Abstract
The prostate transmembrane protein androgen induced 1 (PMEPA1) gene is highly expressed in prostate epithelial cells and is a direct transcriptional target for the androgen receptor (AR). AR protein levels are controlled by the AR-PMEPA1 negative feedback loop through NEDD4-E3 ligase. Reduced expression of PMEPA1 observed in prostate tumors, suggests that loss of PMEPA1 may play critical roles in prostate tumorigenesis. This study focuses on epigenetic mechanisms of reduced PMEPA1 expression in the cancer of the prostate (CaP). Benign (n = 77) and matched malignant (n = 77) prostate epithelial cells were laser capture micro-dissected from optimum cutting temperature embedded frozen prostate sections from 42 Caucasian American (CA) and 35 African American (AA) cases. Purified DNA specimens were analyzed for CpG methylation of the PMEPA1 gene. PMEPA1 mRNA expression levels were evaluated by qRT-PCR. Analysis of PMEPA1 methylation and mRNA expression in the same tumor cell populations indicated a significant inverse correlation between mRNA expression and methylation in CaP (P = 0.0115). We noted higher frequency of CpG methylation within the evaluated first intronic region of the PMEPA1 gene in prostate tumors of CA men as compared with AA. In CaP cell lines, PMEPA1 expression was induced and AR protein levels were diminished in response to treatment with the DNA methyltransferase inhibitor, 5-aza-2'-deoxycytidine (decitabine). Cell culture-based studies demonstrated that decitabine restores PMEPA1 expression in AR-positive CaP cell lines. This report reveals the potential role of PMEPA1 gene methylation in the regulation of AR stability. Thus, downregulation of PMEPA1 may result in increased AR protein levels and function in CaP cells, contributing to prostate tumorigenesis.
Introduction
Androgens and the androgen receptor (AR) play central roles in the normal growth, differentiation and physiological function of the prostate gland.Citation1 It has also been established that AR dysregulation leads to the progression of cancer of the prostate (CaP).Citation2-Citation4 To suppress androgen-dependent CaP growth, currently used therapeutic agents inhibit the binding of androgens to AR or the biosynthesis of androgens.Citation5-Citation8 Although most CaP initially respond to androgen ablation, its therapeutic effect is short lived and patients eventually develop castration resistant CaP.Citation5-Citation9
The AR binds to AR-responsive elements (AREs) and regulates the transcription of androgen responsive genes controlling differentiation and growth.Citation1-Citation4 Dysfunction of the androgen axis contributes to CaP through numerous mechanisms, including increased AR expression, intra-tumoral androgen synthesis, AR splice variants, mutations of the AR and androgen metabolizing enzymes.Citation2-Citation9 In a subset of advanced CaP, AR signaling is bypassed in favor of AR independent pathways.Citation2,Citation3,Citation10,Citation11
The PMEPA1 gene was identified in our laboratory as a highly androgen-induced gene in a screen for androgen regulated genes in CaP cells.Citation12,Citation13 PMEPA1 is predominantly expressed in the prostate gland and is directly regulated by AR.Citation14 PMEPA1 spans 55–60 kb on chromosome 20 (20q13.31-q13.33) and the PMEPA1 protein is highly conserved among vertebrates, suggesting a critical role in the homeostasis of prostate. Functional analysis of PMEPA1 has revealed that it is a NEDD4 E3-ligase binding protein and plays a role in downregulation of AR through a negative feedback loop between AR and PMPEA1.Citation15 Inhibition of PMEPA1 leads to increased AR levels in CaP cells. Thus, decrease or loss of PMEPA1 mRNA expression that is frequently observed in CaP may result in gain of AR function.Citation15,Citation16 Studies also suggest that PMEPA1 is involved in other cancers through regulation of the TGF-β, PI3K and WNT pathways.Citation17-Citation20 These findings highlight the cellular context-dependent role of PMEPA1 in normal and malignant conditions.
Activation of PMEPA1 transcription by decitabine in LNCaP and LAPC4 cells with demethylation of CpG residues within the PMEPA1 promoter downstream sequences suggested a role for DNA methylation in regulating PMEPA1 in CaP.Citation21 The current study focuses on the evaluation of the methylation and expression of the PMEPA1 gene in primary prostate tumors and in CaP cell culture models. The results underscore the role of DNA methylation in silencing the PMEPA1 gene with potential implications in the AR degradation pathway.
Results
PMEPA1 is frequently methylated in prostate cancer
We evaluated the methylation of the PMEPA1 gene in human prostate tumors. As a positive control, we analyzed the promoter methylation of GSTP1, a gene known to be methylated in the majority of CaPs.Citation22 Laser capture microdissection (LCM) was used for the precise isolation of tumor and matched non-adjacent normal cells from 77 CaP cases. Genomic DNA and total RNA were isolated from each specimen.
We optimized the methylated-DNA precipitation and methylation-sensitive restriction enzymes (COMPARE-MS)Citation23 assay for 2 ng of purified genomic DNA obtained from LCM-dissected tumor cells. Methylation analysis was performed for CpG-rich sequences of PMEPA1 gene (20q13.31-q13.33) (). Additionally we analyzed GSTP1 gene (11q13)Citation23 that is highly methylated in CaPCitation22 and a LINE1 repetitive DNA element that has been shown to harbor methylation in the human genome.Citation24
Figure 1. (A) Schematic representation of the PMEPA1 gene structure. Red triangle (+1) marks the transcription initiation site. DNA methylation site (HhaI), PCR primer positions and the sequence of the assayed region of the PMEPA1 gene. (B) Methylation frequencies of PMEPA1 and GSTP1 gene in LCM derived human prostate tumor cells from 77 patients.
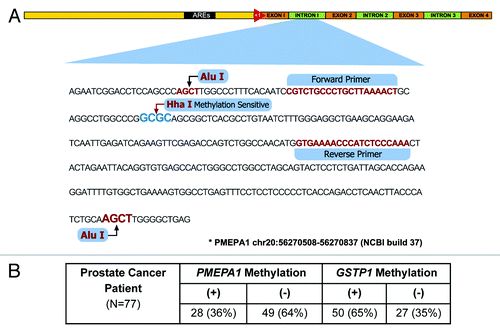
The cohort (42 CA and 35 AA patients) was designed to address the frequency of PMEPA1 methylation. PMEPA1 gene methylation was observed in 28 of 77 cases (36%), whereas GSTP1 methylation was detected in 50 of 77 (65%) cases (). As expected GSTP1 methylation was highly prevalent. In this study, the cancer cells were isolated by LCM yielding low amounts of DNA suitable only for COMPARE-MS, an assay that has been shown to robustly enrich for methylated DNA with high sensitivity and specificity.Citation23 However, limitations in the sensitivity to detect methylated alleles are expected with low amount of DNA input DNA from LCM microdissected primary tumor cells. This likely explains the slightly lower rate of GSTP1 methylation observed in this study compared with previously published studies.Citation22,Citation25
The observed methylation of PMEPA1 prompted us to investigate whether methylation affected the expression level of PMEPA1 in CaP. Thus, we evaluated the level of PMEPA1 gene expression from the same tumor samples that were evaluated for DNA methylation. Patient matched non-adjacent normal epithelial cells were also analyzed for quantitative PMEPA1 expression (n = 77). Matched tumor over normal relative PMEPA1 expressions are summarized in . Consistent with our previous report,Citation15 PMEPA1 expression was reduced in two-thirds of CaP cases. Both CA (65%) and AA (62.9%) groups showed similar frequencies of decreased PMEPA1 mRNA expression in CaP cells in comparison to matched normal epithelial cells.
Figure 2. (A) Quantitative RT-PCR expression analysis of PMEPA1 transcript in LCM derived paired normal and tumor cells of primary CaP revealed decreased expression of PMEPA1 in two-third of CaP patients (Log 10 tumor/normal [T/N]). Cases (tumors) with methylated PMEPA1 gene are highlighted in golden color. (B) Reduced PMEPA1 gene expression correlates with PMEPA1 gene methylation.
![Figure 2. (A) Quantitative RT-PCR expression analysis of PMEPA1 transcript in LCM derived paired normal and tumor cells of primary CaP revealed decreased expression of PMEPA1 in two-third of CaP patients (Log 10 tumor/normal [T/N]). Cases (tumors) with methylated PMEPA1 gene are highlighted in golden color. (B) Reduced PMEPA1 gene expression correlates with PMEPA1 gene methylation.](/cms/asset/32f5770e-9a52-4e95-8494-69464107dcd0/kepi_a_10928710_f0002.gif)
Overall, methylation of the PMEPA1 gene significantly correlated with reduced mRNA expression (P = 0.0115) (). The analysis revealed that 82% of patients, who have methylated PMEPA1 gene, have lower PMEPA1 expression in tumors compared with matched normal epithelium (). Taken together these findings suggest that DNA methylation plays major roles in silencing the PMEPA1 in CaP. In this study, assessment of clinico-pathological data () did not reveal correlation of biochemical recurrence with PMEPA1 methylation status or expression that is likely due to the limited sample size.
Table 1. Clinicopathologic characteristics
Differential PMEPA1 methylation between CA and AA patients
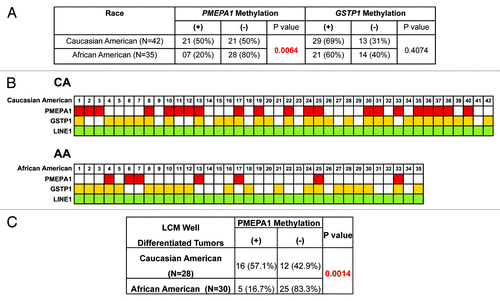
An unexpected findings of this study relates to differential methylation of PMEPA1 between the CA and AA CaP (P = 0.0064) despite similar patterns of PMEPA1 mRNA expression. These data suggested for additional mechanisms involved in downregulation of PMEPA1 in AA CaP.
We noted a higher frequency of PMEPA1 methylation (50%) in CA CaP in comparison to AA (20%) (). The difference in PMEPA1 methylation status between these two groups remained striking (CA: 57%; AA: 17%, P = 0.0014) even when only tumor cells with well-differentiated morphology were compared (). In contrast, the methylation status of the GSTP1 gene showed no significant difference (69% in CA and 60% in AA) (). In the current study design the patient cohort represented nearly equal number of AA and CA cases. While both clinical (2.5-fold higher mortality in AA CaP patients) and cancer genome differences (low frequency of TMPRSS2-ERG in AA patients) have been consistently noted in literature between CA and AA patients,Citation26-Citation28 additional evaluations of differential PMEPA1 methylation between CA and AA are warranted.
Figure 3. (A) Association of PMEPA1 methylation with ethnicity. (B) Methylation status of PMEPA1 and GSTP1 gene in LCM selected prostate tumor DNA. The constitutively methylated LINE1 repetitive element was used as quality control for input DNA. Methylated genes are marked with red (PMEPA1), yellow (GSTP1) and green (LINE1). (C) Correlation analyses of PMEPA1 methylation status in CA and AA CaP patients with well differentiated tumors.
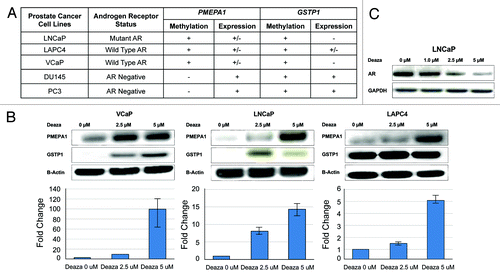
Association of PMEPA1 methylation with AR in prostate cancer cell lines
Association of PMEPA1 methylation with reduced expression of PMEPA1 in CaP specimens provided a rational for testing the methylation status of PMEPA1 in CaP cell lines. We used AR-positive (VCaP, LNCaP, and LAPC4) and AR-negative (DU145 and PC3) cell culture models to analyze the methylation of the PMEPA1 gene. GSTP1 methylation was monitored as an established methylated gene since its methylation status has been established for these CaP cell lines.Citation23,Citation29 The results indicated that PMEPA1 is methylated only in AR positive cell lines and not in AR negatives (). Consistent with previous reports,Citation29,Citation30 this study also showed that GSTP1 is methylated in both AR positive and negative CaP cell lines. These observations suggest a cellular context dependent relationship between PMEPA1 methylation and AR and therefore gain of AR function may be favored by decreased PMEPA1 expression involving epigenetic mechanisms such as DNA methylation.
Figure 4. (A) Methylation (+ or –) and expression (+, – or +/– intermediate) status of PMEPA1 and GSTP1 genes in AR positive and AR negative prostate cancer cell lines. VCaP, LNCaP, and LAPC4 cells harbor low PMEPA1 expression whereas no GSTP1 expression was detected in VCaP and LNCaP cells. (B) PMEPA1 expression is induced by the DNA methyl transferase inhibitor decitabine in VCaP, LNCaP, and LAPC4 cells after 14 d treatment. PMEPA1, GSTP1, and Beta-Actin protein levels were analyzed by immunoblot assays (upper panels). Gene expression was monitored by qRT-PCR (lower panels) and is shown as fold changes normalized to GAPDH control. (C) Reduced AR protein levels in LNCaP cells in response to decitabine treatment.
PMEPA1 expression is induced by the DNA methyltransferase inhibitor decitabine in AR positive CaP cell lines
PMEPA1 methylation may lead to the silencing of PMEPA1 resulting in elevated AR protein levels and AR signaling. To further test this hypothesis, we treated the VCaP, LNCaP, and LAPC4 cell lines with low doses of decitabine. Western blot analysis revealed dose-dependent induction of PMEPA1 protein in response to decitabine treatment (). Consistent with the protein data qRT-PCR analysis also showed decitabine dose-dependent increases of PMEPA1 mRNA expression (). Taken together, these observations suggested that methylation contributes to the silencing of PMEPA1 gene in AR positive CaP cell lines. We also monitored the response to decitabine treatment on AR protein levels. Consistent with the PMEPA1-AR negative feedback model decrease in the expression of AR protein was observed in response to decitabine treatment ().
Silencing of PMEPA1 may result in enhanced AR functions
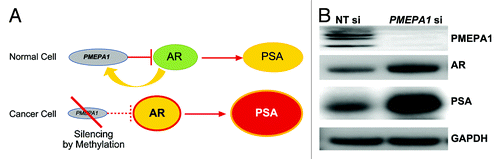
PMEPA1 protein recruits AR to the NEDD4-1 ubiquitin ligase for degradation.Citation16 Thus PMEPA1 modulates levels of AR. Cancer-associated silencing of PMEPA1 may result in elevated levels of AR and increased AR signaling (). We have evaluated the relationship of PMEPA1 levels with AR activity. For monitoring the AR activity, we assessed the expression of the known AR regulated gene, KLK3(PSA) gene by PMEPA1 knockdown in LNCaP cells. Reduced expression of PMEPA1 protein was observed in PMEPA1 siRNA treated LNCaP cells (). As expected, both AR and PSA protein levels were increased in response to PMEPA1 knockdown (). Thus, the silencing of the PMEPA1 gene leads to enhanced AR activity in CaP by eliminating a negative regulatory control of AR protein levels.
Discussion
Emerging data continues to underscore the critical roles of PMEPA1 as an androgen regulated NEDD4 E3 ligase binding protein in maintaining AR protein levels in prostate epithelial cells.Citation14-Citation16,Citation21 Therefore, decrease or loss of PMEPA1 expression and function may have major impact on gain of AR function and CaP progression. The goal of this study was to elucidate the molecular basis for the reduced or lost expression of PMEPA1 in CaP. The results presented here demonstrate that promoter methylation is a major mechanism involved in silencing the PMEPA1 gene in CaP. The potential contribution of genomic methylation in the modulation of PMEPA1 expression was further supported using CaP cell lines. AR positive cells (LNCaP, LAPC4, and VCaP) showed methylation of the PMEPA1 gene, suggesting that the methylation may indeed contribute to the repression of PMEPA1.Citation21 Increased expression of PMEPA1 in response to decitabine treatment of CaP cell lines was reflected both at mRNA and protein levels. The high frequency of methylation silencing of PMEPA1 in CaP samples provides further support for the hypothesis that PMEPA1 may negatively control prostate tumorigenesis through the AR axis (). Similar to other cancers, methylation has been shown to play a major role in the CaP genome.Citation30 In fact, methylation and loss of expression of the GSTP1, a gene involved in the cellular redox maintenance, is present in a majority of CaPs. Moreover, GSTP1 methylation represents one of the earliest genomic alterations in CaP onset.Citation25 Other genes of functional relevance have also been shown to be methylated in CaP.Citation28-Citation32
Studies from our and other groups have shown that PMEPA1 is a multifunctional protein.Citation15-Citation21 However, PMEPA1 functions is likely cellular context dependent and may be different in the presence or absence of AR.Citation20 In normal prostate epithelial cells a negative feedback loop between AR and PMEPA1/NEDD4 regulates AR. In CaP, loss of PMEPA1 appears to be critical in gain of AR function and CaP progression. Out of the AR context, a negative feedback loop between TGF-β and PMEPA1 has been noted through direct interaction of PMEPA1 with SMAD3/4,Citation20 supporting cancer progression. Thus, gain or loss of PMEPA1 expression and function may indeed cancer cell type dependent.
The differential methylation of PMEPA1 gene between CA (50%) and AA patients (20%) was an unexpected finding. Emerging data on CaP genomes is beginning to clarify genomic differences between ethnic groups. CaP-associated somatogenetic and epigenetic differences between various ethnic groups are increasingly recognized.Citation26,Citation27,Citation33-Citation37 However, the distinct biology is not well understood and warrants further investigations, including the observed PMEPA1 methylation difference between CA and AA patients.
A significant effort has been devoted toward developing inhibitors targeting epigenetic modification of the cancer genome including FDA approved DNMT inhibitors (azacitidine and decitabine) for Myelodysplastic Syndrome and HDAC inhibitors (vorinostat and romidepsin) for Cutaneous T Cell Lymphoma.Citation38,Citation39 With the increasing knowledge of epigenetic alterations linked to CaP, such as in GSTP1 or AR axis, more targeted therapeutic strategies may be possible. PMEPA1 may represent a new promising target for epigenetic drugs or new class of drugs complementing AR axis inhibitors.
In conclusion, the PMEPA1 gene is methylated in CaP primary prostate tumors suggesting that methylation may contribute to the silencing of PMEPA1. PMEPA1 gene expression levels correlated with its DNA methylation status. PMEPA1 methylation status displayed an intriguing difference between CA and AA patients. These data, along with our earlier observation, indicate that reduced or lost PMEPA1 expression in CaP cells may lead to elevated AR levels. Our study provides insights into the role of methylation in the CaP-associated silencing of PMEPA1 with potential impact on the AR axis in malignant prostate. Targeting AR for degradation by PMEPA1 may synergize current therapeutic approaches.
Materials and Methods
Prostate cancer specimens and clinico-pathological data
Radical prostatectomy (RP) specimens and clinico-pathological data were obtained from patients enrolled in the Center for Prostate Disease Research (CPDR) from 1996 to 2010 under an institutional review board-approved protocol at the Walter Reed National Military Medical Center (WRNMMC) and Uniformed Services University of the Health Sciences (USUHS) (). Optimum cutting temperature (OCT) embedded RP tissue specimens from 70 seven patients, including 42 CA and 35 AA, were analyzed in this study. None of these patients had received prior androgen deprivation therapy. The biochemical recurrence was defined as two consecutive post-operative PSA values (≥0.2 ng/mL) measured at ≥8 wk post-operatively.
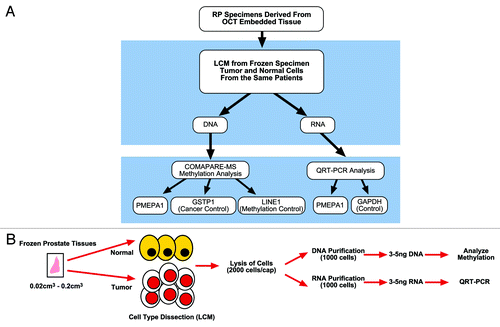
LCM derived normal and malignant prostate epithelial cells were obtained from OCT embedded frozen sections as described previously.Citation40-Citation43 Briefly, specimens obtained immediately after surgical resection of prostate, were OCT embedded and frozen on glass slides placed on dry ice. Benign and malignant cells were isolated by LCM using 6 micron frozen tissue sections archived in CPDR frozen tissue section slide library stored at -80 °C. The strategy for the evaluation of methylation and expression is described in .
Figure 6. (A) Flowchart of DNA methylation and quantitative gene expression analysis in prostate tumor specimens. (B) LCM, determination of methylation by combination of methylated DNA precipitation and methylation-sensitive restriction cleavage (COMPARE-MS), and quantitative gene expression analyses.
Isolation of genomic DNA and total RNA from LCM derived prostate epithelial cells
Total RNA from the LCM derived specimens was isolated, purified and quantified.Citation41,Citation42 For genomic DNA, LCM-derived cells were lysed (buffer containing 10 mM Tris, 1 mM EDTA, 1% Tween 20) and digested with proteinase K (1 mg/ml; Roche Applied Science) at 37 °C overnight.Citation44 DNA was precipitated by adding 1/10th volume of 3M sodium acetate pH 5.2 (Sigma-Aldrich) and equal volume of phenol: chloroform: isoamyl alcohol (25:24:1) (Sigma-Aldrich). For optimal recovery the DNA was precipitated by adding 2 µg of glycogen (Roche Applied Science) and equal volume of chilled isopropanol (Sigma-Aldrich). After centrifugation the DNA pellets were washed with 70% ethanol (Sigma-Aldrich) and resuspended in 25 μl of TE buffer pH 8.0 (Sigma-Aldrich). DNA quantitation of all samples was performed by using Picogreen dye (Quant-iT™ PicoGreen® dsDNA Reagent and Kits, Life Technology) and confirmed with 260/280 ratio by NanoDrop 2000c spectrophotometer (Thermo Scientific).
DNA Methylation assay for genomic DNA isolated from LCM derived prostate cancer cells and prostate cancer cell lines
Based on publicly available genome wide methylation collected by the ENCODE consortium (http://genome.ucsc.edu) as well as by whole genome methylation analyses (Haffner MC and Yegansubramnian S., personal communication) the first intronic region of PMEPA1 was selected for the analyses by the consistent differential methylation between LNCaP and primary prostate epithelium-derived PrEC cells and by the linear detection range in assaying low quantity of input DNA (Fig. S1A and B).
A combination of COMPARE-MS method, followed by Quantitative real time-Polymerase Chain Reaction (Q-PCR) was used for the evaluation of the DNA methylation.Citation23 Two ng of purified genomic DNA from LCM derived prostate tumor cells was digested with 10U each of AluI and HhaI (New England Biolabs) at 37 °C for 3 h followed by heat inactivation of enzymes at 80 °C for 20 min. Methylated genomic DNA fragments were captured by recombinant MBD2 protein derived methyl-binding domain polypeptides immobilized on magnetic beads. In parallel, 2 ng of genomic DNA from normal male white blood cells (WBC), treated with 4U of CpG Methylase (M.SssI) enzyme (Zymo Research) and untreated DNA were also analyzed, serving as positive and negative controls, respectively.
Primers designed for the methylated CpG region of the PMEPA1 gene (forward primer, 5′-CGTCTGCCCT GCTTAAAACT; reverse primer, 5′-TTTGGGAGAT GGGTTTTCAC) and GSTP1 (forward primer, 5′-GTTGGTTTTA TGTTGGGAGT TTTGAGTTTT; reverse primer, 5′-ATCCTCTTCC TACTATCTAT TTACTCCCTA A) were used for SYBR Green-based Q-PCR. LINE1 repetitive elements, which are highly methylated in human genomic DNA, were used as positive control to ensure the recovery of methylated DNA during the COMPARE-MS assay. LINE1 promoter (GenBank accession X58075) was amplified by the following primer set: forward primer 5′-CGCAGAAGAC GGGTGATTTC-3′ and reverse primer 5′-CCGTCACCCC TTTCTTTGAC-3′.
Q-PCR was performed in duplicate with 25 μL reactions containing 1X IQ Sybr-Green Supermix (Bio Rad) and 0.4 μM forward and reverse primers under the following cycling conditions: 95 °C for 10 min, 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s on Stratagene Mx3000P (Agilent Technologies Inc.). The COMPARE-MS assay was used to analyze PMEPA1, GSTP1, and LINE1 methylation in LCM-derived prostate tumor cell and AR positive and AR negative CaP cell lines, VCaP, LNCaP, LAPC4, and DU145, PC3, respectively. The methylation status was categorized binary as present or absent. The methylation levels were normalized to the signal generated by an equal input amount of the positive control.Citation23
Quantitative real-time Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) analysis
TaqMan qRT-PCR using RNA from LCM derived tumor and normal epithelial cells was performed as described previously.Citation40-Citation43 The expression of GAPDH as the RNA input control and the target gene expression were simultaneously analyzed for each sample (in duplicates). A negative control without reverse-transcriptase enzyme was included for each specimen to rule out background signal from potential genomic DNA contamination.
qRT-PCR analysis of PMEPA1 and GAPDH expression was performed on Stratagene Mx3000P (Agilent Technologies, Inc.). The PCR primers for PMEPA1 were 5′-CATGATCCCC GAGCTGCT-3′ (forward) and 5′-TGATCTGAAC AAACTCCAGC TCC-3′, (reverse) and the FAM-labeled probe was 5′-AGGCGGACAG TCTCCTGCGA AAC-3′. GAPDH primers and probe mix were obtained from Life Technologies. All qRT-PCR reactions were performed in triplicate and data were analyzed by using MxPro v.3.2 software (Agilent Technologies Inc.). Amplification plots were evaluated and threshold cycle (CT) was set for each experiment. Multiplex measurements for target gene and GAPDH were averaged across triplicates and used to calculate standard deviation for each set. Subtraction of averaged GAPDH CT from averaged target gene CT yielded the ΔCT. Expression fold-change differences between normal and tumor was calculated by comparing ΔCT values among matched sample sets.
Cell culture
Human prostate tumor cell lines, VCaP, LNCaP were purchased from American Type Culture Collection (ATCC) and LAPC4 kindly provided by Dr Charles L. Sawyers (UCLA, CA). VCaP and LNCaP cells were maintained in DMEM (ATCC), and LAPC4 cells in RPMI-1640 (ATCC) supplemented with 10% of fetal bovine serum (Invitrogen) in a humidified CO2 (5%) incubator at 37 °C. Cell culture and cell line treatment studies have been performed in two independent sets of experiments.
Decitabine (5-aza-2´-deoxycytidine) treatment
One million cells were seeded in T75 flasks in triplicate in the cell growth medium containing 0 µM, 2.5 µM and 5 µM of the DNA methyl transferase inhibitor, decitabine (5-aza-2´-deoxycytidine; Sigma-Aldrich) for 14 d. The media with decitabine were changed every 48 h. After 14 d cells were harvested for isolation of genomic DNA, total RNA and protein.
Transfection and siRNA knock-down of PMEPA1
The LNCaP cells were seeded into 10 cm culture dishes (FALCON, Becton Dicknson) at the density of 1.5 × 106 cells/ dish. The cells were incubated at 37 °C, 5% CO2 for 48 h. After incubation the cells were transfected with 50 nM of control Non-targeting (NT) siRNA and PMEPA1si RNA (5′-GTTATCACCA CGTATATA-3′) (Dharmacon) by using Lipofectamine 2000 (Life Technologies). The cells were incubated with transfection complex at 37 °C, 5% CO2 for 12 h. After incubation the transfection complex was removed and the cells were replenished with fresh complete medium. The cells were harvested 48 h post-transfection for western blot assay.
Western blot analysis
Cells from the cell culture experiments were lysed in Mammalian Protein Extraction Reagent (M-PER) (Pierce from Thermo Fisher Scientific Inc.) in the presence of protease and phosphatase inhibitor cocktails (Sigma-Aldrich). Cell lysates equivalent to 25 μg of total protein were separated on 4–12% Bis-Tris gel (Life Technologies) and transferred to PVDF membrane. Membranes were incubated with primary antibodies: anti-PMEPA1 monoclonal antibody (Novus Biologicals), anti-GSTP1 polyclonal antibody (US Biologicals), anti-β-Actin monoclonal antibody (Cell Signaling), anti-PSA polyclonal antibody (Dako) and anti-AR polyclonal antibody (Santa Cruz Biotech) at 4 °C overnight and were washed before being treated with respective secondary antibodies (GE Healthcare Biosciences). Western blots were visualized by the Amersham ECL western blot detection reagent (GE Healthcare Biosciences).Citation45
Statistical analysis
Seventy-seven CaP patients (42 CA and 35 AA) who underwent RP for primary treatment were analyzed for PMEPA1 and GSTP1 methylation status. The cohort was designed based on our initial observations indicating higher frequency of PMEPA1 methylation in CA patients (42 CA and 8 AA). This minimum sample size (n = 77) was determined by statistical power calculation (90% power with 0.05 α, two sided chi square test). RNA and DNA were analyzed from the same specimens. PMEPA1 mRNA expression was normalized to GAPDH and reported as fold change between matched normal and tumor pairs. PMEPA1 and GSTP1 genomic DNA methylation data were reported as present or absent. Distributions of other clinico-pathological variables were examined by using Chi-square or Fisher exact tests.
Chi-square test was used to evaluate the association of PMEPA1 and GSTP1 methylation and PMEPA1 expression with race/ethnicity. Chi-square test was also used to examine the association of PMEPA1 methylation with PMEPA1 expression. Association of the combination of PMEPA1 methylation and expression groups (methylation-/high expression, methylation+/low expression) with race was tested by using Fisher’s exact test.
| Abbreviations: | ||
| PMEPA1 | = | prostate transmembrane protein androgen induced 1 |
| AR | = | androgen receptor |
| CaP | = | cancer of the prostate |
| CA | = | Caucasian American |
| AA | = | African American |
| LCM | = | laser capture microdissection |
| SMAD | = | Mothers against decapentaplegic homolog |
| TGF-β | = | transforming growth factor-beta |
| PI3K | = | phosphatidylinositol-3-kinase |
Additional material
Download Zip (698.8 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by research grants RO1CA106653 to SS from the National Cancer Institute, by the DoD CDMRP PC100700 to AD and SS, and by the Center for Prostate Disease Research, a program of the Henry M. Jackson Foundation for the Advancement of Military Medicine Program Fund HU0001-10-2-0002 to D.G.M. The authors would like to thank Dr. Jennifer Cullen for advice on evaluation of clinico-pathological data, Mr Stephen Doyle for his assistance with the art and graphics and Ms London Tony for the CPDR laboratory support.
Disclaimer
The views expressed in this manuscript are those of the authors and do not reflect the official policy of the Department of the Army, Department of Defense or the US Government.
References
- Berman DM, Rodriguez R, Veltri RW. Development, Molecular Biology, and Physiology of the Prostate. In: Wein AJ, Kavoussi LR, Novick AC, Partin AW, Peters CA. Campbell-Walsh Urology 10th Edition Review. Philadelphia: Saunders, Elsevier Inc., 2012: 2533-2569.e11.
- Schmidt LJ, Tindall DJ. Androgen receptor: past, present and future. Curr Drug Targets 2013; 14:401 - 7; http://dx.doi.org/10.2174/1389450111314040002; PMID: 23565753
- Dobi A, Sreenath T, Srivastava S. Androgen-Dependent Oncogenic Activation of ETS Transcription Factors by Recurrent Gene Fusions in Prostate Cancer: Biological and Clinical Implications. In: Wang Z. Androgen-Responsive Genes in Prostate Cancer- Regulation, Function and Clinical Applications. New York: Springer Science; 2013:307-28.
- Wen S, Niu Y, Lee SO, Chang C. Androgen receptor (AR) positive vs negative roles in prostate cancer cell deaths including apoptosis, anoikis, entosis, necrosis and autophagic cell death. Cancer Treat Rev 2014; 40:31 40; http://dx.doi.org/10.1016/j.ctrv.2013.07.008; PMID: 23993415
- Madan RA, Shah AA, Dahut WL. Is it time to reevaluate definitive therapy in prostate cancer?. J Natl Cancer Inst 2013; 105:683 - 5; http://dx.doi.org/10.1093/jnci/djt094; PMID: 23615688
- Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009; 324:787 - 90; http://dx.doi.org/10.1126/science.1168175; PMID: 19359544
- Mohler JL, Titus MA, Wilson EM. Potential prostate cancer drug target: bioactivation of androstanediol by conversion to dihydrotestosterone. Clin Cancer Res 2011; 17:5844 - 9; http://dx.doi.org/10.1158/1078-0432.CCR-11-0644; PMID: 21705451
- Chang KH, Li R, Kuri B, Lotan Y, Roehrborn CG, Liu J, Vessella R, Nelson P, Kapur P, Guo X, et al. A gain-of-function mutation in DHT synthesis in CRPC. Cell 2013; 154:1074 - 84; http://dx.doi.org/10.1016/j.cell.2013.07.029; PMID: 23993097
- Eichholz A, Ferraldeschi R, Attard G, de Bono JS. Putting the brakes on continued androgen receptor signaling in castration-resistant prostate cancer. Mol Cell Endocrinol 2012; 360:68 - 75; http://dx.doi.org/10.1016/j.mce.2011.09.038; PMID: 21986558
- Dobi A, Furusato B, Shaheduzzaman S, Chen Y, Vahey M, Nydam T, Sesterhenn IA, McLeod DG, Petrovics G, Srivastava S. ERG expression levels in prostate tumors reflect functional status of the androgen receptor (AR) as a consequence of fusion of ERG with AR regulated gene promoters. The Open Cancer J 2010; 3:101 - 8
- Gsponer JR, Braun M, Scheble VJ, Zellweger T, Bachmann A, Perner S, Vlajnic T, Srivastava M, Tan SH, Dobi A, et al. ERG rearrangement and protein expression in the progression to castration-resistant prostate cancer. Prostate Cancer Prostatic Dis 2014; Forthcoming http://dx.doi.org/10.1038/pcan.2013.62; PMID: 24469092
- Xu LL, Su YP, Labiche R, Segawa T, Shanmugam N, McLeod DG, Moul JW, Srivastava S. Quantitative expression profile of androgen-regulated genes in prostate cancer cells and identification of prostate-specific genes. Int J Cancer 2001; 92:322 - 8; http://dx.doi.org/10.1002/ijc.1196; PMID: 11291065
- Xu LL, Shanmugam N, Segawa T, Sesterhenn IA, McLeod DG, Moul JW, Srivastava S. A novel androgen-regulated gene, PMEPA1, located on chromosome 20q13 exhibits high level expression in prostate. Genomics 2000; 66:257 - 63; http://dx.doi.org/10.1006/geno.2000.6214; PMID: 10873380
- Masuda K, Werner T, Maheshwari S, Frisch M, Oh S, Petrovics G, May K, Srikantan V, Srivastava S, Dobi A. Androgen receptor binding sites identified by a GREF_GATA model. J Mol Biol 2005; 353:763 - 71; http://dx.doi.org/10.1016/j.jmb.2005.09.009; PMID: 16213525
- Xu LL, Shi Y, Petrovics G, Sun C, Makarem M, Zhang W, Sesterhenn IA, McLeod DG, Sun L, Moul JW, et al. PMEPA1, an androgen-regulated NEDD4-binding protein, exhibits cell growth inhibitory function and decreased expression during prostate cancer progression. Cancer Res 2003; 63:4299 - 304; PMID: 12907594
- Li H, Xu LL, Masuda K, Raymundo E, McLeod DG, Dobi A, Srivastava S. A feedback loop between the androgen receptor and a NEDD4-binding protein, PMEPA1, in prostate cancer cells. J Biol Chem 2008; 283:28988 - 95; http://dx.doi.org/10.1074/jbc.M710528200; PMID: 18703514
- Brunschwig EB, Wilson K, Mack D, Dawson D, Lawrence E, Willson JK, Lu S, Nosrati A, Rerko RM, Swinler S, et al. PMEPA1, a transforming growth factor-beta-induced marker of terminal colonocyte differentiation whose expression is maintained in primary and metastatic colon cancer. Cancer Res 2003; 63:1568 - 75; PMID: 12670906
- Watanabe Y, Itoh S, Goto T, Ohnishi E, Inamitsu M, Itoh F, Satoh K, Wiercinska E, Yang W, Shi L, et al. TMEPAI, a transmembrane TGF-beta-inducible protein, sequesters Smad proteins from active participation in TGF-beta signaling. Mol Cell 2010; 37:123 - 34; http://dx.doi.org/10.1016/j.molcel.2009.10.028; PMID: 20129061
- Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011; 19:575 - 86; http://dx.doi.org/10.1016/j.ccr.2011.04.008; PMID: 21575859
- Liu R, Zhou Z, Huang J, Chen C. PMEPA1 promotes androgen receptor-negative prostate cell proliferation through suppressing the Smad3/4-c-Myc-p21 Cip1 signaling pathway. J Pathol 2011; 223:683 - 94; http://dx.doi.org/10.1002/path.2834; PMID: 21341270
- Richter E, Masuda K, Cook C, Ehrich M, Tadese AY, Li H, Owusu A, Srivastava S, Dobi A. A role for DNA methylation in regulating the growth suppressor PMEPA1 gene in prostate cancer. Epigenetics 2007; 2:100 - 9; http://dx.doi.org/10.4161/epi.2.2.4611; PMID: 18174752
- Lin X, Tascilar M, Lee WH, Vles WJ, Lee BH, Veeraswamy R, Asgari K, Freije D, van Rees B, Gage WR, et al. GSTP1 CpG island hypermethylation is responsible for the absence of GSTP1 expression in human prostate cancer cells. Am J Pathol 2001; 159:1815 - 26; http://dx.doi.org/10.1016/S0002-9440(10)63028-3; PMID: 11696442
- Yegnasubramanian S, Lin X, Haffner MC, DeMarzo AM, Nelson WG. Combination of methylated-DNA precipitation and methylation-sensitive restriction enzymes (COMPARE-MS) for the rapid, sensitive and quantitative detection of DNA methylation. Nucleic Acids Res 2006; 34:e19; http://dx.doi.org/10.1093/nar/gnj022; PMID: 16473842
- Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 2004; 32:e38; http://dx.doi.org/10.1093/nar/gnh032; PMID: 14973332
- Nakayama M, Gonzalgo ML, Yegnasubramanian S, Lin X, De Marzo AM, Nelson WG. GSTP1 CpG island hypermethylation as a molecular biomarker for prostate cancer. J Cell Biochem 2004; 91:540 - 52; http://dx.doi.org/10.1002/jcb.10740; PMID: 14755684
- Rosen P, Pfister D, Young D, Petrovics G, Chen Y, Cullen J, Böhm D, Perner S, Dobi A, McLeod DG, et al. Differences in frequency of ERG oncoprotein expression between index tumors of Caucasian and African American patients with prostate cancer. Urology 2012; 80:749 - 53; http://dx.doi.org/10.1016/j.urology.2012.07.001; PMID: 22950997
- Farrell J, Petrovics G, McLeod DG, Srivastava S. Genetic and molecular differences in prostate carcinogenesis between African American and Caucasian American men. Int J Mol Sci 2013; 14:15510 - 31; http://dx.doi.org/10.3390/ijms140815510; PMID: 23892597
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin 2011; 61:212 - 36; http://dx.doi.org/10.3322/caac.20121; PMID: 21685461
- Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, De Marzo AM, Isaacs WB, Nelson WG. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res 2004; 64:1975 - 86; http://dx.doi.org/10.1158/0008-5472.CAN-03-3972; PMID: 15026333
- Aryee MJ, Liu W, Engelmann JC, Nuhn P, Gurel M, Haffner MC, Esopi D, Irizarry RA, Getzenberg RH, Nelson WG, et al. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med 2013; 5:169ra10; http://dx.doi.org/10.1126/scitranslmed.3005211; PMID: 23345608
- Shaheduzzaman S, Vishwanath A, Furusato B, Cullen J, Chen Y, Bañez L, Nau M, Ravindranath L, Kim KH, Mohammed A, et al. Silencing of Lactotransferrin expression by methylation in prostate cancer progression. Cancer Biol Ther 2007; 6:1088 - 95; http://dx.doi.org/10.4161/cbt.6.7.4327; PMID: 17568188
- Nelson WG, De Marzo AM, Yegnasubramanian S. Epigenetic alterations in human prostate cancers. Endocrinology 2009; 150:3991 - 4002; http://dx.doi.org/10.1210/en.2009-0573; PMID: 19520778
- Martin DN, Starks AM, Ambs S. Biological determinants of health disparities in prostate cancer. Curr Opin Oncol 2013; 25:235 - 41; PMID: 23399519
- Enokida H, Shiina H, Urakami S, Igawa M, Ogishima T, Pookot D, Li LC, Tabatabai ZL, Kawahara M, Nakagawa M, et al. Ethnic group-related differences in CpG hypermethylation of the GSTP1 gene promoter among African-American, Caucasian and Asian patients with prostate cancer. Int J Cancer 2005; 116:174 - 81; http://dx.doi.org/10.1002/ijc.21017; PMID: 15800905
- Magi-Galluzzi C, Tsusuki T, Elson P, Simmerman K, LaFargue C, Esgueva R, Klein E, Rubin MA, Zhou M. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate 2011; 71:489 - 97; http://dx.doi.org/10.1002/pros.21265; PMID: 20878952
- Mao X, Yu Y, Boyd LK, Ren G, Lin D, Chaplin T, Kudahetti SC, Stankiewicz E, Xue L, Beltran L, et al. Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res 2010; 70:5207 - 12; http://dx.doi.org/10.1158/0008-5472.CAN-09-4074; PMID: 20516122
- Blattner M, Lee DJ, O’Reilly C, Park K, Macdonald TY, Khani F, Turner KR, Chiu YL, Wild PJ, Dolgalev I, et al. SPOP Mutations in Prostate Cancer across Demographically Diverse Patient Cohorts. Neoplasia 2014; 16:14 - 20; PMID: 24563616
- Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, Luna R, Rashid A, Shen L, Estecio MR, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res 2006; 66:5495 - 503; http://dx.doi.org/10.1158/0008-5472.CAN-05-2385; PMID: 16707479
- Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol 2009; 49:243 - 63; http://dx.doi.org/10.1146/annurev-pharmtox-061008-103102; PMID: 18851683
- Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, Nau M, Ravindranath L, Chen Y, Dobi A, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene 2005; 24:3847 - 52; http://dx.doi.org/10.1038/sj.onc.1208518; PMID: 15750627
- Hu Y, Dobi A, Sreenath T, Cook C, Tadase AY, Ravindranath L, Cullen J, Furusato B, Chen Y, Thangapazham RL, et al. Delineation of TMPRSS2-ERG splice variants in prostate cancer. Clin Cancer Res 2008; 14:4719 - 25; http://dx.doi.org/10.1158/1078-0432.CCR-08-0531; PMID: 18676740
- Sterbis JR, Gao C, Furusato B, Chen Y, Shaheduzzaman S, Ravindranath L, Osborn DJ, Rosner IL, Dobi A, McLeod DG, et al. Higher expression of the androgen-regulated gene PSA/HK3 mRNA in prostate cancer tissues predicts biochemical recurrence-free survival. Clin Cancer Res 2008; 14:758 - 63; http://dx.doi.org/10.1158/1078-0432.CCR-07-1356; PMID: 18245536
- Derosa CA, Furusato B, Shaheduzzaman S, Srikantan V, Wang Z, Chen Y, Seifert M, Ravindranath L, Young D, Nau M, et al. Elevated osteonectin/SPARC expression in primary prostate cancer predicts metastatic progression. Prostate Cancer Prostatic Dis 2012; 15:150 - 6; http://dx.doi.org/10.1038/pcan.2011.61; PMID: 22343836
- Sun M, Srikantan V, Ma L, Li J, Zhang W, Petrovics G, Makarem M, Strovel JW, Horrigan SG, Augustus M, et al. Characterization of frequently deleted 6q locus in prostate cancer. DNA Cell Biol 2006; 25:597 - 607; http://dx.doi.org/10.1089/dna.2006.25.597; PMID: 17132090
- Mohamed AA, Tan SH, Sun C, Shaheduzzaman S, Hu Y, Petrovics G, Chen Y, Sesterhenn IA, Li H, Sreenath T, et al. ERG oncogene modulates prostaglandin signaling in prostate cancer cells. Cancer Biol Ther 2011; 11:410 - 7; http://dx.doi.org/10.4161/cbt.11.4.14180; PMID: 21178489