Abstract
Oxidative stress induces genome-wide remodeling of the chromatin structure. In this study, we identify Methyl-CpG Binding Protein 4 (MBD4), a multifunctional enzyme involved in DNA demethylation, base excision repair, and gene expression regulation, as an essential factor in response to oxidative stress. We provide evidence that MBD4 is upregulated at the protein level upon oxidative stress, and that MBD4 is essential for cell survival following oxidative stress. In these cells, MBD4 and DNMT1 are recruited at sites of oxidation-induced DNA damage, where we speculate they participate in DNA repair. MBD4 and DNMT1 also share genomic targets in unstressed cells. Using genome-wide analysis of MBD4 binding sites, we identified new targets potentially co-regulated by MBD4 and DNA methylation. We identified two new binding sites for MBD4 and DNMT1 at methylated CpG islands of CDKN1A/p21 and MSH4, where they synergistically mediate transcriptional repression. Our study provides evidence that the interaction between DNMT1 and MBD4 is involved in controlling gene expression and responding to oxidative stress.
Introduction
Oxidative deamination of bases is a constant threat to genome integrity. This spontaneous reaction transforms cytosine into uracil, and 5-methylcytosine (5mC) into thymine; these alterations lead to C:G - > T:A transitions after replication, unless they are properly corrected.Citation1 In mammals, only two glycosylase enzymes can repair the T:G mismatch resulting from 5mC deamination: Thymine-DNA Glycosylase (TDG) and Methyl-CpG Binding Protein 4 (MBD4).Citation2
The importance of MBD4 for DNA repair in vivo is demonstrated by the observation that Mbd4−/− mice exhibit an increase in C to T transitions at CpG sites.Citation3,Citation4 In addition to removing spontaneously occurring mismatches, the catalytic activity of MBD4 can potentially be employed in developmentally programmed DNA demethylation.Citation5-Citation9 Aside from its role as a glycosylase, MBD4 has two other described functions.Citation10
First, MBD4 is involved in cell death signaling: it interacts with FADD, a subunit of the death-inducing signaling complex, and the apoptotic response to DNA-damaging agents in the small intestine of Mbd4-deficient mice is severely impaired.Citation4,Citation11,Citation12 In Xenopus as well, MBD4 relays signals that trigger apoptosis.Citation13
Second, MBD4 can function as a transcriptional repressor. This function is well described for the MBD4 paralogs MBD1, MBD2, and MeCP2, all of which recognize methylated DNA using their MBD domain, and then inhibit downstream gene expression via a transcriptional repression domain, which itself recruits co-repressors.Citation10,Citation14 The role of MBD4 in transcriptional repression has not been fully explored, and only 2 target genes are known: MLH1 and p16(INK4a).Citation15 In addition, the mechanisms by which MBD4 represses the expression of its target genes are still poorly understood.Citation16
DNA methyltransferase 1 (DNMT1) is an enzyme, initially characterized for its role in maintaining DNA methylation patterns during replication. It is now clear that DNMT1 has other functions, and in particular that it is involved in transcriptional repression. In certain cases, the co-repressive function of DNMT1 is accompanied by DNA methylation: for instance, upon doxorubicin treatment, TP53 recruits DNMT1, which methylates the p53-repressed promoters of Survivin and Cdc25c.Citation17,Citation18 In contrast, in early Xenopus development, DNMT1 has a general repressive function that does not require its catalytic activity.Citation19 DNMT1 has been linked with MBD4 in the context of apoptotic signaling in Xenopus.Citation13 This raises the possibility that DNMT1 might also be involved in transcriptional repression by MBD4, but this has not yet been tested.
In this work we have investigated the transcriptional role of MBD4 and identified several new genes whose expression it controls. In addition, we show that DNMT1 acts together with MBD4 to repress these genes. Finally, we show that MBD4 is important for human cells to survive oxidative stress. This function correlates with MBD4 being upregulated and recruited to chromatin, along with DNMT1.
Results
MBD4 and DNMT1 bind the methylated CDKN1A/p21 promoter together
In Xenopus, MBD4 and DNMT1 interact,Citation13 and our GST pull-down experiments also support a direct interaction between human MBD4 and DNMT1 (Fig. S1). This observation prompted us to investigate whether the two proteins might share certain genomic targets and regulate gene expression cooperatively in mammals. Only a few promoters are known to be bound by MBD4 or by DNMT1; MBD4 binds the MLH1 promoter in 293T cells,Citation15 so we asked whether DNMT1 was also present at this promoter. Conversely, DNMT1 binds the CDKN1A/p21 promoter,Citation20 and we asked whether MBD4 also does. We performed chromatin immunoprecipitation (ChIP) experiments on the endogenous DNMT1 and MBD4 in various cell types, and obtained the following results. First, we observed that DNMT1 binds the CDKN1A/p21 promoter in 293T cells as expected, but we could not detect its presence at the MLH1 promoter (, gray bars). Second, using the same chromatin samples, we found that MBD4 is bound at the MLH1 and at the CDKN1A/p21 promoters (, black bars). Thus, both MBD4 and DNMT1 bind the CDKN1A/p21 promoter; we scanned the promoter by qPCR and observed that DNMT1 and MBD4 binding sites are centered onto the transcription start site (). Using re-ChIP, we observed a strong enrichment at the CDKN1A/p21 transcription start site, but not at the MLH1 promoter, clearly indicating that DNMT1 and MBD4 are bound together to the CDKN1A/p21 promoter in 293T cells ().
Figure 1. MBD4 binds the methylated CDKN1A/p21 and MLH1 promoters. (A) ChIP analysis of MBD4 and DNMT1 binding to MLH1 and CDKN1A/p21 promoters in 293T cells (n = 3). (B) ChIP analysis of MBD4 and DNMT1 binding at CDKN1A/p21 transcription start site (TSS) and surrounding regions (-2kb/+2kb) in 293T cells (n = 3). (C) Re-ChIP analysis of MBD4 and DNMT1 co-binding at CDKN1A/p21 promoter in 293T cells (n = 3). (D) DNA methylation analysis by MeDIP at TP73, VASH2 and CDKN1A promoters in HeLa and in 293T cells. (E) ChIP analysis of MBD4 binding at MLH1 and CDKN1A/p21 promoters in HeLa cells (n = 3). (F) Summarization of the results. Black circles represent methylated DNA, white circles unmethylated DNA.
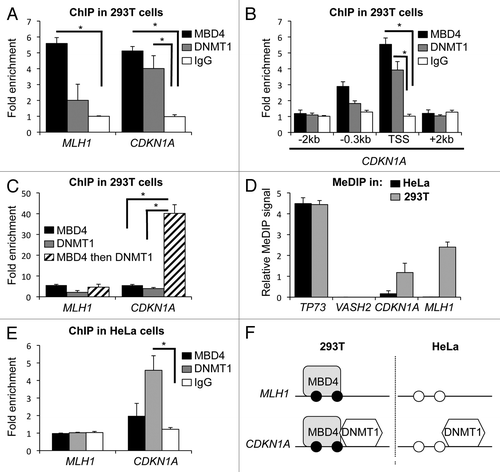
We next investigated whether MBD4 and DNMT1 binding at the CDKN1A/p21 promoter was correlated with its DNA methylation status. We compared MBD4 and DNMT1 binding at the CDKN1A/p21 promoter in 293T cells, in which the promoter is partially methylated, and in HeLa cells, in which the promoter is not methylated () (43). DNMT1 binds the CDKN1A/p21 promoter in both cell lines (), as previously reported,Citation20 whereas MBD4 binds only in 293T cells (). Our results therefore indicate that MBD4 binding at the CDKN1A/p21 promoter correlates with its methylation status, and that MBD4 is present together with DNMT1 ().
MBD4 and DNMT1 synergistically repress the methylated CDKN1A/p21 promoter
To examine whether CDKN1A/p21 is regulated cooperatively by MBD4 and DNMT1, we compared CDKN1A/p21 expression in cells transiently depleted of MBD4, DNMT1, or both (). As reported before,Citation21 the knockdown of DNMT1 tended to decrease the amount of MBD4 protein. This is a posttranscriptional effect, as the MBD4 mRNA was not decreased by DNMT1 siRNA, nor the DNMT1 mRNA affected by MBD4 siRNA (data not shown). This effect, which possibly reflects the functional interaction between the two proteins, was fairly small in 293T cells, especially at higher siRNA concentrations.
Figure 2.CDKN1A/p21 expression is synergistically regulated by MBD4 and DNMT1 in 293T cells. (A) Western blotting (WB) with the indicated antibodies in 293T and HeLa cells mock-depleted (Scr.) or transiently depleted of MBD4, DNMT1, or both. Two different doses of siRNA were transfected: 2 nM, 10 nM, as denoted by the black triangles. (B) Real-time RT-PCR analyses of CDKN1A/p21, TP53, and GADD45beta mRNA expression in 293T cells upon depletion of MBD4, DNMT1, or DNMT1+MBD4 (n = 3). (C) DNA methylation analysis by MeDIP at TP73, VASH2, and CDKN1A promoters in 293T cells upon depletion of MBD4, DNMT1, or DNMT1+MBD4 (n = 3). (D) Summarization of the results. Black circles represent methylated DNA, white circles unmethylated DNA.
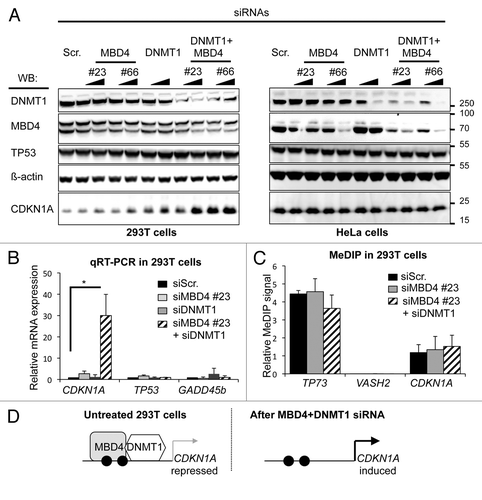
In 293T cells, CDKN1A/p21 protein expression was increased upon MBD4 or DNMT1 knockdown (). CDKN1A/p21 expression was increased more by the combined siRNA knockdown of MBD4 and DNMT1 than the sum of the individual knockdowns, and this effect was seen for two doses of siRNAs (, left panel). In HeLa cells, little or no significant effect on CDKN1A/p21 protein and mRNA expression was detected despite efficient MBD4, DNMT1, and MBD4+DNMT1 knockdown (; right panel and data not shown). Taken together, these results suggest that MBD4 and DNMT1 synergistically repress CDKN1A/p21 expression in 293T cells. To test whether the regulation of CDKN1A/p21 by MBD4 and DNMT1 is transcriptional, we then measured the abundance of the CDKN1A/p21 mRNA by real-time quantitative RT-PCR (). We observed that CDKN1A/p21 expression is increased 2-fold by MBD4 knockdown, 5-fold by DNMT1 knockdown, and 30-fold by the combined knockdown (). These results are in agreement with our observation at the CDKN1A/p21 protein level () and indicate that CDKN1A/p21 regulation by MBD4 and DNMT1 occurs at the mRNA level.
CDKN1A/p21 is a well-known DNA damage response geneCitation22,Citation23; this raises the possibility that its induction is merely a secondary consequence of DNA damage caused by the depletion of MBD4 and DNMT1. However, western blotting showed that the p53 protein level was not modified by the combined knockdown (), which argues against the presence of significant levels of DNA damage in the double knockdown cells. As an additional control, we also measured by quantitative RT-PCR the expression level of a different gene induced in response to DNA damage, GADD45beta; we found that its mRNA level was not measurably increased after the knockdowns (). Therefore the induction of CDKN1A/p21 observed in cells transfected with a combination of DNMT1 and MBD4 siRNAs does not just reflect a DNA damage response.
We next investigated the DNA methylation status of the CDKN1A/p21 promoter after transfection with DNMT1+MBD4 siRNAs, with MBD4 siRNAs and control siRNAs. By MeDIP, we detected a similar level of DNA methylation in each condition () indicating that re-expression of CDKN1A/p21 upon MBD4+DNMT1 depletion is not due to the loss of DNA methylation ().
Taken together, our observations indicate that DNMT1 and MBD4 cooperate for the transcriptional repression of the methylated CDKN1A/p21 promoter in 293T cells.
Genome-wide analysis of MBD4 binding sites identifies new target loci including MSH4
To identify additional genes co-regulated by MBD4 and DNMT1, we analyzed ChIP-Seq data, recently released by the ENCODE consortium.Citation24 We reasoned that MBD4 binding sites that are heavily methylated might represent additional genomic targets co-bound by DNMT1 and MBD4. The ENCODE data set contains 2 independent experiments in the hepatocarcinoma cell line HepG2, so we filtered the data set to keep only the binding sites common to the 2 experiments. We then intersected this list with the list of CpG islands in the human genome, and with the DNA methylation profile identified by ENCODE in HepG2 cells (encodeproject.org/ENCODE). We found that only 15 CpG islands bound by MBD4 are also heavily methylated (; Fig. S3A for the full list of targets). Twelve of these CpG islands overlap with a gene transcription start site: 10 for protein-coding genes (including MSH4, discussed below), and 2 for non-coding RNAs; the remaining 3 CpG islands are intragenic. Of note, the MLH1 and CDKN1A/p21 CpG islands do not fit our selection criteria in HepG2 cells ().
Figure 3. A genome-wide approach identifies the MSH4 promoter as a new MBD4 and DNMT1 target. (A) Venn diagram of MBD4 binding sites, DNA methylation sites and CpG islands in HepG2 cells. (B) DNA methylation analysis by MeDIP at TP73, VASH2 and MSH4 promoters in HeLa and 293T cells. (C) ChIP analysis of MBD4 and DNMT1 binding at MSH4 start site (TSS) and surrounding upstream (+0.8 kb) and downstream (–3 kb) regions in HeLa cells (n = 3). (D) ChIP analysis of MBD4 and DNMT1 binding at the MSH4 start site (TSS) and surrounding upstream (+0.8 kb) and downstream (–3 kb) regions in 293T cells (n = 3). (E) Real-time qRT-PCR analyses of MSH4 mRNA expression in 293T and HeLa cells upon depletion of MBD4, DNMT1, or DNMT1+MBD4 (n = 3). (F) DNA methylation analysis by MeDIP at TP73 and MSH4 promoters in 293T or HeLa cells upon depletion of MBD4, DNMT1, or DNMT1+MBD4. (G) Summarization of the results. Black circles represent methylated DNA, white circles unmethylated DNA.
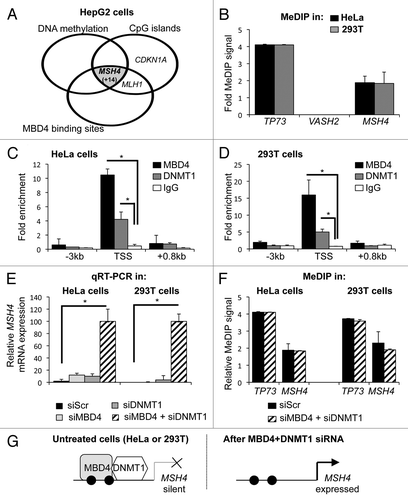
MSH4 encodes a MutS homolog involved in meiotic recombinationCitation25; in mammals this gene is expressed in the male and female germline, but not in somatic cells.Citation26 We conducted MeDIP analysis and ChIP analysis of the MSH4 CpG island in HeLa and 293T cells. We observed that the MSH4 CpG island is methylated in both cell lines (). By ChIP, we could detect the binding of MBD4 and DNMT1 at the MSH4 CpG island in both cell lines (). These observations indicate that MBD4 and DNMT1 both bind the methylated MSH4 CpG island in 293T and HeLa cells.
We next investigated MSH4 mRNA expression in cells treated with control, MBD4, DNMT1 or MBD4+DNMT1 siRNAs. We observed that MSH4 expression is barely detectable in control HeLa and 293T cells, which is consistent with their somatic origin (). Upon depletion of MBD4 or DNMT1, we observed a 10-fold induction of expression; the co-depletion of MBD4 and DNMT1 led to a 100-fold induction (). Interestingly, we observed that the MSH4 promoter was still methylated following co-depletion of MBD4 and DNMT1 ().
Taken together, our observations based on a genome-wide approach indicate that DNMT1 and MBD4 cooperate to repress the methylated MSH4 promoter in 293T cells and in HeLa cells.
MBD4 promotes cell survival upon oxidative stress
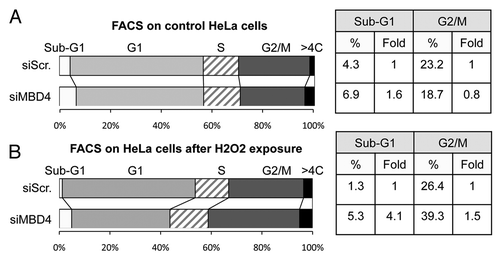
We then investigated the consequences of MBD4 depletion in control cells, and following oxidative stress. After MBD4 siRNA, the cells were treated (or not) with H2O2 for 30 min, and 24 h later analyzed by FACS.
The depletion of MBD4 did not significantly modify the cell cycle distribution of untreated HeLa cells (). At the dose we used (200 µM), the H202 exposure also failed to change the cell cycle parameters of HeLa cells transfected with a scrambled siRNA (). However, the same dose of H2O2 had a clear effect on HeLa cells depleted of MBD4, with significant increases of the sub-G1 and G2/M populations (). The increased sub-G1 fraction is indicative of increased apoptosis when MBD4-depleted HeLa cells are subjected to oxidative stress, and we confirmed this by two independent measures of viability (Fig. S3A and B). In addition, the negative effect of MBD4 depletion on cell survival becomes even more pronounced at higher doses of H202 (Fig. S3A). MBD4-depleted HeLa cells accumulate in G2/M after oxidative stress, whereas control HeLa cells maintain a normal cycle, and this effect was verified in a different cell line, 293T (not shown). We also observed that MBD4−/− immortalized mouse embryonic fibroblasts are more sensitive to H2O2 than their WT counterparts (Fig. S3C).
Figure 4. MBD4 depletion and H2O2 treatment induce G2/M accumulation and cell death in HeLa cells. (A) Cell cycle analysis of HeLa cells treated with siRNA against MBD4 (#23), or a control siRNA (siScr). After 24 h, 20 thousand cells per sample were analyzed by FACS. (B) Cell cycle analysis of HeLa cells treated with siRNA against MBD4 (#23), or a control siRNA (siScr), and then treated with 200µM H2O2. After 24 h, 20 thousand cells per sample were analyzed by FACS.
We conclude from these series of experiments that MBD4 deficiency sensitizes mammalian cells to DNA damage induced by oxidative stress.
The MBD4 protein is stabilized and accumulates upon oxidative stress
After making the observation that MBD4 is necessary for the proper response of human cells to oxidative stress, we sought to identify some of the molecular mechanisms involved. We therefore examined the regulation of MBD4 upon H2O2 exposure.
Western blotting analysis in the HeLa cell line shows that the MBD4 protein is upregulated 30 min after addition of H2O2 (). In contrast, we observed no increase in the expression level of DNMT1 nor in the expression level of MBD-family proteins MBD1, MBD2, and MeCP2 (). As expected after H2O2-induced stress, we observed an increase in DNA damage markers: phospho-H2AX and TP53 (). We also observed the accumulation of MBD4 protein upon H2O2 exposure in other transformed cells including 293T and U2OS, but also in the primary cells MRC5 (). Based on this qualitative observation in 4 different cell lines, we conclude that the MBD4 protein is more abundant after oxidative stress.
Figure 5. MBD4 protein is stabilized upon H2O2 treatment in human cells. (A–D) western blotting analysis in the indicated human cells treated with H2O2 (200 μM for 30 min), or not treated (NT). (E) Analysis of MBD4 and DNMT1 expression by qRT-PCR in HeLa, 293T, MRC5, and U2OS cells treated with H2O2 (200 μM for 30 min) or not. Expression is normalized to a set of 3 house-keeping genes (n = 3). (F) western blotting analysis of MBD4 stability in U2OS cells treated with H2O2 (200 μM for 30 min), or not treated (NT). Right panel: quantification of the MBD4 signal relative to GAPDH.
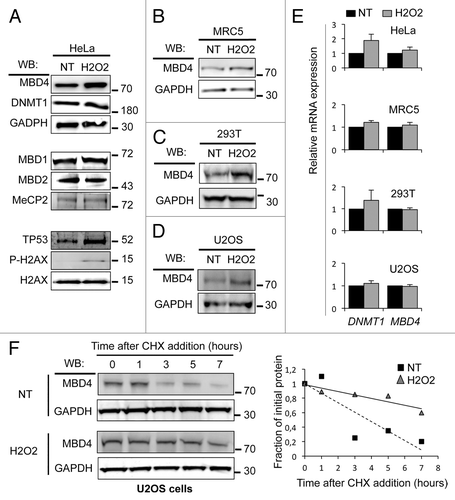
We quantified MBD4 mRNA level in these cells and found no significant increase in H2O2-treated compared with untreated cells (). Thus, our data indicate that MBD4 protein expression is upregulated by addition of H2O2 in the culture medium. This regulation occurs mostly posttranscriptionally, and among MBD-family proteins this effect is specific to MBD4.
To rule out an impact of H2O2 on translation or mRNA stability, we monitored MBD4 protein stability. Cells were treated with cycloheximide, an inhibitor of protein synthesis, and the half-life of MBD4 was studied by western blotting (). In control U2OS cells, MBD4 stability was estimated at less than 3 h. We observed that upon H2O2 treatment the half-life is significantly elevated up to 8 h. Thus, the MBD4 protein amount is higher upon H2O2 treatment in part because of MBD4 protein stabilization.
MBD4 is recruited to sites of DNA damage caused by oxidative stress
We next investigated whether MBD4 was directly involved in the cellular response to H2O2. We first monitored the chromatin loading of MBD4 in H2O2-treated cells. We observed a strong increase in MBD4 protein level in the native chromatin fraction (i.e., DNase resistant fraction) from cells exposed to H2O2 compared with non-treated cells (). We also observed, as described previously,Citation27,Citation28 that DNMT1 and the deacetylase SirT1 become more tightly bound to chromatin after oxidative stress (). The replication protein ORC2, used as a control, was equally present in the chromatin fraction under the control or H2O2 condition.
Figure 6. MBD4 and DNMT1 bind the cMYC CpG island after oxidative damage. (A) Western blotting analysis of MBD4, DNMT1, SirT1, and ORC2 loading onto chromatin. HeLa cells were treated with H2O2 (200 μM for 30 min) or not, and the nuclease-resistant chromatin fraction was purified as previously described.Citation38 (B) ChIP analysis of MBD4 and DNMT1 binding at the indicated regions in control non-treated (gray bars) and H2O2-treated (black bars, 200 μM for 30 min) HeLa cells (n = 3). (C) Western blotting analysis of MBD4, DNMT1, cMYC, and GAPDH expression in HeLa cells treated with control (Scr), MBD4 and DNMT1 siRNAs prior to H2O2 exposure (200 μM for 30 min). (D) Analysis of cMYC, MBD4, and DNMT1 expression by RT-PCR in cells treated with control (Scr), MBD4 and DNMT1 siRNAs prior to H2O2 exposure (200 μM for 30 min) (n = 3). Expression is normalized to a set of 3 house-keeping genes, and set as 1 in the control condition.
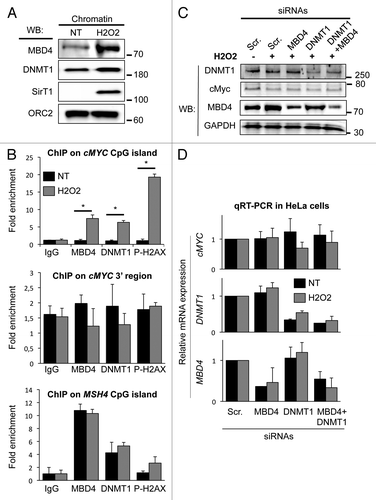
Oxidative stress induces DNA breaks mostly at CpG islands of highly transcribed genes, and DNMT1 is recruited to these damaged CpG islands, such as that of cMYC.Citation28 We therefore hypothezised that MBD4 might also be enriched at damaged CpG islands. By chromatin immunoprecipitation, we investigated MBD4, DNMT1, and phospho-H2AX enrichment at the cMYC CpG island. In untreated HeLa cells, we observed that MBD4 was not enriched at the cMYC CpG island (). Upon exposure to H2O2, we observed a 4- to 6-fold increase in MBD4 recruitment on the cMYC CpG island (). Consistent with previous work,Citation28,Citation29 we also detected a strong increase in DNMT1 binding and an accumulation of phospho-H2AX at these CpG islands (), as well as a reduction of cMyc protein () and mRNA (not shown). Importantly, the increased ChIP signal is not a mere consequence of the increased MBD4 protein abundance, as the MBD4 ChIP signal was not increased at two control regions we tested: a region 3′ to cMyc which does not suffer DNA damage, and the MSH4 promoter, on which MBD4 and DNMT1 binding is neither increased nor decreased by oxidative stress ().
Our results indicate that following oxidative stress MBD4 and DNMT1 are recruited to the cMYC CpG island, which is prone to damage induced by the oxidative treatment. The depletion of MBD4, DNMT1, or DNMT1+MBD4 did not modify cMyc expression following oxidative stress (). We therefore conclude that MBD4 and DNMT1 are jointly recruited to the cMYC CpG island upon oxidative stress, but are not required to affect its transcriptional repression.
Discussion
In this paper, we have explored the role of MBD4 as a transcriptional repressor, the mode by which it represses transcription, and its function during oxidative stress.
MBD4 and DNMT1 jointly repress methylated promoters
We observed that MBD4 and DNMT1 are co-recruited to certain methylated CpG islands and that they jointly repress the expression of the downstream gene. We have demonstrated this effect at the promoters of CDKN1A/p21 and of MSH4, two genes that represent very distinct situations. MSH4 is normally silenced in somatic tissues, and the examination of publically available data (ENCODE/Hudson α RRBS data), shows that its CpG island is methylated in all somatic non-transformed cells, as well as in tumoral cell lines — it is demethylated only in ovarian and testicular cell lines. In the case of MSH4, MBD4, and DNMT1 are thus involved in the programmed repression of a germline gene in somatic cells. In contrast, CDKN1A/p21 is not methylated in healthy cells, but gains methylation and is silenced in certain tumor cells. Therefore, the silencing function of MBD4 and DNMT1 can also be coopted to repress a tumor-suppressor gene in cancer. Data from ES cells shows that MBD4 binding is critically dependent on the density of methylated CpGs,Citation30 so we speculate that MBD4 and DNMT1 could repress transcription at other methylated CpG islands, such as CDKN1A/p21 and MSH4, but not at regions of lesser CpG density.
MBD4 and DNMT1 are also recruited to the cMYC CpG island after oxidative stress, (), but in that case they are not essential for transcriptional repression. The basis for this observation could be the following: after oxidative stress, a protein complex containing multiple repressive activities, including SIRT1 and EZH2, is recruited to the c-MYC CpG island.Citation28,Citation29 These proteins could act in a redundant manner, and we speculate that SIRT1 and EZH2 are sufficient to repress cMYC transcription even in the absence of MBD4 and DNMT1.
MBD4 and DNMT1 are involved in the oxidative stress response
Our experiments indicate that MBD4 plays an important role in the cellular response to oxidative stress.
First, the MBD4 protein is stabilized upon H2O2 exposure, this may be an adaptive response that increases the cellular capacity to remove oxidized bases (). The effect is posttranscriptional and accompanied by increased protein stability. Possible mechanisms include decreased ubiquitination, or increased deubiquitination of MBD4 following oxidative stress; it would be interesting to determine whether these same mechanisms are also used in developmental contexts in which high MBD4 activity is required for DNA demethylation.
We have found that MBD4 depletion has little effect on the cell cycle dynamics and the survival of unstressed cells, but that MBD4 is critical in the context of oxidative stress. In its absence, human cells suffer significantly more death and G2/M accumulation after a treatment with H2O2 (). What is the essential function that this protein serves after oxidative stress? We have observed that it is loaded, concurrently with DNMT1, on the cMYC CpG island that is prone to oxidative damage (). Potential roles for MBD4 at this CpG island (and possibly others) after damage include: removal of damaged bases; restoration of the original chromatin structure; prevention of hypermethylation of CpG islands by DNMT1; re-establishment of CpG methylation pattern in the genome by coordinating base repair and re-methylation. Of note, neither DNMT1 nor MBD4 is necessary for the transcriptional repression of c-Myc that occurs following DNA damage. It could be that their primary role is not to ensure transcriptional repression, or that redundant repressive activities exist.
In summary, our study identified two different contexts where DNMT1 and MBD4 act together. They contribute to gene silencing at certain hypermethylated promoters, such as CDKN1A/p21 and MSH4. They are also recruited at damaged DNA sites after H2O2 exposure and may play an important role at these sites to ensure cell survival.
Materials and Methods
Plasmids and antibodies
All GST-DNMT1, DsRed-DNMT1, and pcDNA 6xHIS-DNMT1 constructs have been described previously,Citation31 and protein production was a previously described.Citation32 The MBD4 isoform 2 clone was obtained from the American Type Culture Collection (ATCC). The GST-MBD4 constructs, the GFP-MBD4 construct and pVIC1-MBD4 constructs were generated by PCR-based cloning procedures using the EcoRI and SalI sites of pGEX-5X-1 (GE healthcare) and peGFP-C2 (Clontech), and the NdeI and SmaI sites of pVIC1 (New England Biolabs). All the constructs were verified by sequencing. Plasmids used in this study are described in Table S1. All PCR amplifications were performed using Phusion DNA polymerase (Finnzyme).
Antibodies used in this study are commercially available: MBD4 (Bethyl Laboratories #A301–634A); MBD4 (Sigma-Aldrich #M9817), DNMT1 (New England Biolabs #M0231L), Beta-Actin (Cell Signaling Technology #4970), HA (Cell Signaling Technology #2367), 6xHIS (Cell Signaling Technology #2366), CDKN1A/p21 (Cell Signaling Technology #2946), TP53 (DO-1, Santa Cruz), GAPDH (Abcam #ab9485), ORC2 (Santa Cruz #28742), cMYC (Santa Cruz #sc-40), phospho-H2AX (Upstate #05-636), H2AX (Bethyl Laboratories #A300–083A), MeCP2 (Abcam #ab3752), MBD1 (Abcam #ab2846) and MBD2 (Santa Cruz #sc-9397). Secondary antibodies coupled with horseradish peroxidase or fluorescent probes were purchased from Jackson ImmunoResearch.
New England Biolabs supplied all the other enzymes and their buffers, protein and DNA markers, plasmids, and competent cells. H2O2 and propidium iodide were purchased from Sigma-Aldrich.
Cell culture and transfection
All cell lines were obtained from the ATCC and maintained in their recommended media supplemented with 10% fetal bovine serum, 2 mM L-glutamine and 1% penicillin, 1% streptomycin (Invitrogen). We performed plasmid transfection into HeLa, 293T, U2OS and COS-7 using Transpass D2 (New England Biolabs) or Lipofectamine 2000 (Life Technologies).
MBD4 siRNAs (HSS113223, HSS189666) and control siRNA (scramble, scr) were purchased from Invitrogen and DNMT1 siRNA from Ambion (s4216). We transfected siRNAs using either Transpass R1 (New England Biolabs), Lipofectamine 2000 (Life Technologies), or Interferin (Polyplus) according to the manufacturer’s recommendations. We collected the cells 24 or 48 h after transfection for western blotting and qRT-PCR analysis.
Western blotting
Cell extracts were prepared as previously describedCitation33 and resolved on NuPage pre-cast SDS-PAGE gels (Invitrogen) and transferred to Immobilon-P membranes (Millipore). The membranes were blocked with 5% fat-free milk in PBS, then incubated overnight at 4 °C with the appropriate primary antibodies. The membranes were incubated with the cognate secondary antibody coupled to HRP, and revealed using the West Dura kit (Pierce, Rockford, USA), in the ChemiSmart 5000 imager (Vilber Lourmat).
Chromatin-immunoprecipitation (ChIP)
ChIP-qPCR were performed as previously described.Citation34 We crosslinked cells at 107 cells/mL for 10 min using 1% formaldehyde at room temperature and stopped the reaction using 125 mM glycine. Nuclear extracts were prepared using the standard hypotonic method. We resuspended the nuclei at 4 × 107 cells/mL in nuclear lysis buffer (50 mM Hepes pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na Deoxycholate, 0.1% SDS) supplemented to 1% SDS and sonicated them using a Bioruptor sonicator (Diagenode).
Chromatin was diluted to 0.1% SDS nuclear lysis buffer and precleared on protein A agarose beads (Diagenode) blocked with BSA. We performed immunoprecipitation (IP) overnight using chromatin from 5 × 107 cells per IP and 3 µg of antibodies (MBD4, DNMT1, phospho-H2AX) or native IgG (Cell Signaling Technology). IPs were collected on protein A-agarose beads and the beads washed successively with nuclear lysis buffer, nuclear lysis buffer supplemented to 0.5 M NaCl, wash buffer (20 mM TRIS-HCl pH 8, 150 mM LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% Na Deoxycholate) and TE buffer. Immunoprecipitated DNA-protein complexes were eluted from protein A beads by boiling in TE supplemented with 1% SDS. We de-crosslinked DNA using pronase (2 h at 42 °C) and heat treatment (6 h at 65 °C), and purified the DNA using Qiagen spin columns. Sequential-ChIP was performed as previously described using an inactivation of the first antibody with dithiothreitol.Citation35
We used the SYBR green mix from Applied Biosystems for quantification. Relative enrichment values are determined using the standard deltaCt method. The average value of 3 random genomic sites is used as the control Ct value. Primer sequences are as follow: GAPDH 5′-CCGGGAGAAG CTGAGTCATG-3′ and 5′-TTTGCGGTGG AAATGTCCTT-3′; Actin 5′-TCCCTGGAGA AGAGCTACG-3′ and 5′-GTAGTTTCGT GGATGCCACA-3′; KLK3 5′-CACACCCGCTC TACGATATG A G-3′ and 5′-GAGCTCGGCA GGCTCTGA-3′; CDKN1A/p21 -2 kb 5′-CACTCCCACT GCTTCATTTA A CTA-3′ and 5′-CTCAAAGTCC AGAACTCAGG TGAT-3′; CDKN1A/p21 +2kb 5′-AGTGGAGTAA GTTCGTCTAG G A-3′ and 5′-TGGCGTAAAG GACCTGAACC-3′; CDKN1A/p21 -0.3kb GAGGAAGAAG ACTGGGCATG TC-3′ and 5′-GCTTGGAGCA GCTACAATTA CT-3′; CDKN1A/p21 (TSS) 5′-GGCTCCACAA GGAACTGACT TC-3′ and 5′-TATATCAGGG CCGCGCTG-3′; cMYC 5′-AAGTTTCCAG CCACCTCCTT-3′ and 5′-GTTTGGCCGT TTTAGGGTTT-3′; MLH1 5′-CGGCCAATAG GAGCAGAGAT GCCG-3′ and 5′-GGCCAATGCT TGGTTGCTAT-3′; MSH4 CpG island 5′-CAAATCGGGT GGTCATTGAT-3′ and 5′-GGTCTATGAC CCTGCTTCCA-3′; MSH4 -3kb 5′-CCCCTGGACA GTGCTTGTAG A-3′ and 5′-GCATGCATCT GCTTTGTTTC-3′; MSH4 +0.8kb 5′-TGCAAGACCT TGACTGTGGA-3′ and 5′- CAGCCAAAGA TCAGCTCACA-3′.
Methyl-DNA-immunoprecipitation (MeDIP)
Methylated DNA was immunoprecipitated using the auto-MeDIP kit on the IP-Star robot according to manufacturer recommandations (Diagenode). In particular, each reaction contains a spike DNA to ensure reproducibility of the IP between biological replicates. Values are normalized to a CpG-free region of the genome. Amplification of the hypermethylated region 1.5kb upstream of the TP73 promoter and amplification of the VASH2 non methylated CpG island are used as control. Primer sequences are as follow: TP73 5′-ACTGACGCGA CTTTCCAAGA-3′ and 5′-GCTGCTTATG GTCTGATGCT T-3′; VASH2 5′-GTCCCGAGGT AGGATCTTGG-3′ and 5′-GAGTTCCAGC GCCTATCACC-3′; CpG-free region 5′-CTGAATCAGC AGACAGAATG G A-3′ and 5′-GGTAGGCAAC ACAGGTTTGG-3′; MLH1 5′-CGGCCAATAG GAGCAGAGAT GCCG-3′ and 5′-GGCCAATGCT TGGTTGCTAT-3′; CDKN1A/p21 5′-GGCTCCACAA GGAACTGACT TC-3′ and 5′-TATATCAGGG CCGCGCTG-3′; MSH4 5′-CAAATCGGGT GGTCATTGAT-3′ and 5′-GGTCTATGAC CCTGCTTCCA-3′.
RNA extraction, cDNA synthesis and quantitative real-time PCR
We extracted RNAs using Trizol (Invitrogen). Reverse Transcription was performed with Superscript III (Invitrogen) and oligo-dT according to the manufacturer’s instructions. Gene expression values are normalized to the average value of three standards: TFRC, MAPK14 and TBP. qRT-PCR primer sequences is as follow: MBD4 (all known isoforms) 5′-CACATCTCTC CAGTCTGC-3′ and 5′-CGACGTAAAG CCTTTAAGAA-3′; DNMT1 5′-CATGAGCACC GTTCTCCAAG G-3′ and 5′-GAATCTCTTG CACGAATTTC TGC-3′; cMYC 5′-GCAGGATAGT CCTTCCGAGT G-3′ and 5′-CCACAGCAAA CCTCCTCACA G-3′; MSH4 5′-CTGGACACCA CAAGTGGGAT A-3′ and 5′-TGGCAAGTCC TCTCCCTTCT A-3′; TFRC 5′-GGCCTTTGTG TTATTGTCAG C AT-3′ and 5′-ACCATTGTCA TATACCCGGT TCA-3′; TBP 5′-CTGCGGTACA ATCCCAGAAC T-3′ and 5′-CCACTCACAG ACTCTCACAA C-3′; MAPK14 5′-TCATAGGTCA GGCTTTTCCA C T-3′ and 5′-TGCCGAAGAT GAACTTTGCG A-3′; TP53 5′-AATCATCCAT TGCTTGGGAC G-3′ and 5′-CCGCAGTCAG ATCCTAGCG-3′; GADD45beta 5′-ATGAGCGTGA AGTGGATTTG C-3′ and 5′-ACAGTGGGGG TGTACGAGTC-3′; CDKN1A/p21 5′-GCGTTTGGAG TGGTAGAAAT C T-3′ and 5′-CCTGTCACTG TCTTGTACCC T-3′.
Genome-wide data analysis
MBD4 ChIP-sequencing data sets in the hepatocellular carcinoma HepG2 cell line were downloaded from the ENCODE portal (GSE32465; encodeproject.org/ENCODE). Reads were aligned using Bowtie (version 0.12.2)Citation36 to build version hg19 of the human genome. We used the MACS version 1.4.1 (model-based analysis of ChIP-sequencing)Citation37 peak finding algorithm to identify regions of MBD4 enrichment over background. A p-value threshold of enrichment of 10−9 was used. Biological replicates were treated independently and intersected to generate the list of MBD4 enriched regions common to both replicates. This list was used for the rest of the analysis. We then assigned MBD4 enriched regions to CpG islands of genes (GRCh17, hg19). A strict overlap of MBD4 enriched regions and CpG islands was used to create MBD4 CpG-island-bound regions. The status of methylation of these CpG islands was then assessed using the HepG2 methyl-CpG-sequencing data sets downloaded from the ENCODE portal (GSE41304). We retrieved from the data sets the methylation status of each CpG dinucleotide present in MBD4 CpG-island-bound regions. A given CpG dinucleotide was considered methylated when over 50% of the molecules sequenced were still CpG (i.e., protected from bisulfite conversion). We then compiled these individual CpG score to access CpG island methylation. CpG islands with over 90% of their individual CpGs methylated were considered methylated.
Additional material
Download Zip (435.7 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
ChIP-Sequencing and DNA methylation profile used in this study were produced as part of the ENCODE consortium and the data are past the nine-month moratorium (http://genome.ucsc.edu). We are grateful to Adrian Bird for the gift of MBD4 antibodies and mutant cells, and we thank Thierry Grange for his help in the initial stages of this work. Work in the lab of PAD is supported by “Centre National de la Recherche Scientifique” (CNRS), by “Association pour la Recherche contre le Cancer” (ARC), by “Ligue contre le Cancer,” by Institut National du Cancer (INCa), by “Groupement des Entreprises Françaises dans la Lutte contre le Cancer” (GEFLUC), and by ANR-11-LABX-0071 under program ANR-11-IDEX-0005-01. B.M. is the recipient of a Marie Curie “International Reintegration Grant” Fellowship.
Author Contributions
S.L., B.M., H.G.C., and P.O.E. performed experiments. S.L., S.P., R.J.R., and P.A.D. supervised the study and interpreted the data. S.L., B.M., and P.A.D. wrote the manuscript.
References
- Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature 1980; 287:560 - 1; http://dx.doi.org/10.1038/287560a0; PMID: 6999365
- Sjolund AB, Senejani AG, Sweasy JB. MBD4 and TDG: multifaceted DNA glycosylases with ever expanding biological roles. Mutat Res 2013; 743-744:12 - 25; http://dx.doi.org/10.1016/j.mrfmmm.2012.11.001; PMID: 23195996
- Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, Keightley PD, Bishop SM, Clarke AR, Bird A. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science 2002; 297:403 - 5; http://dx.doi.org/10.1126/science.1073354; PMID: 12130785
- Wong E, Yang K, Kuraguchi M, Werling U, Avdievich E, Fan K, Fazzari M, Jin B, Brown AM, Lipkin M, et al. Mbd4 inactivation increases Cright-arrowT transition mutations and promotes gastrointestinal tumor formation. Proc Natl Acad Sci U S A 2002; 99:14937 - 42; http://dx.doi.org/10.1073/pnas.232579299; PMID: 12417741
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 2008; 135:1201 - 12; http://dx.doi.org/10.1016/j.cell.2008.11.042; PMID: 19109892
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 2010; 11:607 - 20; http://dx.doi.org/10.1038/nrm2950; PMID: 20683471
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 2012; 40:4841 - 9; http://dx.doi.org/10.1093/nar/gks155; PMID: 22362737
- Hashimoto H, Zhang X, Cheng X. Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation. Nucleic Acids Res 2012; 40:8276 - 84; http://dx.doi.org/10.1093/nar/gks628; PMID: 22740654
- Moréra S, Grin I, Vigouroux A, Couvé S, Henriot V, Saparbaev M, Ishchenko AA. Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Res 2012; 40:9917 - 26; http://dx.doi.org/10.1093/nar/gks714; PMID: 22848106
- Defossez PA, Stancheva I. Biological functions of methyl-CpG-binding proteins. Prog Mol Biol Transl Sci 2011; 101:377 - 98; http://dx.doi.org/10.1016/B978-0-12-387685-0.00012-3; PMID: 21507359
- Screaton RA, Kiessling S, Sansom OJ, Millar CB, Maddison K, Bird A, Clarke AR, Frisch SM. Fas-associated death domain protein interacts with methyl-CpG binding domain protein 4: a potential link between genome surveillance and apoptosis. Proc Natl Acad Sci U S A 2003; 100:5211 - 6; http://dx.doi.org/10.1073/pnas.0431215100; PMID: 12702765
- Sansom OJ, Zabkiewicz J, Bishop SM, Guy J, Bird A, Clarke AR. MBD4 deficiency reduces the apoptotic response to DNA-damaging agents in the murine small intestine. Oncogene 2003; 22:7130 - 6; http://dx.doi.org/10.1038/sj.onc.1206850; PMID: 14562041
- Ruzov A, Shorning B, Mortusewicz O, Dunican DS, Leonhardt H, Meehan RR. MBD4 and MLH1 are required for apoptotic induction in xDNMT1-depleted embryos. Development 2009; 136:2277 - 86; http://dx.doi.org/10.1242/dev.032227; PMID: 19502488
- Joulie M, Miotto B, Defossez PA. Mammalian methyl-binding proteins: what might they do?. Bioessays 2010; 32:1025 - 32; http://dx.doi.org/10.1002/bies.201000057; PMID: 20886526
- Kondo E, Gu Z, Horii A, Fukushige S. The thymine DNA glycosylase MBD4 represses transcription and is associated with methylated p16(INK4a) and hMLH1 genes. Mol Cell Biol 2005; 25:4388 - 96; http://dx.doi.org/10.1128/MCB.25.11.4388-4396.2005; PMID: 15899845
- Fukushige S, Kondo E, Gu Z, Suzuki H, Horii A. RET finger protein enhances MBD2- and MBD4-dependent transcriptional repression. Biochem Biophys Res Commun 2006; 351:85 - 92; http://dx.doi.org/10.1016/j.bbrc.2006.10.005; PMID: 17049487
- Estève PO, Chin HG, Pradhan S. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc Natl Acad Sci U S A 2005; 102:1000 - 5; http://dx.doi.org/10.1073/pnas.0407729102; PMID: 15657147
- Le Gac G, Estève PO, Ferec C, Pradhan S. DNA damage-induced down-regulation of human Cdc25C and Cdc2 is mediated by cooperation between p53 and maintenance DNA (cytosine-5) methyltransferase 1. J Biol Chem 2006; 281:24161 - 70; http://dx.doi.org/10.1074/jbc.M603724200; PMID: 16807237
- Dunican DS, Ruzov A, Hackett JA, Meehan RR. xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function. Development 2008; 135:1295 - 302; http://dx.doi.org/10.1242/dev.016402; PMID: 18305009
- Kim JK, Estève PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res 2009; 37:493 - 505; http://dx.doi.org/10.1093/nar/gkn961; PMID: 19056828
- Loughery JE, Dunne PD, O’Neill KM, Meehan RR, McDaid JR, Walsh CP. DNMT1 deficiency triggers mismatch repair defects in human cells through depletion of repair protein levels in a process involving the DNA damage response. Hum Mol Genet 2011; 20:3241 - 55; http://dx.doi.org/10.1093/hmg/ddr236; PMID: 21636528
- Mirzayans R, Andrais B, Scott A, Murray D. New insights into p53 signaling and cancer cell response to DNA damage: implications for cancer therapy. J Biomed Biotechnol 2012; 2012:170325; http://dx.doi.org/10.1155/2012/170325; PMID: 22911014
- Vurusaner B, Poli G, Basaga H. Tumor suppressor genes and ROS: complex networks of interactions. Free Radic Biol Med 2012; 52:7 - 18; http://dx.doi.org/10.1016/j.freeradbiomed.2011.09.035; PMID: 22019631
- ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 2011; 9:e1001046; http://dx.doi.org/10.1371/journal.pbio.1001046; PMID: 21526222
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell 2004; 15:437 - 51; http://dx.doi.org/10.1016/j.molcel.2004.06.040; PMID: 15304223
- Paquis-Flucklinger V, Santucci-Darmanin S, Paul R, Saunières A, Turc-Carel C, Desnuelle C. Cloning and expression analysis of a meiosis-specific MutS homolog: the human MSH4 gene. Genomics 1997; 44:188 - 94; http://dx.doi.org/10.1006/geno.1997.4857; PMID: 9299235
- Her C, Wu X, Griswold MD, Zhou F. Human MutS homologue MSH4 physically interacts with von Hippel-Lindau tumor suppressor-binding protein 1. Cancer Res 2003; 63:865 - 72; PMID: 12591739
- O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 2011; 20:606 - 19; http://dx.doi.org/10.1016/j.ccr.2011.09.012; PMID: 22094255
- Bosch-Presegué L, Raurell-Vila H, Marazuela-Duque A, Kane-Goldsmith N, Valle A, Oliver J, Serrano L, Vaquero A. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol Cell 2011; 42:210 - 23; http://dx.doi.org/10.1016/j.molcel.2011.02.034; PMID: 21504832
- Baubec T, Ivánek R, Lienert F, Schübeler D. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell 2013; 153:480 - 92; http://dx.doi.org/10.1016/j.cell.2013.03.011; PMID: 23582333
- Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J 2002; 21:4183 - 95; http://dx.doi.org/10.1093/emboj/cdf401; PMID: 12145218
- Sasai N, Nakao M, Defossez PA. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res 2010; 38:5015 - 22; http://dx.doi.org/10.1093/nar/gkq280; PMID: 20403812
- Yamada D, Pérez-Torrado R, Filion G, Caly M, Jammart B, Devignot V, Sasai N, Ravassard P, Mallet J, Sastre-Garau X, et al. The human protein kinase HIPK2 phosphorylates and downregulates the methyl-binding transcription factor ZBTB4. Oncogene 2009; 28:2535 - 44; http://dx.doi.org/10.1038/onc.2009.109; PMID: 19448668
- Miotto B, Struhl K. Differential gene regulation by selective association of transcriptional coactivators and bZIP DNA-binding domains. Mol Cell Biol 2006; 26:5969 - 82; http://dx.doi.org/10.1128/MCB.00696-06; PMID: 16880509
- Miotto B, Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell 2010; 37:57 - 66; http://dx.doi.org/10.1016/j.molcel.2009.12.012; PMID: 20129055
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10:R25; http://dx.doi.org/10.1186/gb-2009-10-3-r25; PMID: 19261174
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9:R137; http://dx.doi.org/10.1186/gb-2008-9-9-r137; PMID: 18798982
- Miotto B, Struhl K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev 2008; 22:2633 - 8; http://dx.doi.org/10.1101/gad.1674108; PMID: 18832067
