, and TDRD1 genes determined by sodium bisulfite modification coupled to pyrosequencing. (B) mRNA expression levels of the PIWIL1, PIWIL2, PIWIL4, and TDRD1 genes determined by quantitative reverse transcription PCR.
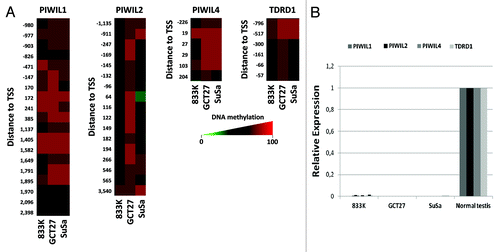
We further tightened the link between CpG island hypermethylation of the studied PIWI-class protein genes and transcriptional inactivation by the analyses of testicular cancer cell lines. Using a DNA methylation microarrayCitation14 that contains numerous CpG sites located in the PIWIL1, PIWIL2, PIWIL4, and TDRD1 CpG islands (), we found that the human testicular germ cell tumor lines 833K, GCT27 and SuSa showed dense promoter CpG island hypermethylation for the described genes. Most importantly, we did not observe expression of the four studied PIWI/piRNA pathway genes in any of the three studied cancer cell lines, while normal testis expressed the PIWIL1, PIWIL2, PIWIL4 and TDRD1 transcripts ().
Figure 2. Epigenetic inactivation of genes encoding piRNA-related proteins in testicular germ cell tumor lines. (A) DNA methylation levels at the 5′end CpG islands of the PIWIL1, PIWIL2, PIWIL4 and TDRD1 genes determined by sodium bisulfite modification coupled to hybridization to a DNA microarray (450K Illumina). DNA methylation levels are color-coded (red: high, green: low). Probe distances to the transcription start site (TSS) are indicated. (B) mRNA expression levels of the PIWIL1, PIWIL2, PIWIL4 and TDRD1 genes determined by quantitative reverse transcription PCR.
Epigenetic inactivation of PIWI-class protein genes is associated with diminished piRNA expression and hypomethylation events at LINE-1 loci
Once we had determined the existence of promoter CpG island hypermethylation events in the described PIWI-class protein genes and the diminished expression of their corresponding transcripts in testicular tumorigenesis, both in primary tumors and cultured transformed cells, we wondered about the downstream impact of the described epigenetic inactivation. We first analyzed the expression levels of piRNAs in the same samples studied for PIWIL1, PIWIL2, PIWIL4, and TDRD1 CpG island methylation and transcription. piRNAs show a high diversity in their genomic sequences and are transcribed from a relatively small number of genomic regions called piRNAs clusters.Citation15 After a primary RNA is generated, piRNA accumulation requires amplification by the above mentioned ping-pong mechanism involving at least two distinct Piwi proteins, a process that occurs in the cytoplasm.Citation1,Citation2 Herein, we randomly selected three piRNAs transcribed from different genomic loci to sample the global levels of expression of these small regulatory ncRNAs: DQ598918 (chr7:99 691 656–99 691 686), DQ589977(chr17:79 479 330–79 479 358) and DQ601609 (chr1:179 557 005–179 557 036). Using quantitative RT-PCR to study the expression levels of the three piRNAs, we found diminished expression of all of them in primary testicular tumors in comparison to normal testis (). The diminished expression of the piRNAs DQ598918, DQ589977 and DQ601609 occurred both in seminoma and non-seminoma tumors ().
Figure 3. Molecular environment of the epigenetic loss of PIWI-protein genes in primary testicular tumors: diminished piRNA expression and LINE1 hypomethylation. (A) Expression levels of the piRNAs DQ598918, DQ589977 and DQ601609 determined by quantitative reverse transcription PCR. (B) DNA methylation levels at the LINE1 sequence determined by sodium bisulfite modification coupled to pyrosequencing.
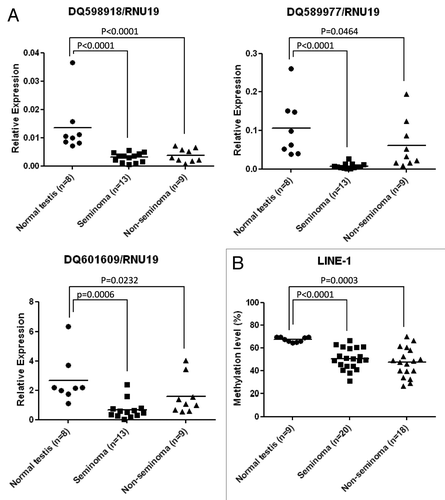
Interestingly, the detected aberrant hypermethylation and diminished expression of PIWI-family genes, together with the downregulated piRNAs, might have the ability to provoke further DNA methylation changes of additional loci. In this regard, genetic and molecular analyses have identified interactions between DNA methyltransferases (DNMTs) and piRNA pathway members. The PIWI/DNMT3L complex targets genomic loci, sequence-guided by small RNAs.Citation16 DNMT3LCitation16 as well as PIWIL2Citation17 and TDRD1Citation11 null models revealed a loss of DNA methylation at LINE-1 and intracisternal A-particle (IAP) transposons, leading to reactivation of the repetitive sequences. Thus, we proceeded to determine in our studied set of primary testicular tumors that harbor PIWIL1, PIWIL2, PIWIL4, and TDRD1 promoter CpG island methylation-associated silencing and piRNA diminished levels, had also undergone DNA methylation changes at the LINE1 sequence. Using bisulfite modification of DNA coupled with pyrosequencing, we observed that the LINE1 sequences experimented hypomethylation events in testicular cancer in comparison to normal testis that show a more methylated LINE1 sequence (). The loss of LINE1 methylation occurred both in primary seminoma and non-seminoma tumors ().
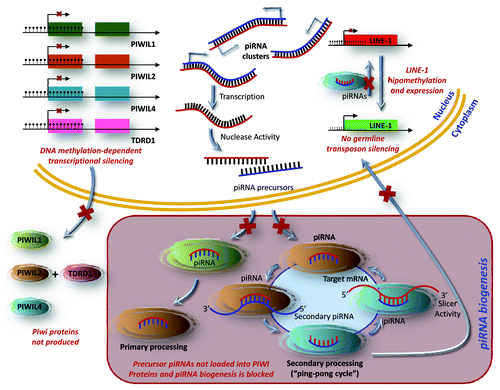
Overall, our results show the existence of cancer specific hypermethylation events in the CpG islands of genes associated with piRNAs that leads to their transcriptional inactivation in testicular cancer. Most importantly, the epigenetic inactivation of PIWI-class protein genes (PIWIL1, PIWIL2, and PIWIL4) and its associated protein TDRD1 in human testicular tumorigenesis occurs in a molecular context characterized by a diminished expression of the piRNAs and the DNA hypomethylation of LINE1, a PIWI/piRNA target sequence. Interestingly, epigenetic disruption of PIWI proteins also occurs in non-genetic infertility syndromes in malesCitation7 and there is an epidemiological association between male infertility and testicular cancer.Citation18,Citation19 Thus, the epigenetic disruption of the PIWI/piRNA pathway could be the missing common link in the genesis of both pathologies. A model for the PIWI/piRNA pathway in normal testis and its disruption in testicular tumors is also shown in . Although much work lies ahead to understand the role of the PIWI/piRNA machinery in cellular transformation, the current work provide intriguing clues for further developments and discoveries in this area.
Figure 4. A model for the PIWI/piRNA pathway in germline cells and its disruption in testicular tumors. piRNAs are small non-coding RNAs that are mainly transcribed as single-stranded intergenic RNAs from well-conserved mono- and bi-directional clusters of repetitive elements. These piRNA precursors translocate into the cytoplasm, where they mature into functional piRNAs. The PIWI proteins catalyze a self-amplification loop, “ping-pong” cycle. Their incorporation into the PIWI ribonucleoprotein (piRNP) complex targets repetitive elements through target degradation and epigenetic silencing. In testicular cancer types, piRNA biogenesis and function are disrupted by DNA hypermethylation mediated transcriptional silencing of PIWI-proteins, leading to the expression of germline transposons.
Methods
Cell lines and patient samples
833K, SuSa and GCT27 testicular germ cell tumor linesCitation20 were cultured in RPMI 1640 medium supplemented with 20% (w/v) fetal bovine serum and penicillin/streptomycin (all from Invitrogen). All the cell lines were cultured at 37 °C in a humidified atmosphere with 5% (v/v) CO2. A representative subset of primary testicular germ cell tumors were obtained from the the IDIBELL Bank of tumors, supported by the Xarxa de Bancs de Tumors de Catalunya sponsored by Pla Director d'Oncología de Catalunya (XBTC). The study was approved by the relevant institutional review boards and ethics committees and informed consent was obtained from each patient. Seventeen primary pure seminomas (SE) and 19 non-seminomas (NSE) including either pure tumors (embryonal carcinomas, choriocarcinomas and yolk-sac tumors) as mix tumors composed by two or more components were included in the study. RNA and DNA from cell lines and primary tumor RNA and DNA from cell lines, testicular cancer samples and healthy controls were extracted using TRIzol (Invitrogen) and Phenol:Chloroform:Isoamylalcohol (Sigma), respectively. Bisulphite modification of genomic DNA was performed with the EZ DNA Methylation Kit (Zymo) following the manufacturer's protocol. Total RNA from adult normal testis (R1234260-50-BC) was purchased from BioCat.
Pyrosequencing
The set of primers for PCR amplification and sequencing were designed using a specific software pack (PyroMark assay design version 2.0.01.15). PIWIL1: FF 5′-GTTTGYGGGG TAGTTAATGA GGG, RV 5′-ACCTCCCAAA ACCTCCTT, SEQ 5′-CCCAAAACCT CCTTC; PIWIL2: FF 5′-ATGGGTTAAT TAGATAGTTT GTTTTTGTGA, RV 5′- AACCCACATA CTCCAAAACC AATTTC, SEQ 5′- AATTAGATAG TTTGTTTTTG TGAA; PIWIL4: FF 5′-AAGTAATGGG AAGAAAAAGG AAGTT, RV 5′-CCAAAATAAC CACAAAAACT ACAAAATCTC, SEQ 5′-CAACATCCAC AACCA; TDRD1: FF 5′-AAGGAATTTT TTGAGTTTGT AATTAGAGTA, RV 5′-ATACAAACCC TCTCCCTCCC CTATA, SEQ 5′-TTGTAATTAG AGTATAAGTT GTTT. LINE-1 was quantified using the PyroMark Q96 LINE-1 assay (Qiagen) that target four CpG sites in positions 331 to 305 (GenBank accession no X58075) within the human LINE-1 transposon DNA consensus sequence. PCR was performed with primers biotinylated to convert the PCR product to single-stranded DNA templates. We used the Vacuum Prep Tool (Biotage) to prepare single-stranded PCR products according to manufacturer’s instructions. Pyrosequencing reactions and methylation quantification were performed in a PyroMark Q24 System version 2.0.6 (Qiagen). Graphic representation of DNA methylation values shows the averaged values over multiple CpG sites.
Real-time qRT-PCR
Total RNA was reverse transcribed using ThermoScript RT-PCR System (Invitrogen, Life Technologies) under standard conditions with hexanucleotide random primers, according to the manufacturer's instructions. cDNA of protein coding genes was amplified by real-time PCR using Taqman assays: PIWIL1 (Hs01041737), PIWIL2 (Hs00216263), PIWIL4 (Hs00381509) and TDRD1 (Hs00229805) and expression values were normalized to GAPDH (AB Assay ID 4333764F). Reverse transcription and amplification of 3 randomly chosen piRNAs was performed using a custom-designed TaqMan assays (Applied Biosystems), providing specificity for the mature RNA target. Expression values of piRNAs DQ598918 (CSQJAPI), DQ589977 (CSMSF6U) and DQ601609 (CSVI3FS) were normalized to RNU19 (AB Assay ID 001003).
Infinium 450K Methylation array
All DNA samples were assessed for integrity, quantity and purity by electrophoresis in a 1.3% agarose gel, picogreen quantification, and nanodrop measurements. All samples were randomly distributed into 96 well plates. Bisulfite conversion of 500 ng of genomic DNA was performed using the EZ DNA methylation kit (Zymo Research) following the manufacturer’s instructions. 200 ng of bisulfite-converted DNA were used for hybridization on the HumanMethylation450 BeadChip (Illumina). Briefly, samples were whole-genome amplified followed by enzymatic end-point fragmentation, precipitation and resuspension. The resuspended samples were hybridized onto the BeadChip for 16 h at 48 °C, and then washed. A single nucleotide extension with labeled dideoxy-nucleotides was performed and repeated rounds of staining were applied with a combination of labeled antibodies differentiating between biotin and DNP. Chip analysis was performed using Illumina HiScan SQ fluorescent scanner. The intensities of the images were extracted and the data was normalized using GenomeStudio (2010.3) Methylation module (1.8.5) software.
Statistical analysis
Statistical analyses were performed using GraphPad PRISM (v5.04). The nonparametric Mann-Whitney U test was used to analyze differences in absolute expression and methylation level in cancer patient groups compared with controls. A value of P < 0.05 was considered significant.
Acknowledgments
The research leading to these results has received funding from the European Research Council under the European Community's Seventh Framework Programme (FP7/2007–2013)/ERC grant agreement no. 268626—EPINORC project, the Spanish Ministry of Economy and Competitiveness (MINECO Project no. SAF2011-22803), the Cellex Foundation and the Health and Science Departments of the Catalan Government (Generalitat de Catalunya). HJF is a Portuguese Foundation for Science and Technology (FCT) Ph.D. Fellow. ME is an ICREA Research Professor.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12:861 - 74; http://dx.doi.org/10.1038/nrg3074; PMID: 22094949
- Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev 2012; 26:2361 - 73; http://dx.doi.org/10.1101/gad.203786.112; PMID: 23124062
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 2013; 14:447 - 59; http://dx.doi.org/10.1038/nrg3462; PMID: 23732335
- Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2002; 2:819 - 30; http://dx.doi.org/10.1016/S1534-5807(02)00165-X; PMID: 12062093
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 2004; 131:839 - 49; http://dx.doi.org/10.1242/dev.00973; PMID: 14736746
- Krausz C, Sandoval J, Sayols S, Chianese C, Giachini C, Heyn H, Esteller M. Novel insights into DNA methylation features in spermatozoa: stability and peculiarities. PLoS One 2012; 7:e44479; http://dx.doi.org/10.1371/journal.pone.0044479; PMID: 23071498
- Heyn H, Ferreira HJ, Bassas L, Bonache S, Sayols S, Sandoval J, Esteller M, Larriba S. Epigenetic disruption of the PIWI pathway in human spermatogenic disorders. PLoS One 2012; 7:e47892; http://dx.doi.org/10.1371/journal.pone.0047892; PMID: 23112866
- Bamezai S, Rawat VP, Buske C. Concise review: The Piwi-piRNA axis: pivotal beyond transposon silencing. Stem Cells 2012; 30:2603 - 11; http://dx.doi.org/10.1002/stem.1237; PMID: 22996918
- Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem 2009; 284:6507 - 19; http://dx.doi.org/10.1074/jbc.M809104200; PMID: 19114715
- Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway?. Genes Dev 2010; 24:636 - 46; http://dx.doi.org/10.1101/gad.1899210; PMID: 20360382
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 2009; 16:639 - 46; http://dx.doi.org/10.1038/nsmb.1615; PMID: 19465913
- Esteller M. Epigenetics in cancer. N Engl J Med 2008; 358:1148 - 59; http://dx.doi.org/10.1056/NEJMra072067; PMID: 18337604
- Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 2012; 13:679 - 92; http://dx.doi.org/10.1038/nrg3270; PMID: 22945394
- Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 2011; 6:692 - 702; http://dx.doi.org/10.4161/epi.6.6.16196; PMID: 21593595
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007; 128:1089 - 103; http://dx.doi.org/10.1016/j.cell.2007.01.043; PMID: 17346786
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 2008; 31:785 - 99; http://dx.doi.org/10.1016/j.molcel.2008.09.003; PMID: 18922463
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 2007; 316:744 - 7; http://dx.doi.org/10.1126/science.1142612; PMID: 17446352
- Hotaling JM, Walsh TJ. Male infertility: a risk factor for testicular cancer. Nat Rev Urol 2009; 6:550 - 6; http://dx.doi.org/10.1038/nrurol.2009.179; PMID: 19724246
- Peng X, Zeng X, Peng S, Deng D, Zhang J. The association risk of male subfertility and testicular cancer: a systematic review. PLoS One 2009; 4:e5591; http://dx.doi.org/10.1371/journal.pone.0005591; PMID: 19440348
- Fung MK, Cheung HW, Ling MT, Cheung AL, Wong YC, Wang X. Role of MEK/ERK pathway in the MAD2-mediated cisplatin sensitivity in testicular germ cell tumour cells. Br J Cancer 2006; 95:475 - 84; http://dx.doi.org/10.1038/sj.bjc.6603284; PMID: 16880791
