Abstract
Recently, Dclk1 expression was identified to be an intestinal cancer stem cell specific biomarker in mouse models, implicating a potential role for targeting the DCLK1-postive cancer cells as a treatment for colorectal cancer. Using quantitative methylation specific PCR (qMSP) we here demonstrated that the DCLK1 promoter is hypermethylated in the vast majority of colorectal cancers (134/164; 82%), with no methylation in the normal mucosa samples (0/106). We further showed by Affymetrix exon arrays that DCLK1 is significantly downregulated in human colorectal cancer (n = 125) compared with normal colonic mucosa (n = 15), which was further confirmed by real-time RT-PCR of a subgroup of the samples. Additionally, a significant negative correlation was observed between methylation and DCLK1 expression in 74 cancer cell lines derived from 15 different tissues, and gene expression increased significantly after epigenetic drug treatment of initially methylated cancer cell lines. These findings underscore the potential of DCLK1 as a colorectal cancer biomarker for early detection, but may also have clinical implications regarding the previously proposed therapy toward DCLK1-positive cancer cells. This therapy would at best affect the cancer stem cell population, but will, based on the present results, not be efficient to treat the bulk of the tumor.
Keywords: :
Introduction
Different roles have been suggested for doublecortin-like kinase 1 (DCLK1), including as an intestinal and adenoma stem-cell marker,Citation1,Citation2 as well as a marker for differentiated tuft cells.Citation3 Recently, it was elegantly demonstrated in mouse models that Dclk1 could be a marker for intestinal cancer stem cells,Citation4 and it was further suggested that targeting the DCLK1-positive tumor cells could be a promising treatment for colorectal cancer.Citation4,Citation5
Four different reference transcript sequences have been identified for DCLK1, including NM_004734.4, which is the only variant with a CpG island at its core promoter. CpG island hypermethylation is a frequent event in cancer and is associated with transcriptional repression and subsequent reduction or loss of gene function.Citation6 In this study, the methylation status of the DCLK1 promoter was investigated in several cancer cell lines, as well as in a large series of colorectal cancer and normal mucosa tissue samples. We further analyzed the mRNA expression levels, and evaluated whether there was a correlation between promoter methylation status of DCLK1 and gene expression.
Results and Discussion
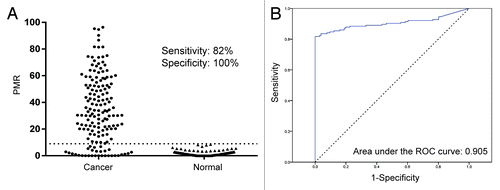
Using quantitative methylation specific PCR (qMSP), hypermethylation of the DCLK1 core promoter was observed in 134 out of 164 analyzed colorectal cancers and in none of 106 normal colorectal mucosa samples, resulting in a sensitivity and specificity of 82% and 100%, respectively (). Receiver operating characteristics (ROC) curve analysis further supported the ability of hypermethylated DCLK1 to separate colorectal cancers from normal mucosa samples with an area under the ROC curve (AUC) of 0.905 (P = 3.09E-29; 95% CI [0.865, 0.944]; ). Interestingly, promoter hypermethylation of DCLK1 is not restricted to colorectal cancer. We recently demonstrated that it was frequent also in other gastrointestinal malignancies, including cholangiocarcinomasCitation7and pancreatic and gastric cancer (data not shown).
Figure 1. Promoter DNA methylation of DCLK1 in colorectal cancer and normal colonic mucosa. (A) PMR values for colorectal cancer and normal mucosa samples. The scoring threshold is marked by a dotted line (PMR = 9), and outliers are excluded for better visualization (n = 6). (B) ROC curve for DCLK1 in colorectal cancer vs. normal mucosa samples.
From gene expression analyses using Affymetrix exon microarrays, we observed that DCLK1 was consistently downregulated in 125 analyzed colorectal cancer samples compared with normal mucosa (n = 15; independent samples t test, fold change 0.5, P = 0.001). This was further confirmed by real-time reverse transcriptase PCR (RT-PCR) of a subgroup of the samples, where a significant reduction in DCLK1 expression was observed among colorectal cancers (n = 50) compared with normal mucosa (n = 20; ). The reduced expression was seen for all three assays analyzed: two assays targeting the CpG island containing transcript variant NM_004734.4 and one assay targeting three transcript variants, including the NM_004734.4 variant (Mann-Whitney U test, P < 0.0001). This downregulation was also seen in an independent and large sample series, curated by the In Silico Transcriptomics (IST) database,Citation8 where DCLK1 was found to be significantly more expressed in the normal samples (n = 64) compared with the cancer samples (n = 991; P = 1.00E-05).
Figure 2. Expression and promoter methylation status of DCLK1. (A) Relative expression of DCLK1 in colorectal cancer (C; n = 50) and normal mucosa (NM; n = 20) samples assessed by three real-time RT-PCR assays. The assay Hs00178027_m1 covers three transcript variants; one with and two without a CpG island in the core promoter, whereas the two assays Hs00973863_m1 and Hs00973865_m1 cover the transcript variant with a CpG island in the core promoter (NM_004734.4). (B) PMR values (red) for DCLK1 (NM_004734.4; assay ID: Hs00973863_m1) and expression values (green) for 74 cell lines derived from 15 different cancer types. The expression levels are displayed as ratios between the median quantity of DCLK1 and the average quantity of the two controls VDAC2 and PES1.
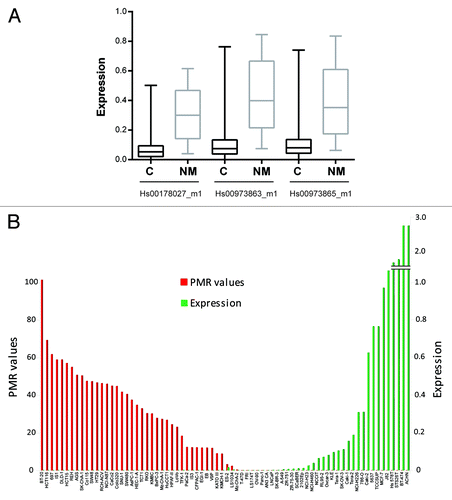
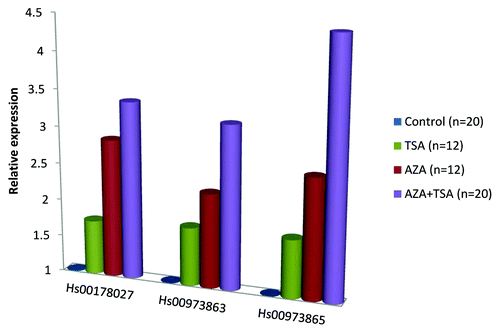
Promoter DNA methylation is commonly associated with gene downregulation, and the methylation at the DCLK1 promoter could potentially explain the reduced expression seen in colorectal cancers. In support of this, by analyzing 74 cell lines derived from 15 different cancer types with qMSP and real-time RT-PCR, we observed a significant and strong negative correlation between the presence of DCLK1 promoter hypermethylation and expression levels of the transcript with a CpG island in its promoter (; ). As expected, for the assay covering three reference transcripts (including two that do not have CpG islands in their core promoters), only a weak correlation was observed (). In further support for the association between methylation and reduced gene expression, in vitro treatment of 20 cancer cell lines with epigenetic drugs (5-aza-2’deoxycytidine, AZA and trichostatin A, TSA alone and in combination) resulted in a significantly increased DCKL1 expression (). The highest fold difference in expression was observed after a combination of both AZA and TSA (Wilcoxon Signed Ranks Test, P = 1.91E-06 for all three assays), but was also significant after treating the cell lines with AZA (P = 4.88E-04 for all assays) or TSA (P < 0.003 for all assays) alone. However, the negative correlation observed from the cancer cell lines could not be confirmed in a small sample set of colorectal cancers with methylated (n = 40) and unmethylated (n = 10) DCLK1 promoters (Fig. S1), indicating that promoter hypermethylation is not the primary cause for reduced DCLK1 expression. This is also the case for promoter methylation of VIM, one of the most promising non-invasive biomarkers for early detection of colorectal cancer.Citation9 Although VIM promoter hypermethylation is fairly specific for colorectal tumors, the gene is transcriptionally silent also in the unmethylated normal mucosa.Citation9 Since increased expression in cancer cells lines was observed also after treatment with TSA alone, histone modifications may be another cause for the reduced DCLK1 expression in colorectal cancers.
Table 1. Correlation between promoter methylation and expression of DCLK1 across 74 cancer cell lines
Figure 3. Relative expression of DCLK1 in cancer cell lines treated with epigenetic drugs. The expression levels are displayed as median fold difference across cell lines treated with TSA (0.5 mM for 12 h; n = 12), AZA (1 mM for 72 h; n = 12) or a combination of both drugs (n = 20) relative to the untreated ones.
In conclusion, we show that promoter hypermethylation of DCLK1 is a promising novel epigenetic biomarker for colorectal cancer. We further demonstrate that the progeny of the cancer stem cells, constituting the bulk of the tumor, have reduced expression of DCLK1 compared with normal mucosa. This is highly relevant when considering whether DCLK1-positive cells could be a promising therapeutic target and represent an Achilles heel for colorectal cancer treatment.Citation4,Citation5 Since this therapy at best would affect the cancer stem cell population, additional treatment would be needed for the bulk of the tumor.
Materials and Methods
Cancer tissue samples and controls
A total of 164 primary colorectal cancer samples were analyzed by quantitative methylation specific PCR (qMSP), and were divided into a test set (n = 59; median age 71; range 33–92 y) and a validation set (n = 105; median age 71; range 29–93 y). The test and validation sets were obtained from seven hospitals in the southeast region of Norway in the period 1987–1989, and from patients who underwent surgical resection at the Oslo University Hospital (OUH)- Aker from 2005- 2007, respectively. Normal mucosa samples (50 samples from 47 individuals; test set; median age 55; range 22–86 y) were obtained from deceased colorectal cancer- free individuals (Institute of Forensic Medicine, University of Oslo), and from a population-based sigmoidoscopy screening study (Telemark, NorwayCitation10; n = 56; validation set; median age 67; range 63–72 y).
A total of 70 tissue samples were analyzed for expression by real-time RT-PCR, including 50 cancer samples and 20 normal mucosa samples taken from the resection margin of cancer patients. The samples were obtained from OUH, and all cancer samples overlapped with the samples included in the qMSP analysis. All colorectal samples were fresh frozen.
Cancer cell lines
The 74 cancer cell lines included in this study were derived from 15 different cancer types, comprising six bile duct, four urinary bladder, eight breast, 19 colon, two gall bladder, four gastric, four kidney, three leukemia, four lung, one MPNST, four ovarian, six pancreatic, one prostate, four testis, and four uterus cancer cell lines. Twenty of these cancer cell lines were, in parallel with standard culturing, subjected to in vitro treatment with the demethylating agent 5-aza-2’deoxycytidine (AZA; 1 mM for 72 h; Sigma-Aldrich), the histone deacetylase inhibitor trichostatin A (TSA; 0.5 mM for 12 h; Sigma-Aldrich) and a combination of both drugs (1 mM AZA for 72 h and 0.5 mM TSA added the last 12 h). All commercially available cell lines were authenticated using the AmpFLSTR Identifiler PCR Amplification Kit (Life Technologies). Results for non-commercial cell lines will be given upon request.
DNA extraction and bisulfite treatment
DNA from cancer cell lines and tissue samples were isolated using either a standard phenol/chloroform extraction protocol or magnetic beads. The EpiTect Bisulfite Kit (Qiagen) was used for bisulfite treatment following the manufacturer’s protocol. For each sample 1.3 µg DNA was used as input and the bisulfite converted DNA was purified using the QIAcube (Qiagen) automated pipetting system.
Quantitative methylation-specific polymerase chain reaction (qMSP)
qMSP assays for DCLK1 and ALU (control) were available from a previous study.Citation7 The DCLK1 assay covered 11 CpG sites, and was designed to amplify a region located -96 to -10 relative to the transcription start site (RefSeq NM_004734.4, hg19 assembly). The primers were purchased from BioNordica (BioNordica, Medprobe), whereas the probes were purchased from Life Technologies.
The qMSP reactions included 1xTaqMan Universal PCR Mastermix No AmpErase UNG (Life Technologies), 0.9 µM of each primer, 0.2 µM probe, and approximately 32.5ng bisulfite treated template DNA in a final volume of 20µl. Bisulfite treated DNA, isolated from whole blood (leucocytes) obtained from healthy individuals, and water were included as methylation negative and template negative controls, respectively. Bisulfite-converted completely methylated DNA (IVD Chemicon; Millipore) was used as a methylation positive control, as well as for generating a standard curve from 1:5 serial dilutions (32.5ng - 0.052 ng). The EpMotion 5075 pipetting robot (Eppendorf) was used to automatically distribute both template and master mix to the 384-well plates.
Amplifications were performed in triplicates in a 7900HT Real-Time PCR System (Life Technologies), and thermal cycling was initiated with a denaturation step at 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s and 60 °C for 60 s.
Samples amplified after cycle 35 were censored in accordance with the recommendations from Life Technologies, and the median quantity value of the triplicates was used for data analysis. To normalize for DNA input the ALU-C4 element was used as a control.Citation11 The qMSP results were calculated as percent of methylated reference (PMR) by dividing the normalized quantity of the samples by the normalized quantity of the positive control (IVD) and multiply by 100, giving values ranging from 0–100. In the present study, six of the analyzed samples had a PMR value above 100, which may be explained by several minor factors, including a positive control that may not be 100% methylated at all CpG sites covered by the qMSP assay.
To ensure high specificity, the threshold for scoring the samples as methylated was set according to the highest PMR value across the test series of normal mucosa samples. Accordingly, the threshold was set to 9 and samples with PMR values equal to or higher than this threshold were considered to be methylated.
Affymetrix exon array data
The expression of DCLK1 in colorectal cancers and normal mucosa was calculated from previously published Affymetrix exon array data (refs. Citation12‒Citation14 accession numbers GSE24550 and GSE29638; GEO).
cDNA synthesis and real- time quantitative gene expression analysis
cDNA from the colorectal cancer samples was synthesized from total RNA using the High Capacity cDNA Reverse Transcription Kit (Life Technologies) according to the manufacturer’s protocol. The reverse transcription reactions were performed in an MJ Mini Personal Thermal Cycler (Bio-Rad) using 2 µg RNA as input.
Commercially available TaqMan gene expression assays for DCLK1 (Hs00178027_m1; Hs00973863_m1; Hs00973865_m1) and the two controls VDAC2 (Hs00748551_s1) and PES1 (Hs00362795_g1) were purchased from Life Technologies.
The gene expression reactions had a final volume of 20 µl, and included 1x TaqMan Universal Mastermix with UNG (Life Technologies), 1× TaqMan Gene Expression Assay (Life Technologies), and 10 ng of cDNA. RNase free water (Sigma-Aldrich) was included as negative control, whereas the human universal reference RNA (containing a mixture of total RNA from ten different cell lines; Agilent) was used as both positive control and for generating a standard curve (50 ng - 0.05 ng). The samples were run in triplicates in 384 plates in a 7900HT Real- Time PCR System (Life Technologies) initiated with 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Samples amplified after cycle 35 were censored. For the tissue samples and the untreated cell lines, relative expression levels were calculated by the relative standard curve method, by dividing the median quantity of the sample by the average median quantity of the two controls VDAC2 and PES1. For the cell lines treated with epigenetic drugs, relative expression levels were calculated by the comparative CT method (∆∆CT), and the average median CT of the two controls was used as reference. All samples amplified after cycle 35 were automatically set to CT 35.
Ethics
The research biobanks for colorectal cancers have been registered according to national legislation (numbers 2781 and 236-2005-16141). The study is part of a project approved by the Regional Committee (REC) for Medical and Health Research Ethics (numbers 1.2005.1629 and S-09282c 2009/4958).
Statistics
The statistical analyses were performed using PASW 18.0 (SPSS) and GraphPad Prism 6 (GraphPad Software, Inc.). A receiver operating characteristics (ROC) curve analysis was used to evaluate the performance of DCLK1 as a biomarker for colorectal cancer. To measure the strength of correlation between the PMR values and the expression for the three assays in cancer cell lines, a Spearman correlation analysis was performed, whereas the Mann-Whitney U- and independent sample t tests were applied to compare the expression of DCLK1 in the normal and cancer tissue samples. Wilcoxon Signed Ranks Test (exact) was used to compare relative expression of DCLK1 in cancer cell lines treated with or without epigenetic drugs. All P values were derived from two-sided tests, and P ≤ 0.05 was considered statistically significant.
| Abbreviations: | ||
| AUC | = | area under the ROC curve |
| AZA | = | 5-aza-2’deoxycytidine |
| PMR | = | percent methylated reference |
| qMSP | = | quantitative methylation specific PCR |
| ROC | = | receiver operating characteristics |
| RT-PCR | = | reverse transcription PCR |
| TSA | = | trichostatin-A |
Additional material
Download Zip (106.2 KB)Disclosure of Potential Conflicts of Interest
A US provisional patent application has been filed describing DCLK1 methylation as a biomarker for detection of gastrointestinal cancers (61/451,198, INVEN-31899/US-1/PRO).
Acknowledgments
This work was supported by a grant from the Norwegian Cancer Society, PR-2008-0163.
References
- Giannakis M, Stappenbeck TS, Mills JC, Leip DG, Lovett M, Clifton SW, Ippolito JE, Glasscock JI, Arumugam M, Brent MR, et al. Molecular properties of adult mouse gastric and intestinal epithelial progenitors in their niches. J Biol Chem 2006; 281:11292 - 300; http://dx.doi.org/10.1074/jbc.M512118200; PMID: 16464855
- May R, Riehl TE, Hunt C, Sureban SM, Anant S, Houchen CW. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells 2008; 26:630 - 7; http://dx.doi.org/10.1634/stemcells.2007-0621; PMID: 18055444
- Gerbe F, Brulin B, Makrini L, Legraverend C, Jay P. DCAMKL-1 expression identifies Tuft cells rather than stem cells in the adult mouse intestinal epithelium. Gastroenterology 2009; 137:2179 - 80, author reply 2180-1; http://dx.doi.org/10.1053/j.gastro.2009.06.072; PMID: 19879217
- Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M, et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet 2013; 45:98 - 103; http://dx.doi.org/10.1038/ng.2481; PMID: 23202126
- Metcalfe C, de Sauvage FJ. A tumor-specific stem cell. Nat Genet 2013; 45:7 - 9; http://dx.doi.org/10.1038/ng.2502; PMID: 23268130
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011; 11:726 - 34; http://dx.doi.org/10.1038/nrc3130; PMID: 21941284
- Andresen K, Boberg KM, Vedeld HM, Honne H, Hektoen M, Wadsworth CA, Clausen OP, Karlsen TH, Foss A, Mathisen O, et al. Novel target genes and a valid biomarker panel identified for cholangiocarcinoma. Epigenetics 2012; 7:1249 - 57; http://dx.doi.org/10.4161/epi.22191; PMID: 22983262
- Kilpinen S, Autio R, Ojala K, Iljin K, Bucher E, Sara H, Pisto T, Saarela M, Skotheim RI, Björkman M, et al. Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol 2008; 9:R139; http://dx.doi.org/10.1186/gb-2008-9-9-r139; PMID: 18803840
- Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst 2005; 97:1124 - 32; http://dx.doi.org/10.1093/jnci/dji204; PMID: 16077070
- Thiis-Evensen E, Hoff GS, Sauar J, Langmark F, Majak BM, Vatn MH. Population-based surveillance by colonoscopy: effect on the incidence of colorectal cancer. Telemark Polyp Study I. Scand J Gastroenterol 1999; 34:414 - 20; http://dx.doi.org/10.1080/003655299750026443; PMID: 10365903
- Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005; 33:6823 - 36; http://dx.doi.org/10.1093/nar/gki987; PMID: 16326863
- Sveen A, Agesen TH, Nesbakken A, Rognum TO, Lothe RA, Skotheim RI. Transcriptome instability in colorectal cancer identified by exon microarray analyses: Associations with splicing factor expression levels and patient survival. Genome Med 2011; 3:32; http://dx.doi.org/10.1186/gm248; PMID: 21619627
- Agesen TH, Sveen A, Merok MA, Lind GE, Nesbakken A, Skotheim RI, Lothe RA. ColoGuideEx: a robust gene classifier specific for stage II colorectal cancer prognosis. Gut 2012; 61:1560 - 7; http://dx.doi.org/10.1136/gutjnl-2011-301179; PMID: 22213796
- Sveen A, Ågesen TH, Nesbakken A, Meling GI, Rognum TO, Liestøl K, Skotheim RI, Lothe RA. ColoGuidePro: a prognostic 7-gene expression signature for stage III colorectal cancer patients. Clin Cancer Res 2012; 18:6001 - 10; http://dx.doi.org/10.1158/1078-0432.CCR-11-3302; PMID: 22991413
