Abstract
It was recently shown that duplications of the RevSex element, located 0.5 Mb upstream of SOX9, cause XX-disorder of sex development (DSD), and that deletions cause XY-DSD. To explore how a 148 kb RevSex duplication could have turned on gonadal SOX9 expression in the absence of SRY in an XX-male, we examined the chromatin landscape in primary skin fibroblast cultures from the index, his RevSex duplication-carrier father and six controls. The ENCODE project supports the notion that chromatin state maps show overlap between different cell types, i.e., that our study of fibroblasts could be of biological relevance. We examined the SOX9 regulatory region by high-resolution ChIP-on-chip experiments (a kind of “chromatin-CGH”) and DNA methylation investigations. The RevSex duplication was associated with chromatin changes predicting better accessibility of the SRY-responsive TESCO enhancer region 14–15 kb upstream of SOX9. Four kb downstream of the TESCO evolutionary conserved region, a peak of the enhancer/promoter-associated H3K4me3 mark was found together with a major dip of the repressive H3K9me3 chromatin mark. Similar differences were also found when three control males were compared with three control females. A marked male/female difference was a more open chromatin signature in males starting ~400 kb upstream of SOX9 and increasing toward the SOX9 promoter. In the RevSex duplication-carrier father, two positions of DNA hypomethylation were also found, one corresponding to the H3K4me3 peak mentioned above. Our results suggest that the RevSex duplication could operate by inducing long-range epigenetic changes. Furthermore, the differences in chromatin state maps between males and females suggest that the Y chromosome or X chromosome dosage may affect chromatin conformation, i.e., that sex-dependent gene regulation may take place by chromatin modification.
Introduction
SRY and SOX9 are master regulators of male sexual development. SRY-stimulated SOX9 expression in the undifferentiated fetal gonad causes Sertoli cell formation, and these cells orchestrate further testicular development, ultimately leading to fetal testosterone production and male differentiation of the external genitalia and probably also male-specific patterning of the brain.Citation1-Citation5 The most important, but not the only, downstream effect of SRY is enhancement of SOX9 transcription in the genital ridge.Citation6 Both SRY and SOX9 are high mobility group box (HMGB) domain transcription factors with partly overlapping DNA binding profiles.Citation7,Citation8 SOX9 plays an important role in, for example, branchial arch, skeletal, and brain development, in addition to its sex-determining function in the gonads.Citation9,Citation10 The regulation of SOX9 gene expression is complex, governed by a ~2 Mb genomic region upstream of the gene itself, which is quite small and occupies only 5–6 kb of genomic sequence.Citation11 Translocations and copy number changes in the large SOX9 regulatory region can cause a Pierre-Robin sequence type of cleft palate, and disturbed chondrocyte differentiation with skeletal dysplasia and/or sex reversal, all dependent on the position and size of the genomic aberrations.Citation9,Citation12-Citation15 This variability suggests that different domains of the upstream “gene desert” regulate SOX9 expression in a time- and tissue-specific manner, and that the genomic architecture is a critical factor for proper regulation.
The HMGB domain group of transcription factors, including SRY and SOX9, are known to cause DNA bending and to interact with other transcription factors (e.g., members of the OCT family) and probably also chromatin modifying proteins.Citation7,Citation16,Citation17 SRY acts synergistically with steroidogenic factor-1 (SF1, encoded by NR5A1) at an enhancer element 14–15 kb upstream of the SOX9 transcription start site.Citation6 This ~1.4 kb enhancer element is called TESCO (testis enhancer sequence core element) and it contains a 176 nt highly conserved motif in its 5′ half; the TESCO evolutionaryly conserved region (TESCO-ECR, 70 103 197–70 103 373 bp from 17pter, hg19).Citation18 Mutations in TESCO were not observed in 66 patients with XY-gonadal dysgenesis without an SRY mutation (i.e., SRY-mutation negative Swyer syndrome patients).Citation19 This indirectly suggests that if malfunction of TESCO should be an important cause of XY-gonadal dysgenesis, it is probably due to other factors than SRY binding-site mutations.
It was therefore of interest to investigate if the genomic architecture of the SOX9 regulatory region could represent an additional and superior level of SOX9 regulation. For this, a family with a 46,XX ovotesticular DSD (DSD; disorder of sex development) male with a 148 kb duplication of the so-called RevSex element 0.5 Mb upstream of SOX9 was well suited. This family and three other RevSex duplication/deletion families was published by Benko et al. in 2011, and the family was labeled DSD2 in this article.Citation12 In this family, the RevSex duplication was inherited from a normal grandmother through the XX-DSD male’s father (Fig. S1). This gave us a chance to investigate the epigenetic profile of the duplicated strand on 46,XX and 46,XY genetic backgrounds. The results led us to explore if there also could be epigenetic differences between three control males and three control females. We found clear indications that the presence of a Y chromosome and/or only one X chromosome affects the conformation of the regulatory region from SOX9 and approximately 0.7 Mb upstream, and in particular the 15 kb region from the TESCO enhancer to the SOX9 transcription start site. Since these results are based on one XX and one XY RevSex duplication carrier and 3 female + 3 male controls, they should be considered as preliminary, especially since chromatin investigations were done in skin fibroblast primary cultures. In support of our study of skin fibroblasts as a proxy for the chromatin state in biologically more relevant but also inaccessible human tissues, it has recently been shown that the chromatin landscape of human primary fibroblasts have qualitative similarities to human embryonic stem cells, the major difference being an expansion of repressive chromatin blocks in the differentiated cell type.Citation20 Whether chromatin data from skin fibroblasts also have relevance for Sertoli cells in early embryonic development is unknown but conceivable, especially since high-order heterochromatin formation and epigenetic remodeling of the genome can be discrete events, as was recently shown in senescent fibroblast.Citation11 Another indication of architectural conservation on a large-scale chromatin level is the preservation of topology-associated domains (TADs) across different cell types.Citation21
If the chromatin differences between males and females in the SOX9 regulatory region are Y chromosome or X-dosage related, a potential epigenetic mechanism for disturbed sex development has been found, with relevance for the large number of unexplained DSD-cases.Citation5,Citation22-Citation24 Our findings also give reason to explore whether SRY may regulate gene expression through regional epigenetic modification.
Results
Since RevSex duplications apparently can bypass the need for SRY to induce testicular SOX9 expression (i.e., cause XX-DSD) and RevSex deletions are associated with insufficient SOX9 response to SRY stimulation (i.e., cause XY-DSD), normal males can be carriers of duplications and normal females can be carriers of deletions. It was therefore unexpected to find a RevSex duplication inherited from a normal female in our previously reported family, i.e., that this was not a de novo or grandpaternal duplication as had been expected (Fig. S1).Citation12 Potential causes of such non-penetrance could be mosaicism for the RevSex duplication in the carrier grandmother, stochastic differences during embryonic development, e.g., random epigenetic differences between grandmother and XX-DSD grandson, or the inheritance of different modifier alleles in grandmother and grandson. A SNP-array profile of blood-DNA did not show signs of mosaicism in the grandmother, but such mosaicism could still have been present in tissues types other than blood.
To explore if the duplication affected the chromatin architecture of the SOX9 regulatory region, we performed chromatin immunoprecipitation studies on cultured dermal fibroblasts from the index patient (an XX-DSD male with the RevSex duplication on his paternal chromosome 17), his normal XY brother, his father (a carrier of the RevSex duplication on his maternal chromosome 17) and his mother (Fig. S2 show all the raw signal-to-input data). Unfortunately, dermal fibroblasts from the duplication-carrier grandmother could not be obtained. The antibodies used were against histone H3 trimethylated at lysine 4 (H3K4me3), histone H3 trimethylated at lysine 27 (H3K27me3), histone H3 trimethylated at lysine 9 (H3K9me3) and histone H3 acetylated at lysine 9 (H3K9ac); the latter two modifications cannot coexist, as they target the same residue (lysine 9) of histone H3. H3K4me3 and H3K9ac are euchromatic marks, i.e., associated with open chromatin, while H3K27me3 and H3K9me3 are heterochromatic marks, associated with two different types of closed chromatin.Citation25 Sometimes the H3K4me3 and H3K27me3 mark coexist on a promoter (bivalent promoters), which may be a sign of dynamic and developmental regulation.Citation26
The chromatin pattern of immunoprecipitated DNA was determined by comparative hybridization to input (non-immunoprecipitated) DNA on custom-designed high-density oligonucleotide arrays, i.e., a kind of “chromatin-CGH.” To supplement the chromatin data, whole genome CpG methylation was determined using Infinium HumanMethylation450 BeadChips. In our main region of interest (69.2–70.2 Mb from 17pter, hg19), the BeadChip interrogates 69 CpG positions, a small subset of the total number in this region. In addition, 4 CpG positions were investigated by methylation-sensitive MLPA.
The first region of interest was the RevSex duplication itself. This was fine-mapped and the breakpoints were sequenced after long-PCR. It was found to be a direct duplication of the segment from 69 521 863 to 69 670 036 nucleotides from 17pter (hg19), i.e., a 148 kb direct duplication 0.5 Mb upstream of SOX9 without loss of genomic sequence, as previously published.Citation12 The raw ChIP-on-chip data suggested that this duplication had a more repressive chromatin signature in the XX-DSD male than in his duplication-carrier father (). To ease data visualization and to attempt to eliminate the confounding effect of the other (normal) SOX9 locus, the XY father’s and the XX-son’s chromatin profiles were compared after subtraction of the profile of the XY-brother of the index male. This brother had not inherited the RevSex duplication from his father, and he shared the normal SOX9 locus with his father and the maternal SOX9 locus with his XX-DSD brother. The latter was determined by haplotyping using a selection of small tandem repeat PCR primers (data not shown). Both before and after such correction, it could be seen that both the repressive chromatin marks (H3K27me3 and H3K9me3) of the RevSex duplication region were enriched in the XX-DSD case compared with his XY-father (). This did not correspond to differences in the degree of DNA methylation (), but one should note that the Infinium BeadChip array contains oligonucleotides against only two CpG positions within the duplicated segment. However, a similar result was obtained with custom designed methylation-sensitive MLPA testing of two other CpG positions within the RevSex duplication ().
Figure 1. An UCSC genome browser panel with custom tracks showing the differences in H3K4me3 (top), H3K27me3 (middle) and H3K9me3 (bottom) chromatin profiles between the 46,XX ovotesticular DSD male and his father in the area containing the RevSex duplication (marked with a black line). The curve displays the ChIP-on-chip signal of the index case after subtraction of the signal of his XY-father. The red lines mark the position of predicted SRY and SOX9 binding sites (“HMR Conserved Transcription Factor Binding Sites”), all with high Z-scores for SRY binding: 3.29 for SRY-1 and 3.13 for SRY-2. SRY-2 had in addition a high degree of genomic sequence conservation beyond the human/mouse/rat comparison that the Z-score calculations are based on; See the UCSC browser “Vertebrate Multiz Alignment & Conservation (46 Species)” track for details. A position of difference commented in the text is marked with “A.”
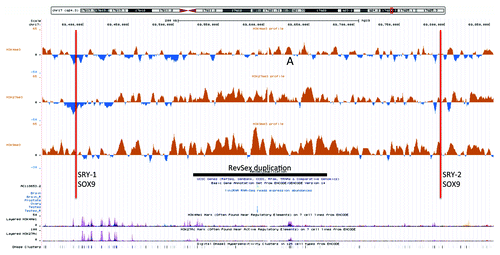
Table 1. Selected methylation values of the SOX9 regulatory region in RevSex duplication carriers (the XX-DSD index case and his father) compared with an average of male (n = 5) and female (n = 4) controls
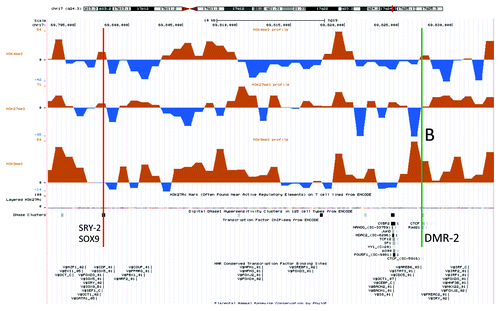
When focusing on a region 158 kb downstream of the duplication (0.3 Mb upstream of SOX9) that contains conserved transcription factor and CTCF binding sites (annotated in the “Transcription Factor ChIP-seq from ENCODE” and “HMR Conserved Transcription Factor Binding Sites” tracks in the UCSC genome browser), a similar but milder tendency of increased heterochromatinization was found in the index compared with his father (). Close to the CTCF binding site itself the difference was striking: a H3K9me3 peak in the index had the same position as an H3K27me3 peak in the father (marked B in ). That suggests different chromatin dynamics at this position in father and XX-DSD son. These peaks corresponded to a site of differential methylation (DMR2) that was hypomethylated in the father (, P < 0.01), in line with the chromatin immunoprecipitation results.
Figure 2. An UCSC genome browser panel with custom tracks displaying the differences in H3K4me3 (top), H3K27me3 (middle) and H3K9me3 (bottom) chromatin profiles between the XX-DSD male and his XY-father in a small region 158 kb downstream of the RevSex duplication containing a transcription factor/CTCF binding sites (UCSC genome browser track “HMR Conserved Transcription Factor Binding Sites”) called DMR2 in and marked B in the figure. The upstream binding site (red line) corresponds to a DNaseI hypersensitivity site and is also predicted to bind SRY and SOX9 (corresponds to the SRY-2/SOX9 site in ). The downstream binding site (green line) contains a CTCF binding site (ENCODE immunoprecipitation data) and a putative SRY binding site 3 kb downstream. It is important to note that SRY and SOX9 binding sites are computationally predicted based on human, mouse and rat conservation of binding sites as described in the Transfac database and have not been validated experimentally (see UCSC genome browser table “HMR Conserved Transcription Factor Binding Sites” description for more details).
To explore how these chromatin differences between father and index could affect SOX9 regulation, we looked for chromatin profile differences in a 120 kb region upstream of SOX9, including a known SOX9 enhancer called TESCO (70 103 kb from 17pter) and a position of hypomethylation in the father (70 107 kb from 17pter, called DMR3 in ). In addition to this point of paternal hypomethylation (DMR3, P < 0.05), the only striking difference between father and son was a stronger H3K27me3 profile in the XX-DSD son from the TESCO element and about 100 kb upstream ().
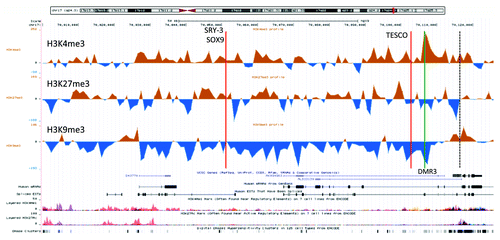
Figure 3. A UCSC genome browser panel with custom tracks displaying the signal-to-input chromatin profiles (H3K4me3, H3K27me3, and H3K9me3) of the XX-DSD case (upper profiles) compared with his RevSex duplication-carrier father (lower profiles) in the area from SOX9 and 120 kb upstream (70 000–70 126 kb from 17pter, hg19). The red line called TESCO corresponds to the position of the TESCO-ECR (70 103 197–70 103 373 bp from 17pter, hg19), the green line marked C to DMR3 in , and the dashed line the start of the SOX9 gene. The same position marks and more detailed explanations can be found in .
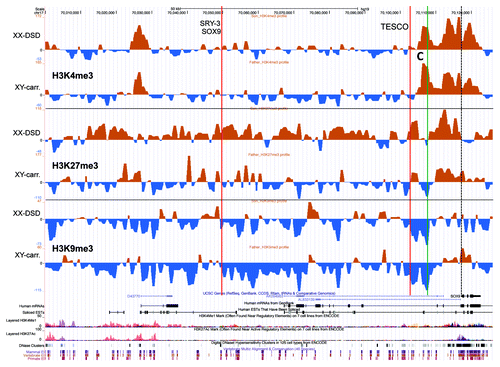
Conceivably, these differences could be due to the presence of a Y chromosome in the father, or different X chromosome dosage. To investigate this possibility, dermal fibroblast chromatin profiles of three unrelated males and three unrelated females were compared (). In the 736 kb region stretching from SOX9 and upstream and including the RevSex duplication, interesting male/female differences were found. Corresponding to a potential regulatory element of ~10 kb at position 69 630 000 (hg19), i.e., in the RevSex duplication region, the females had a stronger H3K4me3 signal flanked by stronger H3K9me3 signals than males (marked A in ), i.e., a similar tendency as in the XX-DSD case when compared with his father (marked A in ). This site corresponds to a “regulatory” H3K4me1/H3K27ac peak in the ENCODE data, indicating a putative enhancer element ( and , bottom). Of interest was also a clearly more pronounced decrease in H3K9me3 signal toward the SOX9 gene in males compared with females (marked D in ). The H3K4me3 “TESCO-peak” found in males was almost invisible in females (marked C in ), while the DSD-case had a peak of intermediate size (marked C in ). A direct XX-DSD-case/female and RevSex-carrier-male/male comparison can be seen in Figure S3. Of note, the XX-DSD case had a stronger H3K4me3 signal than control females corresponding to the known SOX9 enhancer/promoter region, suggesting more open chromatin and increased potential for SOX9 activation. In addition, hypomethylation of two CpG positions in the SOX9 gene body was found (, P < 0.05). These positions were hypermethylated in his RevSex duplication carrier father (P < 0.01). Unfortunately, using highly sensitive RT-PCR we did not detect any SOX9 expression in leukocytes or fibroblasts from any of the investigated individuals, and correlating our findings with mRNA expression levels was therefore not possible.
Figure 4. An UCSC genome browser panel with custom tracks showing the average signal-to-input chromatin profiles (H3K4me3, H3K27me3, and H3K9me3) of control females (upper profiles, n = 3) and control males (lower profiles, n = 3) in the 736 kb region from 69,390,000 to 70,126,000 (hg19) after 1 kb binning of data. A, C, and D mark positions of differences commented in the text. The dashed line marks the start of the SOX9 gene.
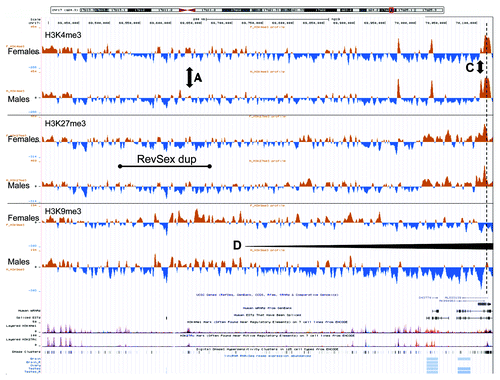
These chromatin profile differences between females and males were not reflected in differences in DNA methylation (). There were only four positions of differential methylation (DMRs) among the 50 CpG positions interrogated by the Illumina BeadChip upstream of SOX9. In addition, 4 positions of potential differential methylation were assayed using methylation-sensitive MLPA; these positions were chosen based on data from the “UCSF Brain DNA methylation” table of the UCSC genome browser. In none of these did males and females have different degrees of methylation (). For comparison, the chromatin profiles of the whole region and of the DMR positions described in were quantitated, and, in this case, marked differences between females and males were detected. Overall, the female SOX9 locus had a higher level of heterochromatin protein 1 (HP1)-associated repressive chromatin marks (H3K9me3) than males (P < 0.01, ). The DMR1 region, close to a lincRNA start site, appeared more “active” in females (stronger H3K4me3 and H3K9ac profiles). DMR2 appeared more polycomb repressive complex 2 (PRC2)-protein associated in males (more H3K27me3) than females, which had a stronger signal of the HP1-associated repressive mark H3K9me3. DMR4, corresponding to two CpGs in the SOX9 gene body, showed no significant differences in chromatin profiles, while DMR3, close to the SOX9 promoter and 4 kb downstream the TESCO element, displayed the marked differences described above (). The differences in male and female chromatin profiles in the 120 kb region upstream of SOX9 can be seen more clearly in , where the average female profile has been subtracted from the average male profile. The opposing H3K4me3 (high in males) and H3K9me3 (high in females) peaks at DMR3 just downstream of the TESCO enhancer are striking. Of note, the female/male differences correspond to the XX-DSD-case/XY-father differences described above, the main exception being a more open chromatin state of the TESCO enhancer region in the RevSex duplication carriers.
Table 2. Male/female difference: Average male H3 chromatin profiles (n = 3) subtracted by average female H3 chromatin profiles (n = 3)
Figure 5. An UCSC genome browser panel with custom tracks displaying the difference between control male (n = 3) and control female (n = 3) chromatin profiles of the part SOX9 locus from the gene itself and 120 kb upstream (70,000 – 70,126 kb from 17pter, hg19). The profiles shown are H3K4me3 (top), H3K27me3 (middle), and H3K9me3 (bottom). The red line called TESCO marks the position of the TESCO-ECR (70,103,197–70,103,373 bp from 17pter, hg19) and the green line the position of DMR3 in . The predicted SRY-3/SOX9 binding site had a Z-score of 3.45 for SOX9 and 2.41 for SRY, while the TESCO enhancer had a Z-score of 2.58 for SOX9 and 2.66 for SRY (UCSC, “HMR Conserved Transcription Factor Binding Sites”). In addition, the TESCO enhancer had a high degree of genomic sequence conservation beyond the human/mouse/rat comparison used to calculate the Z-score (UCSC, “Vertebrate Multiz Alignment & Conservation (46 Species)”). The TESCO SRY binding site has been experimentally verified.Citation31
Discussion
It has so far been unknown how RevSex duplications or deletions may affect SOX9 expression in the primordial gonads.Citation12 We hypothesized that this genomic region with many HCNEs (highly conserved non-coding elements, see Ancora database, http://ancora.genereg.net) affect the chromatin state of the SOX9 TESCO enhancer and promoter region, e.g., through interaction between a regulatory element around 69 630 kb from 17pter (marked A in ) and TESCO. Recently, data on such genomic interactions became publicly available from Bing Ren’s laboratory at University of California San Diego (UCSD): http://chromosome.sdsc.edu/mouse/hi-c/.Citation21 These genome-wide Hi-C-type crosslinking data on mouse and human cell lines suggest the existence of alternative large size topology associated domains (TADs) in the SOX9 regulatory region, including potential interactions between the RevSex duplication and the SOX9 enhancer/promoter about 0.5 Mb downstream. The existence of many alternative long-range SOX9 interactions was supported by recent 4C data from lymphoblastoid and Sertoli cell lines.Citation11 Of note, and rather unexpectedly, the RevSex duplication region was one of the few regions upstream of SOX9 devoid of such long-range promoter interactions in a Sertoli cell line—but not in lymphoblastoid cells.Citation11
We explored our alternative-interactions-affect-chromatin-regulation hypothesis further by comparing the chromatin profiles of the SOX9 regulatory region in dermal fibroblasts from a XY father and his 46,XX ovotesticular DSD son, who both had a small RevSex duplication 0.5 Mb upstream of SOX9. The XX-DSD case had a more repressive chromatin profile corresponding to the duplicated region than his father, with an increase of repressive chromatin marks in both (PRC2-associated H3K27me3 mark and the HP1-associated H3K9me3 mark; ).Citation20 In contrast, a qualitative difference was found for a transcription factor/CTCF-binding site 290 kb upstream of SOX9 (marked B in ), corresponding to a site of differential methylation hypomethylated in the father (DMR2 in ). Here, an apparent “switch” from a H3K27me3 signal in the father to a H3K9me3 signal in the son could be seen ().
From the TESCO enhancer and downwards both the XX-DSD case and his father had a more open chromatin state than controls (stronger H3K4me3 and weaker H3K9me3 signals), the father to a greater degree than his son (). A difference between father and son can also be seen for H3K27me3 in the 100 kb region upstream of TESCO, with a stronger signal in the XX-DSD son than his XY father (). The most notable difference between the XX-DSD case and three female controls was a stronger H3K4me3 signal just downstream of TESCO in the RevSex duplication carrier (marked C in Fig. S3).
In addition to different sex chromosome complements, there is another potentially significant difference between the father and son: the father’s RevSex duplication is on the maternal chromosome 17, while the son’s duplication is on the paternal chromosome. An alternative explanation for non-penetrance in the grandmother could be imprinting, i.e., that only the maternal SOX9 locus is responsive to SRY in the undifferentiated gonads. There are, however, reported instances of a paternal translocation/deletion or just a deletion that caused XY-DSD,Citation15,Citation27 showing that also the paternal allele is needed for appropriate SOX9 transcription in the gonads. Therefore, the hypothesis that imprinting renders the maternal allele responsive and the paternal allele irresponsive to SRY seems unlikely.
Our data support a model where the RevSex duplication facilitates SOX9 activation in the primordial gonads by inducing a conformational change to the SOX9 regulatory region that may or may not lead to SOX9 transcription in the genital ridge in the absence of SRY, establishing a self-stimulatory SOX9 feedback loop.Citation6 Stochastic or genetic background-based epigenetic changes may be a superimposed regulatory element explaining expression differences. This could be the reason for non-penetrance in the grandmother and partly non-penetrance in the XX-DSD case (only one of his gonads developed into a testis). Conceivably, the tiny ~500 nt spliced lincRNA in the RevSex region (, Ensembl ENSG00000225818) could also play a role. This lincRNA is expressed only in the testes (UCSC genome browser track “Human Body Map lincRNAs and TUCP Transcripts Tracks”) with expression quantified by Cufflinks to be 237- 413 FPKM (out of a maximum of 1000) in the two analyzed testicular libraries.
Conceivably, the difference in chromatin profiles between XX and XY RevSex duplication carriers could be due to the Y chromosome and/or X chromosome dosage. In a mouse model, X chromosome dosage has been found to be at least as important as SRY for sex-dependent modulation of general gene expression.Citation28 However, the fact that SOX9 is the major SRY-responsive gene in the primordial gonads, and the existence of three strongly predicted and one verified SRY binding sites upstream of SOX9 ( and ), makes it more likely that it is the presence of a Y chromosome and not different X chromosome complements that causes the father-son difference. To investigate this further, the average chromatin profiles of three unrelated males were compared with three unrelated females (). In the 120 kb region upstream of SOX9 the clearest differences were found: females tended to have a more HP1-associated repressive chromatin profile of this region than males (). A similar and statistically highly significant tendency was found when the chromatin profile of the whole SOX9 regulatory region was quantified (). This suggests that females and males have different chromatin state maps of the SOX9 regulatory region. We do not know how this affects tissue specific SOX9 expression as patient tissues or cells in which this gene is normally expressed were not available. Our study was conducted on skin fibroblast primary cultures, which we assume to recapitulate aspects of the SOX9 locus chromatin conformation in more relevant tissues (see Introduction for supportive argumentation), but we do not know if this is really true. Maybe the difference in chromatin states between males and females would have been even greater if a relevant tissue for sex-dependent SOX9 expression had been investigated.
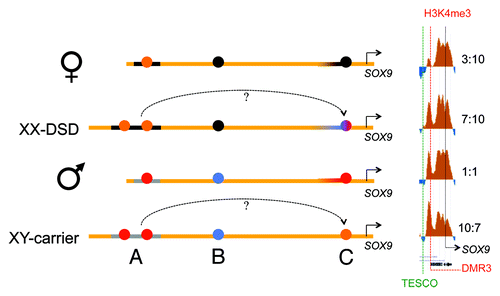
A summary of our findings is displayed in . Between TESCO and SOX9, males had a more open chromatin profile than females, and the XX-DSD case had a profile that was in-between. The clearest marker for this RevSex influence and sex difference is the DMR3 locus ( and ), marked “C” in , , and Figure S3. In the H3K4me3 peak signal from this DMR site has been compared with an internal control, the peak signal from the SOX9 promoter. The female H3K4me3 DMR3/SOX9 ratio was 3:10 and the male ratio was 1:1. In the XX-DSD case, the ratio was 7:10, while his RevSex duplication carrier father had a ratio of 10:7. This indicates that the RevSex duplication and Y chromosome might have additive effects on the SOX9 activation potential.
Figure 6. A schematic drawing of our major findings: In females the RevSex duplication (black line, region A) has more repressive chromatin with a more “active” putative enhancer element (orange dot) than in males. In the XX-DSD case similar findings are done with the exception of a more “open” SOX9 enhancer/promoter (region C). In males a downstream CTCF binding site is enriched in H3K27me3 (PRC2-related) chromatin compared with females (region B), and the SOX9 enhancer/promoter region (region C) is more open (red dot). This region is even more open in the XY RevSex carrier (orange dot). On the right side a comparison of the DMR3 H3K4me3 peaks (region C) can be seen with internal comparisons to the H3K4me3 peaks of the SOX9 promoter with peak ratios shown on the right. Chromatin color codes are follows: black, H3K9me3; blue, H3K27me3; red, H3K4me3; orange, stronger H3K4me3.
As far as we know, the question of whether the presence or absence of a Y chromosome can affect the chromatin landscape in the regulatory region of the main SRY target gene (SOX9) has not been addressed before. Since both SRY and SOX9 are HMGB domain group transcription factors known to cause DNA bending,Citation16 and since the DNA bending property of SRY is essential for its sex-determining function,Citation17 modification of chromatin architecture is likely to be a SRY function. This adds to the complexity of sex determination, where a large number of genes and pathways have been found to be of importance, and variability between closely related species is the rule rather than an exception.Citation24 Foremost, among these genes are the SRY/SOX9 and FOXL2/ESR pairs encoding transcription factors that promote male or female differentiation of the gonads, respectively. FOXL2 ablation in female mice lead to Sertoli cell-like differentiation of ovarian granulosa cells, despite absence of SRY.Citation29 It appears that FOXL2, together with the estrogen receptor (ESR), is needed to keep SOX9 in an inactive state in the mouse ovaries.Citation29 Notably, the SRY/SOX9/SF1 responsive TESCO element was also able to bind FOXL2/ESR.Citation29 Since FOXL2 mutations cause the autosomal dominant blepharophimosis-ptosis-epicanthus-inversus syndrome (BPES) in humans, a condition associated with premature ovarian failure in females, it is likely that FOXL2/ESR and SRY/SOX9 act antagonistically also on the human TESCO enhancer. In our XX-DSD index case, the RevSex duplication must be able to overcome or bypass SOX9 inhibition by FOXL2/ESR, maybe through a conformational change that makes the TESCO element more accessible to SOX9, potentially creating a positive feedback-loop for SOX9 transcription.Citation6 It is also noteworthy that another female differentiation-associated pathway, namely canonical WNT signaling stimulated by WNT4 and R-spondin (RSPO1), may cause chromatin modification via β-catenin’s interaction with another type of high-mobility group (HMG) transcription factors; the TCF/LEF family.Citation7,Citation30 Taken together, the shaping of the chromatin landscape of the SOX9 locus may be a common pathway for important sex determination factors, i.e., SRY/SOX9, FOXL2/ESR, and WNT4/RSPO1. This is also in line with the DSD-susceptibility (hypogonadism and ambiguous genitalia) found in boys with mutations in the X-linked genes ARX or ATRX, encoding a homeobox-containing transcription factor and a chromatin remodeling protein, respectively.Citation24 SRY might just be a facilitator for establishment of a SOX9 positive feedback loop in the male gonads through an influence on chromatin conformation, and a similar feedback loop may be switched on by the RevSex duplicationCitation12 or a lack FOXL2.Citation29
Taken together, we have shown that the RevSex duplication 0.5 Mb upstream of SOX9 is associated with a more open chromatin landscape at the TESCO enhancer and further downstream. This may be a cause of testicular differentiation of primordial gonads despite the absence of SRY. We have also found indications that the sex chromosome complement, most likely the presence of a Y chromosome, influences the chromatin state of the SOX9 regulatory region. If reproduced, our data provides a new angle for understanding sexual differentiation and unexplained DSD cases. Such epigenetic investigations may also lead to a better understanding of how the Y chromosome or SRY may affect the regulation of other genes, e.g., genes involved in autism, which for unexplained reasons is a male-predominant trait.
Patients and Methods
Family and case controls
The family with an XX-DSD male was recently described together with three other families with XX- or XY-DSD individuals due to duplications or a deletion of the RevSex element.Citation12 Of note, the index patient was born with hypospadias, rudimentary anlagen for vagina and uterus, and a scrotum containing a testis on one side and an ovarian remnant on the other side, all findings confirmed histologically. For the ChIP-on-chip experiments, dermal fibroblast samples from three unrelated males and three unrelated females were used as controls (see Table S1 for an overview of all individuals investigated by ChIP-on-chip). For the methylation testing, the number of controls was extended to five males and four females.
All procedures were in accordance with the Helsinki declaration of 1975. After genetic counseling, written informed consent from family members and control individuals were obtained before the experiments were undertaken. Since this started 15 y ago as a routine diagnostic genetic investigation to find the cause of XX-DSD and determine recurrence risks, there is no Regional Ethics Committee file number associated with this investigation. The family has been informed of all results obtained.
Cell cultures
Dermal fibroblasts from family members and controls were grown from skin biopsies under standard conditions, frozen in batches in liquid nitrogen, and later used for short-term fibroblast cultures. T-lymphocyte cultures were also grown under standard conditions and used for genomic mapping of the RevSex duplication and studying mRNA expression.
Molecular characterization of the RevSex duplication
After fine-mapping of the 17q24.3 duplication on the Cytogenetics Whole-Genome 2.7 M array (Affymetrix), following the manufacturer’s instructions, the junction fragment of the duplicated region was PCR amplified using primers between markers rs11871027 and rs11871770 in the 5′ end, and rs2429978 and rs2430557 and in the 3′ end, followed by sequencing of the PCR product.
SOX9 haplotyping
To determine SOX9 haplotypes and allele segregation, simple-tandem repeat markers were PCR amplified and size determined, and there were two informative markers upstream and two downstream of the SOX9 gene. Primer sequences can be provided upon request.
SOX9 mRNA expression
The SOX9 mRNA expression levels were investigated by qRT-PCR on cDNA from patient and control blood T-lymphocytes and skin fibroblasts. Primers and probes were from an AssayOnDemand Hs00165814_m1 kit from Applied Biosystems (Life technologies). Assessments of GAPDH and ACTB mRNAs (Applied Biosystems, Part number 4333764F and 4352935E) served as endogenous controls.
Chromatin DNA crosslinking and isolation
For each sample, a total of 4 × 107 dermal fibroblasts were cultured to about 90% confluence before crosslinking DNA with 1% formaldehyde at 37 °C for 15 min. Thereafter, glycine was added to a final concentration of 125 mM followed by 5 min incubation at room temperature. Next, cells were washed 3 times with ice-cold PBS before harvesting by scraping and centrifugation. To isolate the chromatin-associated DNA, the cell pellet was resuspended in 4 ml cell lysis buffer (5 mM PIPES pH8; 85 mM KCL; 0,5% NP-40; 1× protease inhibitors) followed by 10 min incubation on ice, centrifugation and resuspension in 1 ml of nuclear lysis buffer (50 mM Tris, pH8; 10 mM EDTA; 1% SDS; 1×protease inhibitors). After incubation on ice for 30 min, the homogenate was fragmented in a S220 Focused-ultrasonicator (Covaris) according to the manufacturer’s recommendations. The level of DNA fragmentation was examined on a 1% agarose gel, and fragmentation was continued until the major part of the DNA fragments had sizes of approximately 0.6 kb. After centrifugation, 50 µl of the supernatant was removed to determine the chromatin-DNA concentration (for input DNA determination), and the rest of the supernatant was stored in 100 µl batches at −80 °C.
Chromatin Immunoprecipitation (ChIP)
Per ChIP experiment, 50 µl lysate (on average containing 13.9 µg DNA), 25 µl protein A/G PLUS beads (Santa Cruz Biotechnology), and 425 µl incubation buffer (0.2% SDS, 1% Triton, 150 mM NaCl, 2 mM EDTA, 0,5 mM EGTA, 10 mM Tris pH 8,5, 1× protease inhibitors) were mixed and incubated with rotation for 1 h at 4 °C. Protein-DNA complexes were immunoprecipitated with a ChIP-validated antibody over night at 4 °C. The antibodies used were against histone H3 (histone H3, #4620; Cell signaling), histone H3 trimethylated at lysine 4 (H3K4me3, ab8580; Abcam), histone H3 trimethylated at lysine 27 (H3K27me3, ab6002; Abcam), histone H3 trimethylated at lysine 9 (H3K9me3, ab8898; Abcam), histone H3 acetylated at lysine 9 (H3K9ac, ab4441; Abcam), and rabbit IgG (#2729; Cell signaling). The amount of antibody added was according to the manufacturers’ recommendations. Next day, 25 µl protein A/G PLUS beads were added to each ChIP and incubated for 2 h at 4 °C. Thereafter the protein-DNA complexes were subjected to a series of washes in four different buffers: 2 × (0.1% SDS, 0.1% DOC, 1% triton, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris pH 8.5), 1 × (0.1% SDS, 0.1% DOC, 1% triton, 500 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris pH 8.5), 1 × (0.25% LiCl, 0.5% DOC, 0.5% NP-40, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris pH 8.5), and 2 × (1 mM EDTA, 0.5 mM EGTA, 10 mM Tris pH 8.5). The formaldehyde-induced crosslinks of the ChIPs were reversed by incubating the samples for 30 min at room temperature in alkaline solution (0.1 M NaHCO3, 1% SDS), and the DNA was recovered by standard procedures. The ChIP yield was controlled by PCR amplification of two genes (FBXO33 and LRFN5) in the final samples. In all ChIP-PCR verifications, the amount of immunoprecipitated DNA in input lysate was compared with a positive control (a histone H3 precipitate) and a non-immune control (rabbit IgG precipitate) for determination of the immunoprecipitation efficiency and specificity.
High Resolution ChIP-on-Chip assay
For ChIP-on-CHIP analysis, 60% of each of 2 replicate ChIP samples (from H3K9me3, H3K4me3, H3K27Mme3, and H3K9ac ChIPs) were concentrated using Microcon YM-30 spin columns (Merck Millipore) and amplified using the GenomePlex Complete Whole Genome Amplification kit (Sigma-Aldrich). The ChIP-on-Chip hybridization was done on a custom-designed high-resolution NimbleGen 3 × 720K arrays (Roche NimbleGen). The array probes (50-mers; positions according to GRCh37/hg19) were designed for uniform “tile-path” coverage of a 10 Mb regions of chromosome 2 (168.5–178.5 Mb from 2pter), an 18 Mb region of chromosome 13 (94–113 Mb from 13pter), a 41.5 Mb region of chromosome 14 (21 475–62 975 kb from 14pter), and a nearly 40 Mb region of chromosome 17 (49 370–90 870 kb from 17pter), with a median probe spacing of 0.1 kb. DNA labeling, array hybridization, post-hybridization washes and scanning were performed according to the manufacturer's protocol for ChIP-chip assays, version 6.2 (Roche NimbleGen). In short, the ChIP and non-precipitated (input) DNA were labeled with Cy5- and Cy3-conjugated random nonamers, respectively. The labeled samples were purified, combined, denatured and hybridized for 16 h at 42 °C to the array. After stringent washing the array was scanned using an Axon 4200AL Scanner (Molecular Devices) at 5-μm resolution. The acquired images were analyzed using DEVA v1.2 software (Roche NimbleGen) creating pair reports, including raw intensities for each probe and per image. From these data, ratio files were generated. For data visualization, the average ratios of two replicate experiments were binned per kb, adjusted for the number of probes per bin, and plotted against their chromosomal position.
ChIP-on-chip data analysis and presentation
As mentioned, 1 kb binning of the oligonucleotide signal-to-input-DNA ratios was performed, each 1 kb bin representing on average 8 oligonucleotide signals (range 6–9) from two replicate experiments (i.e., the signal from 12–18 oligonucleotides). After such binning, there were 97 374 data points per ChIP experiment per individual (from selected regions of chromosomes 2, 13, 14, and 17). Thereafter, the average ratio of these 97 374 data points was calculated, and a minor adjustment of all ratio values was done to make an average signal equal to the value of zero (Table S1). The H3K4me3 ratios ranged from −1.22 to +2.65, the H3K27me3 ratios ranged from −1.98 to + 2.28, the H3K9me3 ratios ranged from −1.51 to +1.15, and the H3K9ac ratios ranged from −1.54 to +2.00. To ease visualization of this large amount of ratios on Excel spread sheets, all values were multiplied by 100 and thereafter viewed as whole numbers. These data were used to make signal-to-input profiles (as in Fig. S2), i.e., a kind of comparative genomic hybridization (CGH) comparing immunoprecipitated DNA to input DNA from the same volume of cell lysate. A few chromatin profiles figures were made after subtracting such signal-to-input ratios from one another, e.g., for comparing males to females () or the XX-son to his XY-father ( and ).
Methylation chip assay
Methylation studies of DNA from blood samples were done using the Infinium Human Methylation 450K BeadChip assay (Illumina) according to the manufacturer’s instructions. The work was provided as a service of the Oslo-node of the Norwegian Microarray Consortium (see www.mikromatrise.no). Briefly, from each sample, 500 ng genomic DNA was bisulphite modified using the EZ DNA Methylation Kit (Zumo). Thereafter, the bisulphite converted sample was whole-genome amplified, fragmented, precipitated, and hybridized onto the BeadChip for 16 h at 48 °C. After post-hybridization washing, single-base extension of the probes on the BeadChip, using the captured DNA as a template, was used to identifying the methylation level of the query CpG sites. Labeling was detected by the Illumina iScan System, and signal intensities were estimated by the GenomeStudio Methylation Module v1.9 software (Illumina).
Methylation chip data analysis
The methylation level of each CpG site was calculated as a β value using Illumina’s formula: β = [methylated signal intensity / (methylated signal intensity + unmethylated signal intensity]. Sample independent and sample dependent control probes on the BeadChip were used for data normalization and background subtraction. The degree of methylation of each queried CpG position was calculated as a percentage (from 0–100% methylation), and used for calculation of methylation values in .
MLPA-based methylation analysis
Methylation-sensitive MLPA analysis of four CpG positions in the SOX9 regulatory region was done using the SALSA MLPA P200 Human DNA reference-1 kit (MRC-Holland) following the manufacturer’s instructions. The CpG positions were at 69 531 725, 69 666 757, 69 824 353, and 69 972 353 (hg19). Primer sequences can be provided upon request.
| Abbreviations: | ||
| ChIP | = | chromatin immunoprecipitation |
| CTCF | = | CCCTC-binding Factor |
| DMR | = | differentially methylated region |
| DSD | = | disorder of sex development |
| ENCODE | = | Encyclopedia of DNA Elements |
| ESR | = | estrogen receptor |
| H3 | = | histone 3 |
| H3K4me3 | = | Histone 3 trimethylated at lysine-4 |
| H3K27me3 | = | Histone 3 trimethylated at lysine-27 |
| H3K9me3 | = | Histone 3 trimethylated at lysine-9 |
| H3K9ac | = | histone 3 acetylated at lysine-9 |
| HMGB | = | high-mobility group box |
| LEF | = | lymphoid enhancer-binding Factor |
| MLPA | = | multiple ligation-dependent probe amplification |
| RSPO1 | = | R-Spondin-1 |
| SF1 | = | Stereogenic Factor-1 |
| SOX9 | = | SRY box containing gene 9 |
| SRY | = | sex determining region on the Y chromosome |
| TCF | = | T-Cell Factor |
| TESCO | = | testis enhancer sequence core element |
| TFBS | = | transcription factor binding site |
| UCSC | = | University of California Santa Cruz |
| UCSF | = | University of California San Francisco |
| WNT | = | from Drosophila-wingless |
Additional material
Download Zip (1,023.4 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are most grateful to the family for having contributed with blood samples and skin biopsies, and for their interest in contributing to research. We also thank Peter Koopman for his valuable feedback on the manuscript. This work was supported by HelseVest grants #911459 and #911744.
Author Contributions
Lybæk H designed the experiments and did the experimental work with the help of de Bruijn D, den Engelsman-van Dijk AHA and Brendehaug A,. Vanichkina D and Nepal C helped with computational analysis of the huge amounts of data. Houge G analyzed the data, made the figures, and wrote the manuscript.
References
- Swaab DF. Sexual differentiation of the brain and behavior. Best Pract Res Clin Endocrinol Metab 2007; 21:431 - 44; http://dx.doi.org/10.1016/j.beem.2007.04.003; PMID: 17875490
- Wilhelm D, Koopman P. The makings of maleness: towards an integrated view of male sexual development. Nat Rev Genet 2006; 7:620 - 31; http://dx.doi.org/10.1038/nrg1903; PMID: 16832429
- Parsch J, Ellegren H. The evolutionary causes and consequences of sex-biased gene expression. Nat Rev Genet 2013; 14:83 - 7; http://dx.doi.org/10.1038/nrg3376; PMID: 23329110
- Sekido R, Lovell-Badge R. Sex determination and SRY: down to a wink and a nudge?. Trends Genet 2009; 25:19 - 29; http://dx.doi.org/10.1016/j.tig.2008.10.008; PMID: 19027189
- Kashimada K, Koopman P. Sry: the master switch in mammalian sex determination. Development 2010; 137:3921 - 30; http://dx.doi.org/10.1242/dev.048983; PMID: 21062860
- Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 2008; 453:930 - 4; http://dx.doi.org/10.1038/nature06944; PMID: 18454134
- Malarkey CS, Churchill ME. The high mobility group box: the ultimate utility player of a cell. Trends Biochem Sci 2012; 37:553 - 62; http://dx.doi.org/10.1016/j.tibs.2012.09.003; PMID: 23153957
- Mertin S, McDowall SG, Harley VR. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res 1999; 27:1359 - 64; http://dx.doi.org/10.1093/nar/27.5.1359; PMID: 9973626
- Gordon CT, Tan TY, Benko S, Fitzpatrick D, Lyonnet S, Farlie PG. Long-range regulation at the SOX9 locus in development and disease. J Med Genet 2009; 46:649 - 56; http://dx.doi.org/10.1136/jmg.2009.068361; PMID: 19473998
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 2009; 12:399 - 408; http://dx.doi.org/10.1038/nn.2294; PMID: 19287386
- Smyk M, Szafranski P, Startek M, Gambin A, Stankiewicz P. Chromosome conformation capture-on-chip analysis of long-range cis-interactions of the SOX9 promoter. Chromosome Res 2013; Forthcoming http://dx.doi.org/10.1007/s10577-013-9386-4; PMID: 24254229
- Benko S, Gordon CT, Mallet D, Sreenivasan R, Thauvin-Robinet C, Brendehaug A, Thomas S, Bruland O, David M, Nicolino M, et al. Disruption of a long distance regulatory region upstream of SOX9 in isolated disorders of sex development. J Med Genet 2011; 48:825 - 30; http://dx.doi.org/10.1136/jmedgenet-2011-100255; PMID: 22051515
- White S, Ohnesorg T, Notini A, Roeszler K, Hewitt J, Daggag H, Smith C, Turbitt E, Gustin S, van den Bergen J, et al. Copy number variation in patients with disorders of sex development due to 46,XY gonadal dysgenesis. PLoS One 2011; 6:e17793; http://dx.doi.org/10.1371/journal.pone.0017793; PMID: 21408189
- Cox JJ, Willatt L, Homfray T, Woods CGA. A SOX9 duplication and familial 46,XX developmental testicular disorder. N Engl J Med 2011; 364:91 - 3; http://dx.doi.org/10.1056/NEJMc1010311; PMID: 21208124
- Jakubiczka S, Schröder C, Ullmann R, Volleth M, Ledig S, Gilberg E, Kroisel P, Wieacker P. Translocation and deletion around SOX9 in a patient with acampomelic campomelic dysplasia and sex reversal. Sex Dev 2010; 4:143 - 9; http://dx.doi.org/10.1159/000302403; PMID: 20453475
- Phillips NB, Racca J, Chen YS, Singh R, Jancso-Radek A, Radek JT, Wickramasinghe NP, Haas E, Weiss MA. Mammalian testis-determining factor SRY and the enigma of inherited human sex reversal: frustrated induced fit in a bent protein-DNA complex. J Biol Chem 2011; 286:36787 - 807; http://dx.doi.org/10.1074/jbc.M111.260091; PMID: 21849498
- Koopman P. Sry and Sox9: mammalian testis-determining genes. Cell Mol Life Sci 1999; 55:839 - 56; http://dx.doi.org/10.1007/PL00013200; PMID: 10412367
- Bagheri-Fam S, Sinclair AH, Koopman P, Harley VR. Conserved regulatory modules in the Sox9 testis-specific enhancer predict roles for SOX, TCF/LEF, Forkhead, DMRT, and GATA proteins in vertebrate sex determination. Int J Biochem Cell Biol 2010; 42:472 - 7; http://dx.doi.org/10.1016/j.biocel.2009.07.001; PMID: 19616114
- Georg I, Bagheri-Fam S, Knower KC, Wieacker P, Scherer G, Harley VR. Mutations of the SRY-responsive enhancer of SOX9 are uncommon in XY gonadal dysgenesis. Sex Dev 2010; 4:321 - 5; http://dx.doi.org/10.1159/000320142; PMID: 20838034
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 2011; 12:7 - 18; http://dx.doi.org/10.1038/nrg2905; PMID: 21116306
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012; 485:376 - 80; http://dx.doi.org/10.1038/nature11082; PMID: 22495300
- Eggers S, Sinclair A. Mammalian sex determination—insights from humans and mice. Chromosome Res 2012; 20:215 - 38; http://dx.doi.org/10.1007/s10577-012-9274-3; PMID: 22290220
- Hughes IA, Houk C, Ahmed SF, Lee PA, Lawson Wilkins Pediatric Endocrine Society/European Society for Paediatric Endocrinology Consensus Group. Consensus statement on management of intersex disorders. J Pediatr Urol 2006; 2:148 - 62; http://dx.doi.org/10.1016/j.jpurol.2006.03.004; PMID: 18947601
- Ono M, Harley VR. Disorders of sex development: new genes, new concepts. Nat Rev Endocrinol 2013; 9:79 - 91; http://dx.doi.org/10.1038/nrendo.2012.235; PMID: 23296159
- Ernst J, Kellis M. Interplay between chromatin state, regulator binding, and regulatory motifs in six human cell types. Genome Res 2013; 23:1142 - 54; http://dx.doi.org/10.1101/gr.144840.112; PMID: 23595227
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 2007; 1:299 - 312; http://dx.doi.org/10.1016/j.stem.2007.08.003; PMID: 18371364
- Pop R, Conz C, Lindenberg KS, Blesson S, Schmalenberger B, Briault S, Pfeifer D, Scherer G. Screening of the 1 Mb SOX9 5′ control region by array CGH identifies a large deletion in a case of campomelic dysplasia with XY sex reversal. J Med Genet 2004; 41:e47; http://dx.doi.org/10.1136/jmg.2003.013185; PMID: 15060123
- Wijchers PJ, Yandim C, Panousopoulou E, Ahmad M, Harker N, Saveliev A, Burgoyne PS, Festenstein R. Sexual dimorphism in mammalian autosomal gene regulation is determined not only by Sry but by sex chromosome complement as well. Dev Cell 2010; 19:477 - 84; http://dx.doi.org/10.1016/j.devcel.2010.08.005; PMID: 20833369
- Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, Kress J, Treier AC, Klugmann C, Klasen C, Holter NI, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 2009; 139:1130 - 42; http://dx.doi.org/10.1016/j.cell.2009.11.021; PMID: 20005806
- Mosimann C, Hausmann G, Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol 2009; 10:276 - 86; http://dx.doi.org/10.1038/nrm2654; PMID: 19305417
- Bhandari RK, Haque MM, Skinner MK. Global genome analysis of the downstream binding targets of testis determining factor SRY and SOX9. PLoS One 2012; 7:e43380; http://dx.doi.org/10.1371/journal.pone.0043380; PMID: 22984422
