Abstract
In 2007, the Ubiquitously Transcribed Tetratricopeptide Repeat on chromosome X (UTX) was identified as a histone demethylase that specifically targets di- and tri-methyl groups on lysine 27 of histone H3 (H3K27me2/3). Since then, UTX has been proven essential during normal development, as it is critically required for correct reprogramming, embryonic development and tissue-specific differentiation. UTX is a member of the MLL2 H3K4 methyltransferase complex and its catalytic activity has been linked to regulation of HOX and RB transcriptional networks. In addition, an H3K27me2/3 demethylase independent function for UTX was uncovered in promoting general chromatin remodeling in concert with the BRG1-containing SWI/SNF remodeling complex. Constitutional inactivation of UTX causes a specific hereditary disorder called the Kabuki syndrome, whereas somatic loss of UTX has been reported in a variety of human cancers. Here, we compile the breakthrough discoveries made from the first disclosure of UTX as a histone demethylase till the identification of disease-related UTX mutations and specific UTX inhibitors.
Introduction
A decade ago, the identification of the first histone demethylase Lysine (K)-Specific Demethylase 1 (LSD1) served as a landmark discovery that triggered research on dynamic regulation of histone methylation.Citation1 In the following years, numerous additional histone demethylases, that execute the removal of methyl groups on specific lysine residues of the histone tails and of non-histone substrates, were characterized in more detail (reviewed in ref. Citation2). In 2007, several groups identified Ubiquitously Transcribed Tetratricopeptide Repeat on chromosome X (UTX) and Jumonji D3 (JMJD3) as novel histone demethylases that catalyze the removal of di- and trimethyl groups on histone H3 lysine 27, thereby promoting target gene activation.Citation3-Citation6 Notably, Ubiquitously Transcribed Tetratricopeptide Repeat on chromosome Y (UTY) is a closely related homolog of UTX on the Y-chromosome but, up until now, no enzymatic H3K27me2/3 demethylase activity has been reported for UTY.Citation3,Citation7
The X-linked H3K27me2/3 eraser UTX is a member of the MLL2 histone H3K4 methyltransferase complexCitation8,Citation9 and contributes to animal body patterning by regulation of homeobox (HOX) genes.Citation4,Citation5 In contrast, a histone demethylation-independent role for UTX and JMJD3 has been demonstrated in normal and malignant T-cells through interaction with the BRG1-containing SWI/SNF remodeling complex.Citation10 UTX knockout (KO) studies have unraveled important roles for UTX in many developmental processes, including cardiac development and hematopoiesis, but also suggested that UTX and UTY might have redundant functions during embryonic development.Citation11-Citation17
Upon the establishment of UTX as a histone eraser in the context of normal development, a number of studies started to report genetic defects targeting UTX as the underlying cause of specific diseases. In 2009, a role for the histone H3K27me2/3 demethylase UTX as tumor suppressor was initially postulated in several human tumors including multiple myeloma, esophageal and renal cancer.Citation18 In 2012, specific loss-of-function defects in UTX were identified in patients with a specific hereditary disorder named the Kabuki syndrome.Citation19 In this review, we summarize the current knowledge on UTX in normal development and highlight recent findings on its implication in cancer and hereditary disease.
UTX Drives Context Dependent Transcriptional Regulation Mainly Through its H3K27 Demethylase Activity
H3K27me2/3 demethylation
Methylation of H3K27 is a critical mediator of transcriptional gene repression and contributes to important biological processes including X-inactivation, genomic imprinting, stem cell maintenance, animal body patterning, circadian rhythms and cancer.Citation5,Citation20 Regulation of cellular H3K27me3 levels is mainly mediated by the H3K27 methyltransferase Polycomb Repressor Complex 2 (PRC2) and the H3K27me2/3 demethylases UTX and JMJD3 ().Citation3-Citation6,Citation21-Citation24 Two main classes of histone demethylases have been discovered until now including the flavin-dependent amine oxidases, such as LSD1,Citation1 and the iron and α-ketoglutarate-dependent dioxygenases with a Jumonji C (JmjC) catalytic domain, such as UTX and JMJD3.Citation3-Citation7
Figure 1. The UTX family mediates H3K27me2/3 demethylation. Graphical illustration of open and closed chromatin states that are mediated by the histone demethylases UTX and JMJD3 and the histone methyltransferase PRC2 hereby erasing or writing methyl groups on H3K27 enabling activation or blockage of gene transcription, respectively (graphics from www.somersault1824.com).
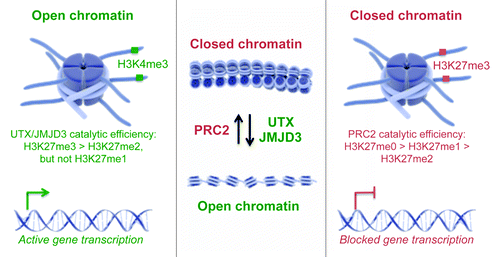
The H3K27me2/3 demethylases UTX and JMJD3 preferentially demethylate H3K27me3 followed by H3K27me2 in vitro and in vivo. This demethylase activity is dependent on the catalytic JmjC domain, which contains conserved residues for binding with the co-factors iron and α-ketoglutarate.Citation3-Citation7 Moreover, a newly identified zinc-binding domain within these H3K27me2/3 erasers provides specificity toward the histone lysine H3K27 and excludes interaction with the near-cognate histone lysine H3K9.Citation25,Citation26 Notably, the family member UTY lacks H3K27me2/3 erasing activity in vitro and in vivo despite a conserved JmjC domain and 88% sequence homology with the UTX protein.Citation3,Citation7,Citation14 Finally, UTX and UTY proteins contain tetratricopeptide repeats (TPRs) at their N-terminal regions that are important for protein-protein interactions. These TPRs are lacking in the JMJD3 protein,Citation3-Citation7 which might suggest lack of redundant functions between the H3K27me2/3 demethylases UTX and JMJD3.
UTX, UTY and JMJD3 are evolutionary conserved from Caenorhabditis elegans (C. elegans) to human.Citation3-Citation7 The mouse genome contains the three H3K27me2/3 demethylase family members Utx, Uty and Jmjd3, whereas the genome of Drosophila melanogaster harbors only one ortholog of the mammalian H3K27me2/3 demethylases called dUTX.Citation27 The C. elegans genome possesses 4 orthologs whereby UTX-1 resembles the mammalian UTX and the three other orthologs are more related to JMJD3.Citation28
UTX is a member of the MLL2 H3K4 methyltransferase complex
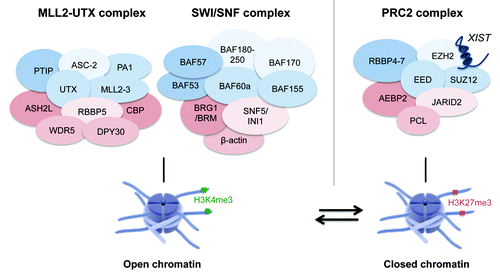
In 2007, an MLL2 (also called ALR and formerly called MLL4) complex was identified in different mammalian cell types containing 12 protein members including MLL2, PTIP, UTX, ASC-2, ASH2L, RBBP5, WDR5, DPY30, matrin3, MGC4606, α- and β-tubulin.Citation8 Furthermore, the 3 complex members MLL2, PTIP and UTX were shown to co-localize at promoter regions and first exons of MLL2 target genes marked with the transcriptional activation mark H3K4me3.Citation8 Another group confirmed the identification of a similar H3K4 methyltransferase complex in which MLL2, MLL3, PTIP, UTX, ASC-2, ASH2L, RBBP5, WDR5, DPY30 and PA1 were also linked to H3K4 methylation activity ().Citation9 Interestingly, the composition of the MLL2 complex was confirmed in the Drosophila ortholog Trithorax related (Trr) complexCitation29 and the C. elegans ortholog SET-16 complex.Citation28 Hence, this conserved MLL2 complex acts like a classical H3K4 methyltransferase complex,Citation8 which suggests a dynamic interplay between H3K27me2/3 demethylation and H3K4 methylation during transcriptional gene activation.
Figure 2. The chromatin complexes MLL2-UTX, SWI/SNF and PRC2 contribute to open and closed chromatin conformations. Schematic representation of the H3K4 methyltransferase complex MLL2-UTX, the SWI/SNF ATPase remodeling complex and the H3K27 methyltransferase complex PRC2 composed out of different protein-coding and non-protein-coding members. The histone eraser UTX is part of the MLL2 complex leading to a dynamic interplay between H3K4 methylation and H3K27me2/3 demethylation. Furthermore, UTX can cooperate with the BRG1-containing SWI/SNF complex where it plays a role in general chromatin remodeling independent of its H3K27 demethylase function. The MLL2-UTX and SWI/SNF complexes both contribute to an open chromatin formation. The PRC2 complex enables efficient methylation of H3K27 thereby promoting gene silencing and chromatin compaction (graphics from www.somersault1824.com).
In flies, dUTX mutant tissues are marked by an increase in global H3K27me3 levels and surprisingly also a reduction in global H3K4me1 levels.Citation20,Citation30 The effect on H3K4me1, a mark enriched at enhancer regions and at gene bodies of actively transcribed genes, seems to be independent of the demethylation capacity of dUTX.Citation20,Citation30 Similarly, loss of the Trr H3K4 methyltransferase complex results in a global profound reduction of H3K4me1 levels in many tissues.Citation30,Citation31 Notably, Trr and dUTX are enriched genome-wide near transcriptional start sites (TSSs) (enriched for H3K4me3 and RNAP II) but also on active enhancers (enriched for H3K4me1) indicating a promoter-proximal role as well as a promoter-distal role for the Trr-dUtx complex in transcriptional regulation.Citation30 Importantly, loss of Trr provoked the most profound reduction in H3K4me1 levels at the enhancer regions compared with dUTX lossCitation30 indicating that the Trr complex itself is the main driver of H3K4 monomethylation with support of dUTX.
UTX interacts with the SWI/SNF remodeling complex and the histone acetyltransferase CBP
Besides regulation of histone modifications like H3K27me3, ATPase-dependent remodeling complexes including the SWI/SNF family contribute to regulation of chromatin accessibility hereby enabling transcriptional activation and repression.Citation32 The SWI/SNF family is mainly linked to transcriptional activation whereby the ATPase complex members BRG1 and BRM interact with acetylated histone tails through their bromodomain ().Citation33,Citation34 An interaction between the Brg1-containing SWI/SNF remodeling complex and the histone H3K27me2/3 erasers Utx and Jmjd3 was demonstrated promoting general chromatin remodeling that was independent of their H3K27me2/3 demethylase potential. Furthermore, UTX and JMJD3 were shown to be functionally required for transcriptional activation of lineage-defining T-box transcription factors (including T-bet, Eomes and Tbx5),Citation10 which represent essential transcriptional regulators in early cell-fate decisions, differentiation and organogenesis.Citation35
An interaction between the histone acetyltransferase Creb-binding protein (CBP) and the SWI/SNF remodeling complex is observed in mammalsCitation10,Citation36 and Drosophila.Citation37 In flies, CBP and the PRC2 complex act antagonistically in the regulation of active and repressed chromatin states at Polycomb target genes by acetylation and methylation of H3K27, respectively ().Citation37 H3K27 acetylation is mainly detected at enhancers, promoters and gene bodies of actively transcribed genes.Citation30 To enable efficient H3K27 acetylation, CBP was shown to associate with dUTX and the ATPase Brm of the SWI/SNF complex.Citation37 The ATPase Brm in flies resembles the 2 mammalian orthologs BRM and BRG1 of the SWI/SNF complexes.Citation32,Citation37 Genome-wide binding studies showed that the chromatin components dUTX, Brm and CBP co-occurred at Polycomb target genes enriched for H3K27ac, suggesting a collaborative effort antagonizing Polycomb mediated gene silencing.Citation37 Of note, loss of dUTX or Trr results in a reduction of global H3K27ac levels in various tissues,Citation30,Citation37 so dUTX might be capable of promoting H3K27 acetylation and H3K4 monomethylation in concert with Trr, Brm and CBP.
UTX regulates dynamic HOX expression
The HOX genes are highly conserved transcriptional regulators essential during development of the anterior-posterior axis (head-tail).Citation38 The HOX family in humans consists of 39 HOX genes that encode 4 clusters of HOX proteins, i.e., HOXA to HOXD.Citation39 Aberrant expression of HOX genes leads to developmental defects and disease.Citation39 Polycomb and Trithorax proteins are implicated in the complex regulation of spatio-temporal expression of HOX genes thereby regulating animal body patterning and cell differentiation.Citation40 Evidence for direct involvement of UTX in regulation of HOX gene activity was demonstrated through UTX knockdown experiments in HEK293T cells in which loss of UTX induced transcriptional repression of HOXA and HOXC clusters.Citation5 Furthermore, UTX loss also resulted in an increase of H3K27me2/3 levels at the promoter regions of HOXA13 and HOXC4, whereas no changes in MLL2 and PRC2 occupancy were observed.Citation5
Chromatin immunoprecipitation (ChIP) of UTX followed by HOX-specific ultra-dense tiling microarrays (ChIP-chip) showed a strong UTX binding to narrow windows within 500 base pairs downstream of the TSSs of many HOX genes in human fibroblasts, but not in mouse embryonic stem cells (ESCs) where HOX genes are silenced. Importantly, H3K27 trimethylation was concomitantly diminished at UTX binding sites.Citation3 Hence, HOX expression is tightly regulated by opposing activities of the H3K27me3 writer PRC2 and the H3K27me2/3 eraser UTX in order to enable context dependent transcriptional regulation.
UTX regulates a retinoblastoma gene network
Genome-wide ChIP-chip analysis of UTX, H3K4me2 and H3K27me3 in human fibroblasts identified UTX binding at 1945 promoters mostly upstream of TSS.Citation41 Out of these, the majority of UTX targets were enriched for H3K4me2 (62%) and showed transcriptional activity. This observation is consistent with UTX being a member of the H3K4 methyltransferase MLL2 complex.Citation8,Citation9,Citation41 Using Ingenuity Pathway analysis (IPA), a RB gene network including RB1, HBP1 and RBBP4, 5, 6 and 9 was discovered as the most significant network of UTX-bound genes enriched for H3K4me2.Citation41 Loss of UTX in human fibroblasts and mouse embryonic fibroblasts (MEFs) confirmed increased levels of H3K27me3 at promoters of genes from the identified RB-network thereby contributing to deregulated RB-dependent cell cycle arrest leading to ectopic cell proliferation.Citation41,Citation42
UTX Escapes X-Inactivation
The female genome harbors 2 X-chromosomes of which one X-chromosome is silenced to compensate the difference in gene dosage with males.Citation43 X-inactivation is mediated by the long intergenic non-coding RNA (lincRNA) XIST, which coats the inactive X-chromosome. Next, the PRC2 complex is guided by XIST to the inactive X-chromosome hereby enabling H3K27 methylation leading to the formation of facultative heterochromatin and gene silencing ().Citation44
Utx is one of the few genes that can escape X-inactivation in female mice and human.Citation45 In agreement, Utx shows higher expression in female tissue of brain, liver, neurons and sexual organs.Citation46,Citation47 In line with this observation, a female-specific function was reported for Utx in the regulation of the X-linked homeobox genes Rhox6 and Rhox9.Citation46 It was shown that Utx is expressed at higher levels in undifferentiated female ESCs as compared with male ESCs, which resulted in a stronger Utx binding in the promoter regions of Rhox6 and Rhox9 in females. Subsequently, Utx mediates a more profound H3K27me3 removal and transcriptional activation of these homeobox genes in female cells.Citation46
The Role of UTX in Cellular Reprogramming
Pluripotency is the unique characteristic of ESCs and the early developmental stage embryo to induce the generation of the three germ layers endoderm, ectoderm and mesoderm finally constituting all tissues of the adult organism.Citation48 Through in vitro reprogramming of somatic cells via induction of the four transcription factors OCT4, SOX2, KLF4 and MYC; pluripotency can be re-established hereby generating induced pluripotent stem cells (iPSC).Citation48,Citation49 During reprogramming, genome-wide changes in the transcriptome and the chromatin structure are induced to achieve the switch to the pluripotent state.Citation48
The histone H3K27me3 demethylase Utx has a critical role in efficient induction or re-establishment of pluripotency during the generation of iPSCs in vitro, but is dispensable for maintenance of pluripotency.Citation50 The requirement for UTX to ensure correct reprogramming depends on its H3K27me3 demethylase activity.Citation50 During iPSC induction, Utx target genes that are enriched with H3K4me3 in Utx wild-type MEFs aberrantly accumulate H3K27me3 in Utx mutant MEFs. These loci, including the stem-cell maintenance genes Sall1, Sall4, and Utf1 as well as validated Klf4 and Oct4 target genes, fail to reactivate during reprogramming due to loss of Utx.Citation50 Hence, Utx seems to play a crucial role during reprogramming through its H3K27me2/3 demethylation activity in correct re-activation of essential pluripotency genes.
A Critical Role for UTX in Embryonic Stem Cells and Embryonic Development
Embryonic stem cells
H3K27 methylation is important in the maintenance of self-renewing ESCs by repressing tissue-specific developmental genes.Citation51-Citation53 During ESC differentiation, the re-activation of developmental genes is associated with loss of H3K27me3.Citation51 Loss of Utx does not influence ESC self-renewal and proliferation, but seems to provoke an effect on the differentiation capacity of ESCs.Citation11,Citation12,Citation50,Citation54 This differentiation defect is reflected by loss of induction of a set of developmental genes including some ectoderm markers (Otx2, Msi1, Sox1, and Pax6),Citation54 endoderm markers (Gata4, Gata6, Foxa2, Sox17, Gsc, and Ncad)Citation12,Citation55 and mesoderm markers (Vegfr2, Wnt3, and Brachyury),Citation12,Citation13,Citation54,Citation55 presumably due to loss of Utx binding to the promoter regions of these genes.Citation54
Contradicting studies report on whether the effect of UTX loss on ESC differentiation depends on its H3K27me3 demethylase activity.Citation12,Citation13,Citation54,Citation55 Namely, male UTX KO ESCs that express a catalytic inactive or wild-type Utx protein both could rescue the induction of expression of the mesoderm genes Wnt3 and Brachyury supporting an H3K27me3-demethylase-independent role of Utx during ESC differentiation.Citation54 Furthermore, Utx binds directly to the promoters of the ectoderm genes Msi1 and Sox1 and the mesoderm gene Brachyury marked with an increase in H3K4me3 levels but without a change in H3K27me3 levels.Citation54 In contrast, a decrease in H3K27me3 levels and an increase in H3K4me3 levels was seen at the promoter of the Utx target Hoxb1 during differentiation of Utx wild-type ESCs confirming that Utx still plays an H3K27me3 demethylase-dependent role in transcriptional regulation of homeotic and developmental genes.Citation54 Therefore, UTX regulates transcriptional activation of UTX target genes during ESC differentiation in different ways dependent or independent of its H3K27me2/3 demethylation activity.
Embryonic development in mice
Several groups have shown that complete loss of Utx in UtxΔ/Δ female mice leads to embryonic lethality between embryonic day (E) 10.5 and E12.5, whereas UtxΔ/+ heterozygous female mice are viable and fertile.Citation11-Citation15 Surprisingly, UtxΔ/Y male mice are marked by a diverse outcome ranging from embryonic lethality to tumor formation in adults.Citation11-Citation15 The viable adult UtxΔ/Y male mice are smaller in size compared with their littermatesCitation12,Citation14 and remain fertile indicating that Utx is not required for male fertility.Citation12 Interestingly, UtxΔ/UtyΔ male mice, in which both Utx and Uty are eliminated, phenocopy the UtxΔ/Δ female mice,Citation14 suggesting that the Y-linked Uty gene can partially rescue the effect of complete loss of Utx.Citation11 In addition, Utx and Uty can both interact with Rbbp5 (member of the Mll2 complex) regulating H3K4 methylation rather than H3K27me2/3 demethylation at Utx and Uty target genes.Citation14
Embryonic development in Drosophila melanogaster
Homozygous dUTX Drosophila mutants are lethal, with only 5% of the animals that survive the pupal stage although not reaching adulthood.Citation20 More specifically, dUTXΔ homozygotes that lack both maternal and zygotic dUTX protein (dUTXΔmat-zyg-) die as larvae, whereas dUTXΔ homozygotes that lack only zygotic dUTX protein (dUTXΔmat+zyg-) can develop further but die quickly after the pupal stage.Citation56 dUTX was shown to be necessary for the controlled expression of 2 specific homeotic genes Ubx and Abd-B in very early fly development.Citation56 Furthermore, sex combs malformation, rough eyes, wrinkled wings and wing vein defects are observed in the dUTX mutants which resembles some characteristics of Trithorax mutants and is in concordance with the effect of these proteins on Hox gene regulation.Citation20,Citation56
Embryonic development in Caenorhabditis elegans
In agreement with Drosophila, UTX-1 mutant worms lacking both maternal and zygotic UTX-1 protein (UTX-1m-z-) die as late stage embryos or malformed L1 larvae. Mutants with loss of only the zygotic UTX-1 (UTX-1m+z-) protein are viable and reach adulthood, but a reduction in fertility is observed due to defects in gonad migration and oocyte organization. The UTX-1m-z- mutants show an increase in global H3K27me2/3 levels at the embryonic stage, but introduction of a catalytic inactive mutant of UTX-1 in UTX-1m+z- mutant animals rescued the fertility indicating that the UTX-1 demethylase activity is not necessary for this developmental process. Interestingly, loss of UTX-1 complex members SET-16 (MLL2–3), PIS-1 (PTIP), WDR-5.1 (WDR5) and F21H12.1 (RBBP5) also resulted in posterior and gonadal defects further confirming that these genes are acting in the same genetic complex.Citation28
A Critical Role for UTX in Tissue-Specific and Developmental Processes
UTX contributes to a variety of tissue-specific and developmental processes including cardiac development,Citation11-Citation15 hematopoiesis,Citation13,Citation15,Citation57,Citation58 myogenesis,Citation16,Citation17,Citation27,Citation59 osteogenic differentiation,Citation60 wound healing,Citation61 and aging.Citation62,Citation63 Cardiac development and hematopoiesis will be discussed in further detail.
Cardiac development
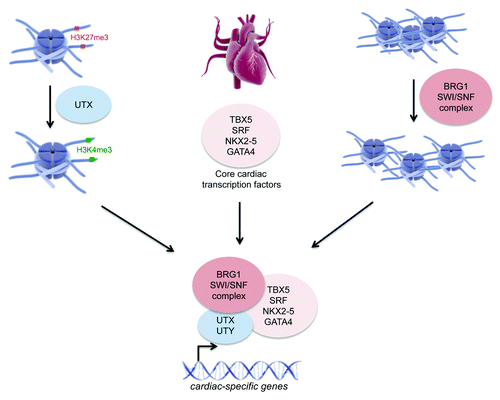
The histone eraser Utx seems to play an important role in cardiac development. UtxΔ/Y ESCs that in vitro differentiate into mature cardiac cells show absence of effective heart-like rhythmic contractions due to a failure of inducing cardiac-specific gene expression.Citation11 Furthermore, UtxΔ/Δ female mice and UtxΔ/UtyΔ male mice exhibited strong cardiac developmental and neural tube closure defects (cranioschisis) at day E9-E10.5.Citation11-Citation15 This cardiac malformation is presumably caused by 2 mechanisms. First, loss of Utx disables the induction of transcriptional activation of cardiac-specific genes mediated by H3K27me2/3 demethylation, whereby the core cardiac transcription factors Nkx2-5, Tbx5, Gata4 and Srf specifically guide Utx to the promoter regions of cardiac-specific genes ().Citation11 Second, loss of Utx disturbs the interaction between the Brg1-containing SWI/SNF chromatin remodeling complex and the core cardiac transcription factor Tbx5, an effect that is thought to be independent from the H3K27me2/3 demethylase activity of UTX ().Citation10,Citation11 This dual role of UTX is further supported by an independent study showing that UTX and UTY can physically interact with BRG1, NKX2-5, SRF and TBX5 hereby inducing expression of cardiac-specific genes.Citation14 Hence UTX enables transcriptional activation of cardiac-specific genes through H3K27me3 demethylase dependent and independent mechanisms during cardiac development.
Figure 3. The involvement of UTX in genetic regulation of cardiac development. During cardiac development UTX, guided by the core cardiac transcription factors NKX2–5, TBX5, GATA4 and SRF, promotes specific gene activation of cardiac-specific genes through demethylation of H3K27me2/3 at their promoter regions. In addition, UTX and UTY can interact with the BRG1-containing SWI/SNF chromatin remodeling complex enabling general chromatin remodeling at these cardiac-specific gene loci (graphics from www.somersault1824.com).
Hematopoiesis
Utx expression levels peak in hematopoietic stem and progenitor cells and drop during hematopoietic differentiation.Citation58 In UtxΔ/Δ female mice embryos (E10.5), a lack of red bloods cells (anemia) was observed indicating a critical role for Utx in hematopoiesis.Citation13 Furthermore, female adult mice in which Utx was homozygously eliminated at 11–14 wk of age showed an enlarged spleen, reduced hemoglobin levels, anemia, thrombocytopenia and mild leukocytopenia compared with normal controls. In addition, hematopoietic transcription factors Tal1, Gata1, and Lyl1 were downregulated in the bone marrow of Utx KO female mice.Citation15 Furthermore, Utx was identified as an essential regulator of stem cell migration since loss of Utx blocked the migration potential of hematopoietic progenitor cells in vitro and of primordial germ cells in vivo.Citation15
Constitutional UTX Defects Cause the Kabuki Syndrome
Constitutional loss-of-function defects in UTX cause the so-called Kabuki syndrome. This rare congenital anomaly syndrome, also called the Kabuki make-up syndrome, was first described by Niikawa and Kuroki in 1981.Citation64,Citation65 The Kabuki syndrome is characterized by moderate-to-severe mental retardation, visceral and skeletal abnormalities, postnatal growth impairment (short stature) and facial abnormalities including large protruding ears.Citation64-Citation66 The Kabuki syndrome has a prevalence of about 1 in 32 000 live births.Citation65 Using whole-exome sequencing, nonsense and frameshift mutations in the MLL2 gene were first identified as a cause of the Kabuki syndromeCitation67 in 74% of these patients.Citation68 Two years later, 3 focal deletions targeting the histone demethylase UTX were identified in 1 male and 2 female Kabuki patients.Citation69 Subsequently, 2 nonsense mutations and a small indel mutation were discovered in 1 female and 2 male Kabuki patients.Citation19
As discussed in the next part, both the MLL2 and the UTX gene have been reported as important histone modifier genes involved in cancer pathogenesis. Until now, 6 Kabuki patients have been described that developed different types of cancer including pre-B-ALL,Citation70 hepatoblastoma,Citation71 neuroblastoma,Citation71,Citation72 Burkitt lymphoma,Citation73 and fibromyxoid sarcomaCitation74 marking the Kabuki syndrome as a cancer predisposition syndrome.
UTX as a Bona Fide Tumor Suppressor Gene in Cancer Biology
Over the last years, deregulated histone methylation became a major theme in cancer biology research including the balance of H3K27 methylation. In 2009, somatic loss-of-function mutations and deletions targeting the UTX gene were identified in multiple cancer types including multiple myeloma, esophageal and renal cancer.Citation18 This notion was further supported by subsequent identification of recurrent inactivating UTX mutations and deletions in several leukemia as well as solid tumor types (; Fig. S1; Tables S1 and 2), mainly through exome- or genome-wide sequencing strategies.Citation75-Citation84 Furthermore, a recent study of The Cancer Genome Atlas (TCGA) in which whole-exome sequencing was performed on 3281 tumors derived from 12 tumor types identified 127 significantly mutated genes including the tumor suppressor gene UTX.Citation85
Figure 4.UTX defects in primary cancer samples. Schematic representation of the presence of UTX defects (%) in primary cancer samples of a broad range of leukemic and solid cancer types investigated in multiple genomic studies.
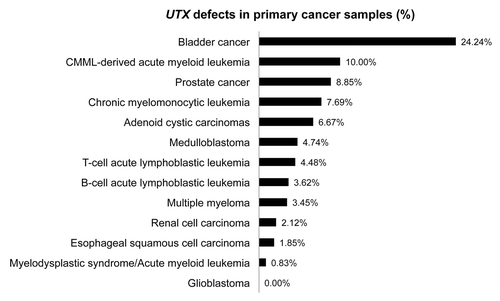
Bladder cancer has one of the highest frequencies of UTX mutations () typically consisting of truncating mutations in the region coding for the functional JmjC domain of UTX. Interestingly, more than 50% of bladder cancer patients harbor genetic aberrations in a bigger set of chromatin remodeling genes including UTX, CREBBP, EP300, ARID1A, CHD6, MLL1, MLL3, and NCOR1.Citation77 Furthermore, multiple myeloma cases harboring mutations in UTX or in the Trithorax complex members MLL1, MLL2, and MLL3 were linked with high expression of HOXA9Citation76 confirming the role of UTX in HOX gene regulation.Citation3
Interestingly, a study in chronic myelomonocytic leukemia (CMML) identified loss-of-function mutations in both UTX and EZH2 despite their opposing roles on H3K27me3 regulation. Notably, these mutations occurred in a mutually exclusive manner in CMML patient samples.Citation86 UTX and EZH2 defects in CMML were associated with ASXL1Citation87 and TET2Citation88 loss-of-function mutations, two genes also implicated in epigenetic regulation of gene expression.Citation86 These studies indicate that the chromatin regulatory machinery is a recurrent mutational target in a broad range of cancer types highlighting the importance of correct regulation of chromatin remodeling in the homeostasis of normal tissues.Citation32
Whole-exome and whole-genome next-generation sequencing studies in medulloblastoma identified mutations in different chromatin regulators including UTX, MLL2, MLL3, SMARCA4 (BRG1), ZMYM3, and CREBBP.Citation78-Citation80 In general, four prognostically relevant subgroups (SHH-subgroup, WNT-subgroup, subgroup 3 and subgroup 4) are defined in this malignant childhood brain tumor.Citation80 Of particular interest, medulloblastoma is marked by a disturbed gender distribution toward males in subgroups 3 and 4Citation89 and mutations targeting the X-linked UTX gene were exclusively identified in these 2 subgroups.Citation79,Citation80,Citation90 In line with the fact that UTX escapes X-inactivation,Citation45 female medulloblastoma patients harboring UTX defects showed bi-allelic UTX inactivation (splice-site UTX mutation and deletionCitation80 or missense mutation and loss of chromosome XCitation79), suggesting that complete loss of UTX is required for malignant transformation. In addition, more than 50% of UTX mutant male medulloblastoma patients had a deletion of the Y-chromosome (including the UTX family member UTY) compared with less than 10% of UTX wild-type males.Citation80 Furthermore, gain or overexpression of the H3K27 methyltransferase EZH2 as well as inactivating mutations in H3K4me3 regulators CHD7 and ZMYM3 were also present in subgroups 3 and 4 of medulloblastoma. All together, these data indicate that deregulation of H3K27 and H3K4 methylation is a core oncogenic component of medulloblastoma. Importantly, mutations targeting the X-linked gene UTX might partially explain the higher male prevalence in medulloblastoma subgroups 3 and 4.Citation80
So far, the functional consequences of cancer related UTX mutations have been poorly characterized. Western blot analyses show lack of UTX protein expression in UTX mutant cancer cell lines.Citation18 Furthermore, re-expression of UTX in UTX mutant cancer cell lines was associated with lower H3K27me3 levels at the promoters of Polycomb target genes and concomitant inhibition of cell proliferation.Citation18 In adenoid cystic carcinomas, the impact of the identified UTX missense mutations was evaluated by introduction of these UTX mutants in HEK293T cells, which led to an increase in cell growth and a decrease in H3K27me3 levels.Citation84 Finally, a Sleeping Beauty based insertional mutagenesis screen using a Kras driven pancreatic mouse model identified 543 candidate cancer driver genes with 10% of the genes involved in chromatin regulation including the Utx gene. This un-biased screening approach provides an independent in vivo confirmation of the potential role of Utx as a bona fide tumor suppressor gene in cancer biology.Citation91
Targeting UTX by Small Molecule Inhibitors
The most selective compound against JMJD3 and UTX, GSK-J1, was discovered based on the structural insights of the human and mouse JMJD3 protein with an IC50 of 60nM for inhibiting JMJD3. Furthermore, GSK-J1 is active against both H3K27me3 erasers UTX and JMJD3 but inactive against a panel of demethylases of the JmjC family. Notably, the cell-penetrating derivative GSK-J4 was able to inhibit the JMJD3-induced loss of total nuclear H3K27me3 levels and to enable specific inhibition of H3K27 demethylation at promoter regions of JMJD3 and UTX target genes.Citation92
Several general inhibitors of JmjC demethylases have been identified including the α-ketoglutaric acid mimics N-oxalylglycine,Citation93,Citation94 methylstatCitation95 and 2,4-dicarboxypyridine; the iron-chelating agent deferoxamineCitation95; the pyridine hydrazone JIB-04Citation96 and catechols.Citation97 Unfortunately, these inhibitors are selective against some or all JmjC enzymes but are unable to target one specific JmjC member.
Conclusions
In this review, we have summarized and discussed the identification of the histone H3K27me2/3 erasers UTX and JMJD3, and the role of UTX in normal development and disease. UTX cooperates with a set of chromatin players, including the histone methyltransferase complex MLL2, the histone acetyltransferase CBP and the BRG1-containing SWI/SNF remodeling complex,Citation8-Citation10,Citation37 to trigger active gene transcription by H3K27me2/3 demethylation or H3K4 methylation.Citation3-Citation6,Citation8-Citation10,Citation30,Citation37 Up until now, the main direct target genes of UTX encompass a broad set of HOX genes important in animal body patterningCitation3,Citation5 and a RB gene network implicated in cell fate control.Citation41 Furthermore, UTX loss can lead to the development of the Kabuki syndromeCitation69 and can contribute to cancer pathogenesis.Citation18
The third UTX family member UTY has no clear enzymatic demethylase activity despite the presence of a conserved JmjC domain and high amino acid conservation.Citation3,Citation7,Citation14 Surprisingly, UTY can partly compensate UTX functions including the cooperation with the MLL2 and BRG1-containing SWI/SNF complexes.Citation14 Also, UTY can partially rescue embryonic lethality in UTX KO male mice.Citation11-Citation15 Furthermore, UTX mutations are frequently found together with deletions encompassing the UTY gene locus in male cancer patients.Citation18,Citation80 Hence, a set of overlapping functions between UTX and UTY in transcriptional regulation seems present despite the lack of H3K27me2/3 demethylase activity for UTY.
All together, the complexity of biological functions assigned to UTX is just starting to emerge and will trigger additional research in a broad range of developmental processes. Multiple studies have highlighted UTX as an important player in cancer biology, but its role as a bona fide tumor suppressor still needs further confirmation using genetically engineered in vivo models. Ultimately, these biological insights will clarify to what extend loss of UTX provides therapeutic opportunities for human disease.
| Abbreviations: | ||
| CBP | = | CREB-binding protein |
| ChIP | = | chromatin immunoprecipitation |
| CMML | = | chronic myelomonocytic leukemia |
| E | = | embryonic day |
| ESC | = | embryonic stem cell |
| H3K27me2/3 | = | histone 3 lysine 27 di- and tri-methylation |
| HOX | = | homeobox |
| IPA | = | Ingenuity Pathway Analysis |
| iPSC | = | induced pluripotent stem cell |
| JmjC | = | Jumonji C |
| JMJD3 | = | Jumonji D3 |
| KO | = | knockout |
| lincRNA | = | long intergenic non-coding RNA |
| LSD1 | = | Lysine-Specific Demethylase 1 |
| MEF | = | mouse embryonic fibroblast |
| PRC2 | = | Polycomb Repressor Complex 2 |
| TCGA | = | The Cancer Genome Atlas |
| TPR | = | tetratricopeptide repeat |
| Trr | = | Trithorax related |
| TSS | = | transcriptional start site |
| UTX | = | ubiquitously transcribed tetratricopeptide repeat on chromosome X |
| UTY | = | ubiquitously transcribed tetratricopeptide repeat on chromosome Y |
Additional material
Download Zip (206.9 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work is supported by the Fund for Scientific Research (FWO) Flanders (postdoctoral grant to P.V.V., PhD grant to J.V.d.M., Odysseus grant to P.V.V, project grants G.0198.08, G.0564.13N, G.0550.13N, G.0869.10N to F.S. and G.A001.13N to P.V.V.); the Flemish Liga against Cancer (VLK) (PhD grant to J.V.d.M.); the GOA-UGent (grant no. 12051203 to F.S.); the Cancer Plan from the Federal Public Service of Health (F.S.); the Children Cancer Fund Ghent (F.S.); the Belgian Program of Interuniversity Poles of Attraction IUAP (F.S.) and the Belgian Foundation Against Cancer (365O9110 to F.S.).
References
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119:941 - 53; http://dx.doi.org/10.1016/j.cell.2004.12.012; PMID: 15620353
- Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 2012; 13:297 - 311; PMID: 22473470
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 2007; 449:689 - 94; http://dx.doi.org/10.1038/nature06192; PMID: 17851529
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007; 449:731 - 4; http://dx.doi.org/10.1038/nature06145; PMID: 17713478
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 2007; 318:447 - 50; http://dx.doi.org/10.1126/science.1149042; PMID: 17761849
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007; 130:1083 - 94; http://dx.doi.org/10.1016/j.cell.2007.08.019; PMID: 17825402
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A 2007; 104:18439 - 44; http://dx.doi.org/10.1073/pnas.0707292104; PMID: 18003914
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 2007; 27:1889 - 903; http://dx.doi.org/10.1128/MCB.01506-06; PMID: 17178841
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 2007; 282:20395 - 406; http://dx.doi.org/10.1074/jbc.M701574200; PMID: 17500065
- Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell 2010; 40:594 - 605; http://dx.doi.org/10.1016/j.molcel.2010.10.028; PMID: 21095589
- Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell 2012; 22:25 - 37; http://dx.doi.org/10.1016/j.devcel.2011.11.009; PMID: 22192413
- Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, Jaenisch R. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci U S A 2012; 109:13004 - 9; http://dx.doi.org/10.1073/pnas.1210787109; PMID: 22826230
- Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C, Ge K. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A 2012; 109:15324 - 9; http://dx.doi.org/10.1073/pnas.1204166109; PMID: 22949634
- Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet 2012; 8:e1002964; http://dx.doi.org/10.1371/journal.pgen.1002964; PMID: 23028370
- Thieme S, Gyárfás T, Richter C, Özhan G, Fu J, Alexopoulou D, Muders MH, Michalk I, Jakob C, Dahl A, et al. The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood 2013; 121:2462 - 73; http://dx.doi.org/10.1182/blood-2012-08-452003; PMID: 23365460
- Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, Blais A, Brand M, Ge K, Dilworth FJ. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J 2010; 29:1401 - 11; http://dx.doi.org/10.1038/emboj.2010.37; PMID: 20300060
- Wang AH, Zare H, Mousavi K, Wang C, Moravec CE, Sirotkin HI, Ge K, Gutierrez-Cruz G, Sartorelli V. The histone chaperone Spt6 coordinates histone H3K27 demethylation and myogenesis. EMBO J 2013; 32:1075 - 86; http://dx.doi.org/10.1038/emboj.2013.54; PMID: 23503590
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O’Meara S, Teague J, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 2009; 41:521 - 3; http://dx.doi.org/10.1038/ng.349; PMID: 19330029
- Miyake N, Mizuno S, Okamoto N, Ohashi H, Shiina M, Ogata K, Tsurusaki Y, Nakashima M, Saitsu H, Niikawa N, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat 2013; 34:108 - 10; http://dx.doi.org/10.1002/humu.22229; PMID: 23076834
- Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, Gupta R, Davuluri R, Shilatifard A, Hariharan IK, Bergmann A. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell Biol 2010; 30:2485 - 97; http://dx.doi.org/10.1128/MCB.01633-09; PMID: 20212086
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298:1039 - 43; http://dx.doi.org/10.1126/science.1076997; PMID: 12351676
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002; 111:185 - 96; http://dx.doi.org/10.1016/S0092-8674(02)00975-3; PMID: 12408863
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 2002; 16:2893 - 905; http://dx.doi.org/10.1101/gad.1035902; PMID: 12435631
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002; 111:197 - 208; http://dx.doi.org/10.1016/S0092-8674(02)00976-5; PMID: 12408864
- Kim E, Song JJ. Diverse ways to be specific: a novel Zn-binding domain confers substrate specificity to UTX/KDM6A histone H3 Lys 27 demethylase. Genes Dev 2011; 25:2223 - 6; http://dx.doi.org/10.1101/gad.179473.111; PMID: 22056667
- Sengoku T, Yokoyama S. Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev 2011; 25:2266 - 77; http://dx.doi.org/10.1101/gad.172296.111; PMID: 22002947
- Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, Shilatifard A. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol 2008; 28:1041 - 6; http://dx.doi.org/10.1128/MCB.01504-07; PMID: 18039863
- Vandamme J, Lettier G, Sidoli S, Di Schiavi E, Nørregaard Jensen O, Salcini AE. The C. elegans H3K27 demethylase UTX-1 is essential for normal development, independent of its enzymatic activity. PLoS Genet 2012; 8:e1002647; http://dx.doi.org/10.1371/journal.pgen.1002647; PMID: 22570628
- Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol 2011; 31:4310 - 8; http://dx.doi.org/10.1128/MCB.06092-11; PMID: 21875999
- Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev 2012; 26:2604 - 20; http://dx.doi.org/10.1101/gad.201327.112; PMID: 23166019
- Kanda H, Nguyen A, Chen L, Okano H, Hariharan IK. The Drosophila ortholog of MLL3 and MLL4, trithorax related, functions as a negative regulator of tissue growth. Mol Cell Biol 2013; 33:1702 - 10; http://dx.doi.org/10.1128/MCB.01585-12; PMID: 23459941
- Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet 2013; 14:765 - 80; http://dx.doi.org/10.1038/nrg3554; PMID: 24105274
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 2002; 111:369 - 79; http://dx.doi.org/10.1016/S0092-8674(02)01005-X; PMID: 12419247
- Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling. Trends Mol Med 2007; 13:373 - 80; http://dx.doi.org/10.1016/j.molmed.2007.07.004; PMID: 17822959
- Wilson V, Conlon FL. The T-box family. Genome Biol 2002; 3:REVIEWS3008; http://dx.doi.org/10.1186/gb-2002-3-6-reviews3008; PMID: 12093383
- Naidu SR, Love IM, Imbalzano AN, Grossman SR, Androphy EJ. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene 2009; 28:2492 - 501; http://dx.doi.org/10.1038/onc.2009.121; PMID: 19448667
- Tie F, Banerjee R, Conrad PA, Scacheri PC, Harte PJ. Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Mol Cell Biol 2012; 32:2323 - 34; http://dx.doi.org/10.1128/MCB.06392-11; PMID: 22493065
- Mallo M, Alonso CR. The regulation of Hox gene expression during animal development. Development 2013; 140:3951 - 63; http://dx.doi.org/10.1242/dev.068346; PMID: 24046316
- Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer 2010; 10:361 - 71; http://dx.doi.org/10.1038/nrc2826; PMID: 20357775
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 2006; 20:1123 - 36; http://dx.doi.org/10.1101/gad.381706; PMID: 16618801
- Wang JK, Tsai MC, Poulin G, Adler AS, Chen S, Liu H, Shi Y, Chang HY. The histone demethylase UTX enables RB-dependent cell fate control. Genes Dev 2010; 24:327 - 32; http://dx.doi.org/10.1101/gad.1882610; PMID: 20123895
- Terashima M, Ishimura A, Yoshida M, Suzuki Y, Sugano S, Suzuki T. The tumor suppressor Rb and its related Rbl2 genes are regulated by Utx histone demethylase. Biochem Biophys Res Commun 2010; 399:238 - 44; http://dx.doi.org/10.1016/j.bbrc.2010.07.061; PMID: 20650264
- Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet 2011; 12:542 - 53; http://dx.doi.org/10.1038/nrg3035; PMID: 21765457
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011; 469:343 - 9; http://dx.doi.org/10.1038/nature09784; PMID: 21248841
- Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, Steiner K, Tam PP, Monaco AP, Willard HF, et al. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet 1998; 7:737 - 42; http://dx.doi.org/10.1093/hmg/7.4.737; PMID: 9499428
- Berletch JB, Deng X, Nguyen DK, Disteche CM. Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A. PLoS Genet 2013; 9:e1003489; http://dx.doi.org/10.1371/journal.pgen.1003489; PMID: 23658530
- Xu J, Deng X, Watkins R, Disteche CM. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J Neurosci 2008; 28:4521 - 7; http://dx.doi.org/10.1523/JNEUROSCI.5382-07.2008; PMID: 18434530
- Rada-Iglesias A, Wysocka J. Epigenomics of human embryonic stem cells and induced pluripotent stem cells: insights into pluripotency and implications for disease. Genome Med 2011; 3:36; http://dx.doi.org/10.1186/gm252; PMID: 21658297
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663 - 76; http://dx.doi.org/10.1016/j.cell.2006.07.024; PMID: 16904174
- Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, Krupalnik V, Zerbib M, Amann-Zalcenstein D, Maza I, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 2012; 488:409 - 13; http://dx.doi.org/10.1038/nature11272; PMID: 22801502
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006; 441:349 - 53; http://dx.doi.org/10.1038/nature04733; PMID: 16625203
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315 - 26; http://dx.doi.org/10.1016/j.cell.2006.02.041; PMID: 16630819
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 2006; 125:301 - 13; http://dx.doi.org/10.1016/j.cell.2006.02.043; PMID: 16630818
- Morales Torres C, Laugesen A, Helin K. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS One 2013; 8:e60020; http://dx.doi.org/10.1371/journal.pone.0060020; PMID: 23573229
- Jiang W, Wang J, Zhang Y. Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res 2013; 23:122 - 30; http://dx.doi.org/10.1038/cr.2012.119; PMID: 22907667
- Copur Ö, Müller J. The histone H3-K27 demethylase Utx regulates HOX gene expression in Drosophila in a temporally restricted manner. Development 2013; 140:3478 - 85; http://dx.doi.org/10.1242/dev.097204; PMID: 23900545
- Chaturvedi CP, Hosey AM, Palii C, Perez-Iratxeta C, Nakatani Y, Ranish JA, Dilworth FJ, Brand M. Dual role for the methyltransferase G9a in the maintenance of beta-globin gene transcription in adult erythroid cells. Proc Natl Acad Sci U S A 2009; 106:18303 - 8; http://dx.doi.org/10.1073/pnas.0906769106; PMID: 19822740
- Liu J, Mercher T, Scholl C, Brumme K, Gilliland DG, Zhu N. A functional role for the histone demethylase UTX in normal and malignant hematopoietic cells. Exp Hematol 2012; 40:487 - 98, e3; http://dx.doi.org/10.1016/j.exphem.2012.01.017; PMID: 22306297
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 2004; 18:2627 - 38; http://dx.doi.org/10.1101/gad.1241904; PMID: 15520282
- Hemming S, Cakouros D, Isenmann S, Cooper L, Menicanin D, Zannettino A, Gronthos S. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 2014; 32:802 - 15; http://dx.doi.org/10.1002/stem.1573; PMID: 24123378
- Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep 2009; 10:881 - 6; http://dx.doi.org/10.1038/embor.2009.102; PMID: 19575012
- Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell 2011; 10:980 - 90; http://dx.doi.org/10.1111/j.1474-9726.2011.00738.x; PMID: 21834846
- Jin C, Li J, Green CD, Yu X, Tang X, Han D, Xian B, Wang D, Huang X, Cao X, et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab 2011; 14:161 - 72; http://dx.doi.org/10.1016/j.cmet.2011.07.001; PMID: 21803287
- Niikawa N, Matsuura N, Fukushima Y, Ohsawa T, Kajii T. Kabuki make-up syndrome: a syndrome of mental retardation, unusual facies, large and protruding ears, and postnatal growth deficiency. J Pediatr 1981; 99:565 - 9; http://dx.doi.org/10.1016/S0022-3476(81)80255-7; PMID: 7277096
- Niikawa N, Kuroki Y, Kajii T, Matsuura N, Ishikiriyama S, Tonoki H, Ishikawa N, Yamada Y, Fujita M, Umemoto H, et al. Kabuki make-up (Niikawa-Kuroki) syndrome: a study of 62 patients. Am J Med Genet 1988; 31:565 - 89; http://dx.doi.org/10.1002/ajmg.1320310312; PMID: 3067577
- Miyake N, Koshimizu E, Okamoto N, Mizuno S, Ogata T, Nagai T, Kosho T, Ohashi H, Kato M, Sasaki G, et al. MLL2 and KDM6A mutations in patients with Kabuki syndrome. Am J Med Genet A 2013; 161:2234 - 43; http://dx.doi.org/10.1002/ajmg.a.36072; PMID: 23913813
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 2010; 42:790 - 3; http://dx.doi.org/10.1038/ng.646; PMID: 20711175
- Hannibal MC, Buckingham KJ, Ng SB, Ming JE, Beck AE, McMillin MJ, Gildersleeve HI, Bigham AW, Tabor HK, Mefford HC, et al. Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome. Am J Med Genet A 2011; 155A:1511 - 6; http://dx.doi.org/10.1002/ajmg.a.34074; PMID: 21671394
- Lederer D, Grisart B, Digilio MC, Benoit V, Crespin M, Ghariani SC, Maystadt I, Dallapiccola B, Verellen-Dumoulin C. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am J Hum Genet 2012; 90:119 - 24; http://dx.doi.org/10.1016/j.ajhg.2011.11.021; PMID: 22197486
- Scherer S, Theile U, Beyer V, Ferrari R, Kreck C, Rister M. Patient with Kabuki syndrome and acute leukemia. Am J Med Genet A 2003; 122A:76 - 9; http://dx.doi.org/10.1002/ajmg.a.20261; PMID: 12949977
- Tumino M, Licciardello M, Sorge G, Cutrupi MC, Di Benedetto F, Amoroso L, Catania R, Pennisi M, D’Amico S, Di Cataldo A. Kabuki syndrome and cancer in two patients. Am J Med Genet A 2010; 152A:1536 - 9; PMID: 20503331
- Merks JH, Caron HN, Hennekam RC. High incidence of malformation syndromes in a series of 1,073 children with cancer. Am J Med Genet A 2005; 134A:132 - 43; http://dx.doi.org/10.1002/ajmg.a.30603; PMID: 15712196
- Ijichi O, Kawakami K, Matsuda Y, Ikarimoto N, Miyata K, Takamatsu H, Tokunaga M. A case of Kabuki make-up syndrome with EBV+Burkitt’s lymphoma. Acta Paediatr Jpn 1996; 38:66 - 8; http://dx.doi.org/10.1111/j.1442-200X.1996.tb03439.x; PMID: 8992864
- Shahdadpuri R, O’Meara A, O’Sullivan M, Reardon W. Low-grade fibromyxoid sarcoma: yet another malignancy associated with Kabuki syndrome. Clin Dysmorphol 2008; 17:199 - 202; http://dx.doi.org/10.1097/MCD.0b013e3282f5f4e3; PMID: 18541969
- De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, Gianfelici V, Geerdens E, Clappier E, Porcu M, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet 2013; 45:186 - 90; http://dx.doi.org/10.1038/ng.2508; PMID: 23263491
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011; 471:467 - 72; http://dx.doi.org/10.1038/nature09837; PMID: 21430775
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 2011; 43:875 - 8; http://dx.doi.org/10.1038/ng.907; PMID: 21822268
- Jones DT, Jäger N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stütz AM, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 2012; 488:100 - 5; http://dx.doi.org/10.1038/nature11284; PMID: 22832583
- Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 2012; 488:106 - 10; http://dx.doi.org/10.1038/nature11329; PMID: 22820256
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 2012; 488:43 - 8; http://dx.doi.org/10.1038/nature11213; PMID: 22722829
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 2010; 463:360 - 3; http://dx.doi.org/10.1038/nature08672; PMID: 20054297
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012; 487:239 - 43; http://dx.doi.org/10.1038/nature11125; PMID: 22722839
- Lindberg J, Mills IG, Klevebring D, Liu W, Neiman M, Xu J, Wikström P, Wiklund P, Wiklund F, Egevad L, et al. The mitochondrial and autosomal mutation landscapes of prostate cancer. Eur Urol 2013; 63:702 - 8; http://dx.doi.org/10.1016/j.eururo.2012.11.053; PMID: 23265383
- Ho AS, Kannan K, Roy DM, Morris LG, Ganly I, Katabi N, Ramaswami D, Walsh LA, Eng S, Huse JT, et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet 2013; 45:791 - 8; http://dx.doi.org/10.1038/ng.2643; PMID: 23685749
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502:333 - 9; http://dx.doi.org/10.1038/nature12634; PMID: 24132290
- Jankowska AM, Makishima H, Tiu RV, Szpurka H, Huang Y, Traina F, Visconte V, Sugimoto Y, Prince C, O’Keefe C, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood 2011; 118:3932 - 41; http://dx.doi.org/10.1182/blood-2010-10-311019; PMID: 21828135
- Fisher CL, Berger J, Randazzo F, Brock HW. A human homolog of Additional sex combs, ADDITIONAL SEX COMBS-LIKE 1, maps to chromosome 20q11. Gene 2003; 306:115 - 26; http://dx.doi.org/10.1016/S0378-1119(03)00430-X; PMID: 12657473
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010; 466:1129 - 33; http://dx.doi.org/10.1038/nature09303; PMID: 20639862
- Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 2012; 123:465 - 72; http://dx.doi.org/10.1007/s00401-011-0922-z; PMID: 22134537
- Dubuc AM, Remke M, Korshunov A, Northcott PA, Zhan SH, Mendez-Lago M, Kool M, Jones DT, Unterberger A, Morrissy AS, et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol 2013; 125:373 - 84; http://dx.doi.org/10.1007/s00401-012-1070-9; PMID: 23184418
- Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, Black MA, Brett BT, Sheetz TE, Dupuy AJ, Chang DK, et al, Australian Pancreatic Cancer Genome Initiative. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A 2012; 109:5934 - 41; http://dx.doi.org/10.1073/pnas.1202490109; PMID: 22421440
- Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 2012; 488:404 - 8; http://dx.doi.org/10.1038/nature11262; PMID: 22842901
- Kristensen JB, Nielsen AL, Jørgensen L, Kristensen LH, Helgstrand C, Juknaite L, Kristensen JL, Kastrup JS, Clausen RP, Olsen L, et al. Enzyme kinetic studies of histone demethylases KDM4C and KDM6A: towards understanding selectivity of inhibitors targeting oncogenic histone demethylases. FEBS Lett 2011; 585:1951 - 6; http://dx.doi.org/10.1016/j.febslet.2011.05.023; PMID: 21575637
- Hamada S, Kim TD, Suzuki T, Itoh Y, Tsumoto H, Nakagawa H, Janknecht R, Miyata N. Synthesis and activity of N-oxalylglycine and its derivatives as Jumonji C-domain-containing histone lysine demethylase inhibitors. Bioorg Med Chem Lett 2009; 19:2852 - 5; http://dx.doi.org/10.1016/j.bmcl.2009.03.098; PMID: 19359167
- Luo X, Liu Y, Kubicek S, Myllyharju J, Tumber A, Ng S, Che KH, Podoll J, Heightman TD, Oppermann U, et al. A selective inhibitor and probe of the cellular functions of Jumonji C domain-containing histone demethylases. J Am Chem Soc 2011; 133:9451 - 6; http://dx.doi.org/10.1021/ja201597b; PMID: 21585201
- Wang L, Chang J, Varghese D, Dellinger M, Kumar S, Best AM, Ruiz J, Bruick R, Peña-Llopis S, Xu J, et al. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat Commun 2013; 4:2035; PMID: 23792809
- Nielsen AL, Kristensen LH, Stephansen KB, Kristensen JB, Helgstrand C, Lees M, Cloos P, Helin K, Gajhede M, Olsen L. Identification of catechols as histone-lysine demethylase inhibitors. FEBS Lett 2012; 586:1190 - 4; http://dx.doi.org/10.1016/j.febslet.2012.03.001; PMID: 22575654
