Abstract
Altered expression of microRNA (miRNA) can significantly contribute to cancer development and recent studies have shown that a number of miRNAs may be regulated by DNA methylation. Through a candidate gene approach, we identified MIR941 and MIR1247 to be transcriptionally silenced by DNA hypermethylation in several gastric cancer cell lines. We confirmed that these miRNAs are also densely methylated in primary gastric cancers but not in normal gastric tissues. In addition, we demonstrated that ectopic expression of these two miRNAs in AGS gastric cancer cells resulted in suppression of growth and migration. Furthermore, we tested genes predicted to be the targets of MIR941 and MIR1247 and identified 7 and 6 genes, whose expressions were significantly downregulated by transfection of MIR941 and MIR1247 mimics, respectively, in gastric cancer cell lines. Some of these genes are known to promote proliferation and invasion, phenotypes we observed upon ectopic expression of the two miRNAs. Thus, we examined these candidates more closely and found that downregulation of mRNA corresponded to a decrease in protein levels (observed by western blot). Our study provides unequivocal evidence that MIR941 and MIR1247 are transcriptionally regulated by DNA methylation in gastric cancer and that they have tumor suppressor properties through their inhibition of key cancer promoting genes in this context.
Introduction
microRNAs (miRNAs) are small 19–25 nucleotide-long, non-coding RNAs that regulate gene expression by inducing translational inhibition or direct degradation of target mRNAs through base pairing to partially complementary sites. It is estimated that 30% of the human genome is under miRNA regulation, with one miRNA able to modulate posttranscriptionally hundreds of target genes.Citation1 miRNAs are first transcribed as long pri-miRNA, then processed into intermediate pre-miRNAs, and finally cleaved by Dicer to form mature miRNAs.Citation2-Citation4 Many miRNAs are highly conserved among species and play critical roles in a variety of biological processes, including development, differentiation, cellular proliferation, apoptosis, and stem cell maintenance.Citation5,Citation6 It has also become increasingly clear that abnormalities in miRNA activity contribute to human disease pathogenesis.Citation5 In particular, dysregulation of miRNAs in cancer has emerged as an important step during carcinogenesis. Significant miRNA expression differences between cancer and normal cells have been reported in chronic lymphocytic leukemia and several solid tumors including colon, lung, breast, gastric, and liver cancers.Citation7-Citation12 Furthermore, a variety of mechanisms, including genomic deletion, mutation, epigenetic silencing, and miRNA processing defects, are implicated in the disruption of miRNAs biogenesis in cancers.Citation7,Citation13-Citation15
Of particular interest to us is the epigenetic regulation of miRNA transcription in gastric cancer. Gastric cancer is the 4th most common cancer and the second-highest cause of death worldwide. Most importantly, it is the leading cause of cancer deaths in Asian countries.Citation16,Citation17 Due to the lack of symptoms in early stages, gastric cancer is usually diagnosed at advanced stages when overall 5-y survival rate is approximately 20%.16Early detection remains a critical challenge for the management of gastric cancer, and we hypothesized that disruptions of miRNA expression by epigenetic mechanisms may be an early event during gastric cancer carcinogenesis. If true, these epigenetic changes may be developed into diagnostic tests to fulfill the unmet clinical need while the affected miRNAs can guide our understanding of the biology. Epigenetic gene silencing due to promoter CpG island hypermethylation is a pervasive mechanism for silencing tumor suppressor genes in gastric cancer.Citation18-Citation21 Several reports have demonstrated that miRNA genes may also become targets of aberrant epigenetic silencing in cancer.Citation14,Citation22,Citation23 Our group has previously classified novel DNA methylation regulated-miRNAs in colon cancer cells by combining unbiased global DNA methylation and miRNA expression analyses.Citation14,Citation24 In the present study, we aimed to test select miRNAs for their epigenetic silencing in gastric cancer and to evaluate the phenotypic impact of such epigenetic silencing on gastric cancer cells.
In short, we found MIR941 and MIR1247 to harbor dense DNA methylation in gastric cancer cell lines and gastric adenocarcinomas and that such aberrant DNA hypermethylation correlated with robust transcriptional silencing of the two miRNAs. We also determined that ectopic expression of MIR941 and MIR1247 resulted in decreased cell proliferation and migration. Finally, we identified several target mRNAs, including the lysine-specific demethylase 6B (KDM6B), TAO kinase 1 (TAOK1), PR domain containing 16 (PRDM16), retinoic acid receptor α (RARA), syntaxin 1B (STX1B), and regulator of chromosome condensation 2 (RCC2), for these two miRNAs in gastric cancer cell lines. These results are the first to define epigenetic disruption for the tumor suppressors MIR941 and MIR1247 in early stage gastric cancer and support future investigations into the functions of these miRNAs in gastric cancer carcinogenesis.
Results
Correlation between DNA methylation and expression of MIR941 and MIR1247 in gastric cancer cell lines
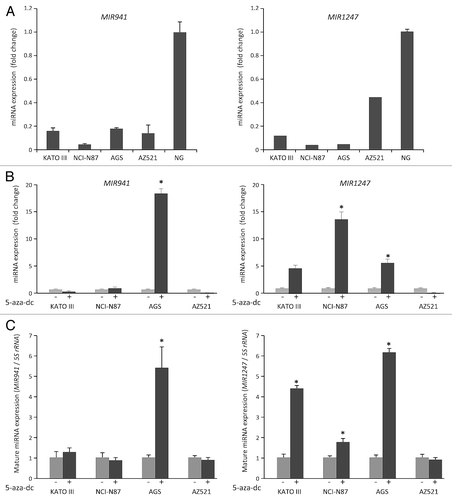
We previously identified a pair of primary miRNAs, pri-MIR941 and pri-MIR1247, to be regulated by DNA methylation in colon cancers using both genome-wide DNA methylation and gene expression profiles for HCT116 cell line and its DNA demethylated isogenic DNA methyltransferase 1 and 3b double knockout (DKO) cell line.Citation24-Citation26 To investigate whether these miRNAs might be regulated similarly in gastric cancer, we first analyzed the expression of MIR941 and MIR1247 in four gastric cancer cell lines and in normal stomach tissue using quantitative real-time RT-PCR (qRT-PCR). Both MIR941 and MIR1247 were downregulated in all cell lines when compared with normal stomach tissue (). We then treated four gastric cancer cell lines, KATO III, NCI-N87, AGS, and AZ521, with the demethylating agent 5-aza-2'-deoxycytidine (5-aza-dC) and performed qRT-PCR analysis for pri-MIR941 and pri-MIR1247 to see if their expressions change after 5-aza-dC treatment in these gastric cancer cells (). We observed increased expression after 5-aza-dC treatment for pri-MIR941 in AGS cells and for pri-MIR1247 in KATO III, NCI-N87, and AGS cells. We also examined whether the expression of mature miRNAs could be restored by 5-aza-dC in the same gastric cancer cell lines (). We confirmed that mature MIR941 and MIR1247 were re-expressed by 5-aza-dC, consistent with the data of primary transcript of MIR941 and MIR1247. Based on the basal expression level and restored expression level by 5-aza-dC, we suspect that CpG islands in promoter regions of both miRNAs are hypermethylated in gastric cancer cell lines.
Figure 1. Analysis of MIR941 and MIR1247 expression in gastric cancer cell lines. (A) Quantitative RT-PCR analysis of expression pattern of pri-MIR941 and pri-1247 in gastric cancer cell lines (KATO III, NCI-N87, AGS, and AZ521) and normal stomach tissues (NG). (B) Expression analysis of miRNAs in gastric cancer cells with 5-aza-dC treatment. RT-PCR was performed to assess pri-miRNAs levels in gastric cancer cell lines (KATO III, NCI-N87, AGS, and AZ521) before and after treatment with 5 μM 5-aza-dC for 72hrs. Dark gray bar indicate 5-aza-dC treatments. (C) Expression of mature MIR941 and MIR1247 in gastric cancer cells with 5-aza-dC treatment. RT-PCR was performed to assess mature miRNAs levels after treatment with 5 μM 5-aza-dC for 72 h. Dark gray bars indicate 5-aza-dC treatments. Relative pri-mRNAs levels are shown as mean ± standard deviation of three independent experiments. *Indicates significant increase in pri-miRNA expression after 5-aza-dC treatment (*P < 0.05).
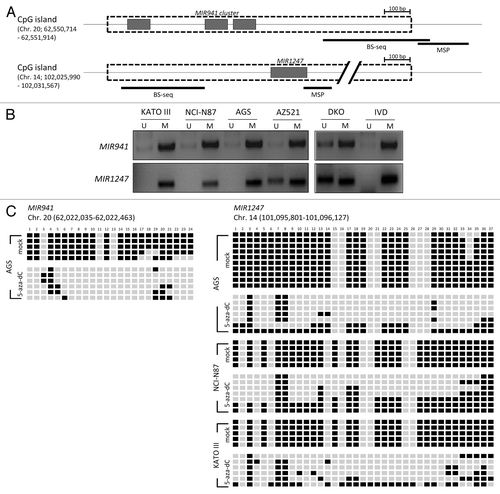
As previously reported, MIR941 and MIR1247 each has a CpG island in the proximal region ().Citation24 We next asked whether DNA methylation in this region is responsible for the silencing of MIR941 and MIR1247 in gastric cancer cells. Methylation specific PCR analysis showed that the CpG island was methylated in most of the gastric cancer cell lines we tested (). To confirm this, we assessed DNA methylation status in the proximal regions of MIR941 and MIR1247 by bisulfite sequencing analysis (). Both miRNAs exhibited dense DNA methylation in gastric cancer cells and demethylation was observed in cells (AGS for MIR941 and KATO III, NCI-N87, and AGS for MIR1247) upon 5-aza-dC treatment, strongly supporting that the re-expression observed was due to DNA demethylation (). Therefore, we concluded that these two miRNAs are likely epigenetically silenced in gastric cancer cells.
Figure 2. Methylation analysis of MIR941 and MIR1247 in gastric cancer cell lines. (A) Schematic representation of MIR941 and MIR1247 CpG island (dotted box). Both miRNAs are embedded into the CpG island (gray box). The regions analyzed using methylation specific PCR (MSP), bisulfite sequencing are indicated by black bars below the CpG island. (B) MSP analysis of MIR941 and MIR1247 in four gastric cancer cell lines. In vitro methylated DNA (IVD) and DKO cells were used as positive and negative controls, respectively. U and M indicates unmethylation and methylation signals, respectively. (C) Representative bisulfite sequencing analysis was performed for MIR941 and MIR1247 in gastric cancer cell lines (AGS, NCI-N78, and KATO III) as well as cells treated with 5-aza-dC. Each box represents a CpG dinucleotide. Black boxes represent methylated cytosines while white boxes represent unmethylated cytosines.
DNA methylation status of MIR941 and MIR1247 in gastric cancer patients
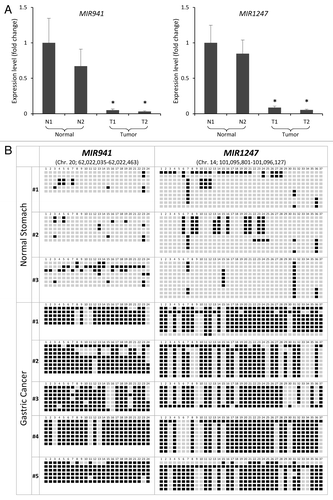
We analyzed the expression of MIR941 and MIR1247 in normal gastric mucosae from healthy individuals comparing with tumors from gastric cancer patients by quantitative RT-PCR (qRT-PCR). We found significant downregulation of MIR941 and MIR1247 expression in the tumor tissues, as compared with normal gastric mucosae (). To verify the correlation between DNA methylation and silencing of miRNAs in vivo, we next analyzed the methylation status in gastric cancer and normal gastric tissue specimens by bisulfite sequencing analysis (). Compared with normal gastric tissues, we observed a significant increase in methylation at the MIR941 and MIR1247 loci in gastric cancer samples. Normal samples showed less than 20% and 17% total DNA methylation for MIR941 and MIR1247, respectively, while cancer samples harbored greater than 80% (MIR941) and 63% (MIR1247) DNA methylation. We expanded our analyses to include an additional 15 gastric cancer patient samples to evaluate the frequency of methylation of MIR941 and MIR1247. Most samples, except for two, showed methylation at MIR941 and MIR1247 (Fig. S1). Since our main interest lies in identifying early events of aberrant methylation and silencing of miRNAs, all cancer specimens were early stage adenocarcinoma (10 adenomas and 5 adenocarcinomas). These data demonstrated cancer-specific DNA methylation of MIR941 and MIR1247 in primary gastric tumor samples and hinted at a cancer-promoting role for the aberrant silencing of these miRNAs in gastric cancer development.
Figure 3. Correlation between expression and methylation of MIR941 and MIR1247 in gastric cancer patient samples. (A) Expression level of pri-MIR941 and pri-1247 in primary gastric tumor tissues compared with normal gastric mucosae from healthy individuals by qRT-PCR. * indicates significant decrease in miRNA expression (*P < 0.05). (B) Representative bisulfite sequencing analysis was performed for MIR941 and MIR1247 in three normal gastric tissues and five gastric adenocarcinomas. Each box represents a CpG dinucleotide. Black boxes represent methylated cytosines while white boxes represent unmethylated cytosines.
Functional analysis of MIR941 and MIR1247 in gastric cancer cells
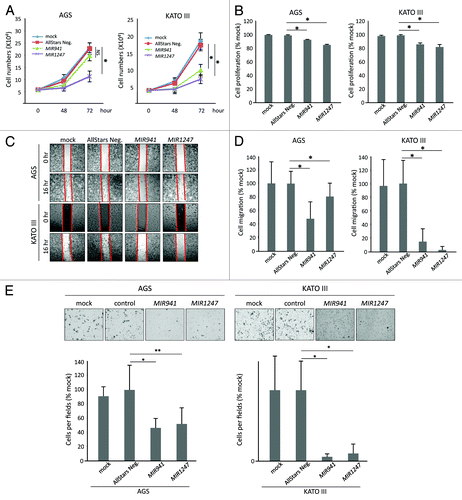
Having established that MIR941 and MIR1247 are hypermethylated in gastric cancer, we wondered if these epigenetically silenced miRNAs have tumor suppressive properties in gastric cancer cells. To test this, we transfected AGS and KATO III gastric cancer cells with miRNA mimics of MIR941, MIR1247, and non-targeting negative controls (AllStars Neg.). First, we performed growth curve analysis () and MTT assay () to assess cell growth and metabolic fitness, respectively. Ectopic expression of MIR1247 significantly decreased growth and metabolism of AGS and KATO III cells when compared with controls. Cell growth was inhibited by 55% in both cell lines at 72 h (), while the MTT assay results showed a 15% (AGS) and 17% (KATO III) decrease compared with control (). In the case of MIR941, cell growth was not significantly hindered () but a measurable decrement in MTT assay readout was observed in AGS cells (). In KATO III cells, ectopic expression of MIR941 inhibited cell growth by 40% at 72 h and resulted in 13% decrease in MTT assay readout when compare with controls (). Furthermore, we performed wound healing (), migration (), and matrigel invasion () assays to analyze the effects of MIR941 and MIR1247 expression on gastric cancer cells migration and invasion. We found that wound closure by AGS and KATO III cells transfected with the negative control was almost complete by 16 h and comparable to the mock-treated controls. On the contrary, MIR941 and MIR1247 mimics transfected cells migrated into the wound at a much slower rate, which is consistent with the migration result (). Finally, we also observed significant inhibition of cell invasion by MIR941 and MIR1247 in AGS cells when compared with the negative controls (). Taken together, these results strongly suggest that MIR941 and MIR1247 have tumor suppressor functions in gastric cancer cells.
Figure 4. Functional analyses of MIR941 and MIR1247 in AGS and KATO III cells. (A) Growth curves and (B) MTT assays of AGS mock treated cells and AGS cells transfected with non-targeting negative control miRNA (All Star neg.), MIR941, or MIR1247 mimics. (C) Wound healing assay for AGS cells transfected with mock, non-targeting negative controls miRNA (All Star neg.), MIR941, or MIR1247 mimics. Photographs were taken at 16 h later after wounding. (D) The bar graph represents the quantity of migrated cells using transwell migration assay with AGS cells transfected with mock, non-targeting negative controls miRNA (All Star neg.), MIR941, or MIR1247 mimics. Quantity of migrated cells represents the mean of 3 random microscopic fields per membrane and error bars represent the SDs. (E) Transwell invasion assay using AGS cells transfected with mock, non-targeting negative controls miRNA (All Star neg.), MIR941, or MIR1247 mimics. Representative field of invasive cells on the membrane are shown. The Bar graph represents the mean of 3 random microscopic fields per membrane and error bars represent the SDs.
Target genes regulated by MIR941 and MIR1247 in gastric cancer
Defining biological targets of miRNAs remains challenging since each miRNA can regulate multiple gene targets through imperfect base pairing.Citation27,Citation28 To help us identify potential target genes of mature MIR941 and MIR1247 in gastric cancer, we utilized the target prediction algorithm “TargetScan” to generate a list of candidates for empirical testing.Citation29 TargetScan predicted 10 and 13 high confidence target genes for MIR941 and MIR1247, respectively, (). We then directly analyzed the mRNA levels of these candidate target genes using real-time RT-PCR in AGS and KATO III gastric cancer cells upon transfection of the respective miRNA mimics (). We found that MIR941 mimics consistently depleted mRNA levels of lysine-specific methyltransferase 2D (KMT2D), TAO kinase 1 (TAOK1), and lysine-specific demethylase 6B (KDM6B) in the two cell lines (). Syntaxin 1B (STX1B), PHD finger protein 21A (PHF21A), and retinoic acid receptor α (RARA), were reproducibly downregulated by MIR1247 mimics in both AGS and KATO III cells (). These candidates are likely to be bona fide targets of MIR941 and MIR1247. Additionally, PR domain containing 16 (PRDM16), immediate early response 5-like (IER5L), nuclear factor I/X (NFIX), and RAS-like family 10 member B (RASL10B) expressions were reduced by MIR941 mimics in either AGS or KATO III, while regulator of chromosome condensation 2 (RCC2), zinc finger-and BTB domain-containing protein 46 (ZBTB46), and molybdenum cofactor synthesis gene 1 (MOCS1) were diminished by MIR1247 mimics in one of the cell lines.
Table1. Target gene list of MIR941 and MIR1247
Figure 5.Candidate target genes are downregulated by MIR941 and MIR1247 mimics. (A and B) Real-time RT-PCR was performed in AGS and KATO III cells transfected with non-targeting negative control miRNA (All Star neg.), MIR941 and MIR1247 mimics with target genes listed in . *Indicates statistically significance decrease of gene expression level compared with control (P < 0.05). (C and D) Western blot analysis of target genes in AGS and KATO III cells transfected with MIR941 and MIR1247, or negative control.
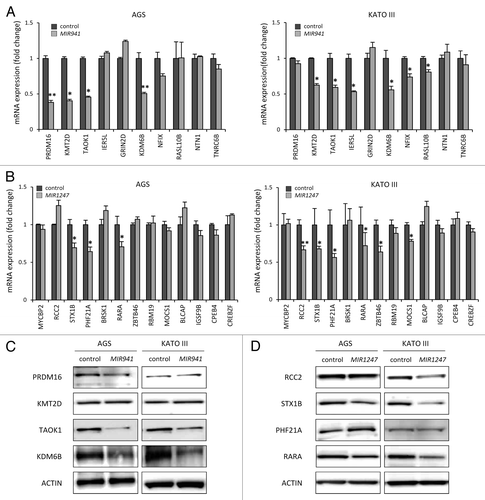
All together, 7 out of 10 predicted targets for MIR941 and 6 out of 13 for MIR1247 showed some evidence of being authentic targets in our study system. To further validate our observations, we focused on KDM6B, KMT2D, TAOK1, and PRDM16 for MIR941, and RARA, STX1B, PHF21A, and RCC2 for MIR1247, based on the significant reduction in their mRNA levels upon exposure to ectopic expression of the respective miRNAs in both AGS and KATO III cell lines. We tested cellular levels of these proteins by western blot analysis in AGS and KATO III cell lines and observed that MIR941 mimic reduced TAOK1 and KDM6B protein levels concordantly with the detected mRNA depletion (). In MIR1247 mimics-transfected cells, decreased protein levels of RARA, STX1B, and RCC2 paralleled the detected mRNA depletion (). These findings support that epigenetically regulated MIR941 and MIR1247 modulate critical pathways that influence gastric cancer phenotypes and have tumor suppressor-like functions.
Although there are a handful of target genes for MIR941 and MIR1247, we were interested in how these genes are associated within the cellular pathway network. Using our target genes, we therefore analyzed well curated databases (GO and KEGG) that include a variety biological processes, molecular functions, and cellular components (Fig. S2). The possible roles of target genes regulated by MIR941 and MIR1247 can be hypothesized by looking at the various signaling pathways predicted by KEGG pathway (listed in Table S1). Using Cytoscape, we could visualize these genes within their cellular pathway network (Fig. S3). Collectively, these data highly suggested that MIR941 and MIR1247 target genes play important roles in regulatory events.
Discussion
microRNAs are increasingly recognized as key players in cancer development and progression. They have been proposed as tumor suppressors, whose expressions have anti-proliferative or pro-apoptotic effects on several different types of cancers. For example, ectopic expression of let-7 in lung cancer cell lines directly suppresses growth in vitro.Citation9 MIR15a and MIR16–1 represent additional miRNAs with tumor suppressing activities that target BCL2 protein expression in chronic lymphocytic leukemia, and expression of these two miRNAs resulted in activation of the intrinsic apoptotic pathway and ultimate cell death.Citation30 In the current study, we focused on testing two previously reported colon cancer-specific DNA hypermethylated miRNAs, MIR941 and MIR1247, to determine if they may be similarly disrupted in gastric cancer.
Dysregulation of miRNA expression is pervasive in many cancers, and epigenetic mechanisms are key mediators underlying the downregulation of tumor-suppressor type miRNA expression. MIR127, MIR9, MIR148, MIR203, MIR340, MIR18, and MIR34b/c are several examples of DNA methylation regulated miRNAs.Citation14,Citation23,Citation31-Citation33 In the case of MIR34b/c, this locus has been reported to be epigenetically regulated in several cancersCitation31,Citation34 Recently, MIR34b/c was shown to be a tumor suppressor mediating p53 activation in both colon and gastric cancers.Citation23,Citation33,Citation35 In addition, methylation of MIR124 families was initially described in colorectal cancer and was subsequently reported in tumors or other origins.Citation22 We postulated that MIR941 and MIR1247, like the examples above, may be commonly DNA hypermethylated in many cancer types, including gastric cancer. Importantly, even though the two miRNAs studied here, MIR941 and MIR1247, were previously identified as hypermethylated miRNAs in colon cancer, to our knowledge, epigenetic regulation by DNA methylation and functional significance of these two miRNAs in other cancers have not yet been investigated. Our observations here would support this hypothesis and suggest further investigation into the regulation and biological functions of these miRNAs in other cancer types.
From a clinical point of view, miRNAs have a great potential at the diagnostic and therapeutic levels. A previous genome-wide study using microarray analysis highlights a general downregulation of miRNAs in tumors when compared with normal tissues.Citation36 In clear cell renal cancer, an miRNA signature can distinguish between metastatic and non-metastatic tumors. The 5-y survival rate of patients with primary metastasis is 10%, as compared with 70–90% in non-metastatic patients, which suggest that the miRNAs signature is a powerful tool for early diagnosis.Citation37 Furthermore, the use of miRNA gene hypermethylation to improve cancer diagnosis has been extended recently to include the use of samples that can be obtained non-invasively, such as serum,Citation38 feces,Citation39 and colorectal mucosal wash fluids.Citation40
In terms of the therapeutic potential of miRNAs, several strategies have been proposed to restore the function of tumor suppressor miRNAs that are downregulated in cancer.Citation41 This reexpression may be achievable with the use of DNA demethylating agents and histone deacetylase inhibitors for epigenetically inactivated tumor suppressor miRNAs.Citation14
In this study, we analyzed five adenoma patient samples, which are early gastric carcinoma obtained by endoscopic mucosal resection or endoscopic submucosal dissection, and found significant DNA methylation in both MIR941 and MIR1247 loci. Such methylation is absent in normal stomach tissues, demonstrating a gastric cancer-specific methylation pattern, as well as suggesting that MIR941 and/or MIR1247 could be useful methylation biomarkers for detecting early stage gastric cancer. Based on MSP analysis, CpG islands of MIR941 and MIR1247 are frequently methylated in a small cohort of gastric tumors. Further studies containing large numbers of gastric cancer patient samples will be needed to accurately determine the utility of MIR941 and MIR1247 methylation as early detection biomarkers.
Using the AGS and KATO III gastric cancer cell models, we determined that ectopic expression of either MIR941 or MIR1247 led to reduced cell growth, proliferation, and invasion, suggesting that these miRNAs behave like tumor suppressors in this context. Finally, we assayed the transcript levels of predicted targets for MIR941 and MIR1247 in AGS and KATO III cells transfected with miRNA mimics. Based on the qRT-PCR results, 7 and 6 distinct target genes could be validated for each miRNA, respectively, in at least one gastric cancer cell line. For some of these target genes, we further validated that the protein levels were reduced upon transfection of MIR941 mimics (KDM6B and TAOK1) or MIR1247 mimics (STX1B, RARA, and RCC) in both AGS and KATO III cells.
We searched the literature to understand our targets of MIR941 and MIR1247 in terms of their cellular functions. KDM6B and RARA as targets of MIR941 and MIR1247 have well-established cancer relevance. KDM6B, also known as JMJ3D, is a histone H3 lysine-27 tri/dimethylation (H3K27me3/2)-specific demethylase.Citation42 H3K27me3/2 are repressive histone modifications important for gene silencing, and removal of such marks by KDM6B allows for activation of genes both during normal development and tissue differentiation as well as during carcinogenesis.Citation43,Citation44 In breast cancer, KDM6B is upregulated by the transforming growth factor β (TGFB) and is necessary to activate gene transcription for TGFB-induced epithelial-mesenchymal transition (EMT).Citation45 We found that KDM6B mRNA and protein levels were depleted in AGS cells transfected with MIR941 mimic. These cells exhibited profound defects in wound healing and matrigel invasion in the absence of significant reduction in cell growth, and such a phenotype is supported by the role of KDM6B in promoting EMT. Our results suggest DNA hypermethylation of MIR941 in gastric cancer as a mechanism to endorse KDM6B expression for propagating a cancer-promoting epigenomic landscape.
RARA belongs to the nuclear retinoic acid receptor family (RAR-α, -β, and -gamma) and is an important regulator of embryonic development and adult cell growth and differentiation.Citation46 The RARA locus is involved in chromosomal translocations in virtually all cases of acute promyelocytic leukemia (APL).Citation47 The most common of which is the t(15:17) translocation that generates the PML-RARA fusion protein that inhibits the function of normal RARs and PML.Citation48-Citation50 All trans retinoic acid (ATRA) and arsenic trioxide (ATO) are the two most important drugs in clinical use for the treatment of early APL, and they work, at least in part, by degrading PML-RARA fusion proteins.Citation51 In our study, ectopic expression of MIR1247 in AGS cells resulted in decreased RARA transcript and protein levels and led to decreased cell proliferation. We also observed diminished migratory/invasion capacity for MIR1247 expressing cells, which may be contributed by reduced cell doubling. These phenotypes are consistent with the biological functions of RARA. Our current work is the first to suggest that RARA can be repressed by epigenetically regulated MIR1247 in gastric cancer.
Among the target genes for which mRNA and protein levels were downregulated by MIR941 or MIR1247, the function associated with cancer or other diseases is not known for TAOK1, STX1B, and RCC2. Taken together, previous reports of cellular function for our target genes are in agreement with our pathway analysis data, which strongly support that target genes epigenetically regulated by MIR941 and MIR1247 are associated with important cellular pathway networks in human cancer. To further clarify the effect of these miRNAs, genome-wide approaches will be necessary in order to assess the global changes in gene expression profiles induced by MIR941 and MIR1247 in gastric cancer cells.
In summary, we have shown that MIR941 and MIR1247 are aberrantly silenced in gastric cancer by DNA hypermethylation, that is, reactivation of these two miRNAs have tumor suppressive effects in gastric cancer cells by our functional analyses. These miRNAs can abrogate known pro-cancer molecular targets, including KDM6B and RARA, and other genes for which a function in cancer has not been yet reported. Molecular targets are associated with multiple biological pathways by our bioinformatics prediction. Based on this, our functional data on MIR941 and MIR1247 target genes is a compelling lead; however, the detailed relationships between target gene candidates and MIR941 and MIR1247 will require further study. More importantly, we demonstrated that such abnormal epigenetic silencing occurs in early gastric cancer development and may be useful in developing new potential therapeutic tools for the management of gastric cancer.
Materials and Methods
Cell culture and 5-aza-dC treatments
Four human gastric cancer cell lines (AGS, KATO III, NCI-N87, and AZ521) were used in this study. NCI-N87 cell line was cultured using RPMI-1640 (WelGENE), AGS was maintained in Ham’s F-12 medium (WelGENE), AZ521 was grown in MEM medium (WelGENE), and KATO III was propagated in IMDM medium (WelGENE). All cell culture media was supplemented with 10% fetal bovine serum (Hyclone) and 1% antibiotic-antimycotic (Gibco). All cell lines were incubated at 37 °C, 20% O2, 5% CO2 condition. To investigate the effect of 5-aza-2’-deoxicytidine (5-aza-dC) treatments, cells were treated with 5 μM 5-aza-dC (Sigma, St. Louis, MO, USA) for 72 h.
Tissue samples
The biospecimens (Tumor DNA samples) for this study were provided by National Biobank of Korea-Kyungpook National University Hospital (KNUH), which is supported by the Ministry of Health, Welfare and affairs. All materials derived from the National Biobank of Korea-KNUH were obtained (with informed consent) under institutional review board (IRB)-approved protocols. For MSP and Bisulfite sequencing analyses, a total 15 primary GC specimens were obtained from endoscopic mucosal resection or endoscopic submucosal dissection (10 male and 5 female; average age 67 y, ranging from 54 to 87; 10 adenomas and 5 adenocarcinomas). For expression analysis, normal gastric mucosae and gastric tumors were obtained by endoscopic mucosal resection from 2 healthy individuals (2 male, 35 y and 58 y) and gastric cancer patients (1 male and 1 female, 56 y and 66 y), respectively. Genomic DNA of normal gastric mucosa (n = 3) from healthy individual were purchased from Biochain (Hayward, USA).
DNA methylation analyses
Primer pairs for methylation analysis were preferentially designed near the putative transcriptional start site (TSS) in the 5′ CpG islands of the genes. All methylation and non-methylation specific primers were designed by MethPrimer. All primer sequences are listed in Table S2. For methylation analyses, DNA was extracted using a phenol-chloroform standard method. A total of 2 µg of DNA was modified by bisulfite with EZ DNA methylation Kit™ (Zymo Research), which promises a > 99% conversion rate (non-methylated C residue to U; protection of methylated cytosines). Methylation analysis of gene promoters was performed using MSP primer pairs located close to the putative transcriptional start site in the 5′ CpG island with 2 μl of bisulfite-treated DNA as template and JumpStart Red Taq DNA Polymerase (Sigma) for amplification as previously described.Citation52 For bisulfite sequencing analysis, Amplicons by PCR were analyzed by 2% agarose gel electrophoresis, purified with Gel Extraction Kit (QIAGEN, GmbH) and cloned using the TOPO TA vector system (Invitrogen). Individual clones were isolated and purified using NucleoSpin Plasmid isolation kit (MN-Macherey-Nagel). Positive clones were sequenced using M13F primer and analyzed for DNA methylation status at each CpG dinucleotide.
Small RNA extraction and cDNA synthesis for mature mRNAs expression
Small RNA containing mature miRNA was extracted by miRNeasy Mini Kit (QIAGEN). RNA quantity was measured using a NanoDrop 2000/2000c instrument (Thermo Scientific), and 1 µg of small RNA was poly A tailing using by Poly(A) Tailing Kit (Ambion). Poly A tailed small RNA was reverse-transcribed using by M-MLV reverse transcriptase kit (Promega) with 10 pmol oligo-dT-Adaptor.
Quantitative real-time RT-PCR
Total RNA was isolated from human gastric cancer cell lines and tissues using TRI-Solution (Bio Science Technology) following the manufacturer’s protocol. RNA quantity was measured using a NanoDrop 2000/2000c instrument (Thermo Scientific), and 1 µg of RNA was reverse-transcribed into cDNA using the iScriptTMcDNA Synthesis kit (BioRad). For expression studies using RT-PCR, primers were designed using the open access program Primer3 (http://frodo.wi.mit.edu/primer3). qPCR was performed on a C1000 Thermal Cycler (BioRad) using the PCR primers listed in Table S2. The expressions of pri-miRNAs and target genes were normalized by actin. The expression of mature miRNAs was normalized by 5S-rRNA.
miRNAs transfection
To ectopically express MIR941 and MIR1247 in gastric cancer cell lines, cells were transfected with 20 nM hsa-miR941 miScript mimic (MSY0004984, QIAGEN), hsa-miR-1247 miScript mimic (MSY0005899, QIAGEN), or Allstars Negative Control siRNA (1027281, QIAGEN) using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions.
Cell proliferation assay and viability assays
Cell proliferation was analyzed using the MTT assay. At 24 h after transfection of miRNA mimics or the negative control, AGS and KATO III cells (2 × 105 cells/well) were re-plated in 6-well plates, and incubated at 37 °C. After 72 h, cells were washed twice with PBS, and 5 mg/ml MTT in PBS was added to each well for 4 h. After removing the MTT solution, a solubilization solution (DMSO/EtOH, 1:1 ratio) was added to each well to dissolve the formazan crystals. The absorbance at 570 nm was measured using a ParadigmTM microplate reader (Beckman Coulter). At 48 h and 72 h after re-plating (5 × 104 cells/well) in 6 well plates, cell numbers were counted.
Wound-healing assay
Cells were plated overnight to achieve a subconfluent cell monolayer in 6-well plates. Then, a scratch was made on the cell layer with a sterile 200 μl pipette tip, and cultures were washed twice with serum-free medium to remove floating cells. Cells were incubated in culture medium. After 16 h, wound healing was visualized by comparing photographs using a Qimaging QI Click Camera system mounted on a phase-contrast Nikon microscope TS100 (Nikon). The distance traveled by the cells was determined by measuring the wound width at time 16 h and subtracting it from the wound width at time 0.
Migration and invasion assay
Cell migration was determined using transwell plates (24well, 8 μm pore size, Corning Costar), and invasion assay was carried using Matrigel-coated invasion chamber (24 well, 8 μm pore size, Corning Costar). The upper chamber contained cells in specific medium with 1% FBS, and the lower chamber contained medium with 10% FBS. Cells were incubated for 16 h at 37 °C in 20% O2, 5% CO2.condition. Non-migrated or non-invasive cells were scraped off the upper membrane with a cotton swab. Migrated or invasive cells remaining on the bottom membrane were counted after staining with Giemsa (Sigma). Photographs were taken using a Qicam image camera system mounted on Nikon ECLIPSE 80i microscope (Nikon).
Western blot analysis
Total cell lysates (20 μg) were loaded onto SDS–PAGE and transferred to PVDF (GE Healthcare Life Sciences). The membranes were blocked with 5% milk dissolved in TBS containing 0.02% Tween 20, and incubated overnight at 4 °C with specific primary antibodies. The membranes were subsequently incubated with specific horseradish peroxidase conjugated secondary antibodies. Protein bands were visualized using a Fusion FX5 system (Vilber Lourmat). The following primary antibodies were used: anti-KDM6B (Abcam), anti-RARα (Santa Cruz Biotechnology), anti-TAOK1 (Abcam), anti-PRDM16 (Abcam), anti-STX1B (Abcam), anti-RCC2 (Abcam), anti-PHF21A (Abcam), anti-KMT2D (Abnova) and anti-β-actin (Sigma) antibodies.
Bioinformatics analysis of miRNA target gene
Target genes for MIR941 and MIR1247 were identified by TargetScanHuman 6.2 (http://www.targetscan.org). Target genes with poorly conserved sites are not shown. Function and signal pathway of target genes indicated by Super-pathway category within GeneCard database (http://www.genecards.org/cgi-bin) that included KEGG (http://www.kegg.jp/kegg/pathway.html) (Table S1).
Statistical analyses
Quantified data are expressed as the mean ± SE values. Significance testing was conducted via the Student t test.
| Abbreviations: | ||
| miRNA | = | microRNA |
| 5-aza-dC | = | 5-aza-2'-deoxycytidine |
| GC | = | gastric cancer |
| qRT-PCR | = | quantitative real-time RT-PCR |
| RARA | = | retinoic acid receptor, alpha |
| KDM6B | = | lysine-specific demethylase 6B |
| TAOK1 | = | TAO kinase 1 |
| STX1B | = | syntaxin 1B |
| PHF21A | = | PHD finger protein 21A |
| PRDM16 | = | PR domain containing 16 |
| IER5L | = | immediate early response 5-like |
| NFIX | = | nuclear factor I/X |
| RASL10B | = | RAS-like family 10 member B |
| RCC2 | = | chromosome condensation 2 |
| ZBTB46 | = | zinc finger-and BTB domain-containing protein 46 |
| MOCS1 | = | molybdenum cofactor synthesis gene 1 |
Additional material
Download Zip (90.8 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was supported by the National R&D program (50596-2014) through the Dongnam Institute of Radiological and Medical Sciences (DIRAMS) funded by the Korean Ministry of Education, Science, and Technology. This work was also supported by the National Research Foundation (NRF) and Ministry of Education, Science, ICT and Future Planning (MSIP), Korean government, through its National Nuclear Technology Program (2013M2A2A7043665).
References
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120:15 - 20; http://dx.doi.org/10.1016/j.cell.2004.12.035; PMID: 15652477
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004; 23:4051 - 60; http://dx.doi.org/10.1038/sj.emboj.7600385; PMID: 15372072
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11:597 - 610; PMID: 20661255
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010; 328:1694 - 8; http://dx.doi.org/10.1126/science.1190809; PMID: 20448148
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281 - 97; http://dx.doi.org/10.1016/S0092-8674(04)00045-5; PMID: 14744438
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5:522 - 31; http://dx.doi.org/10.1038/nrg1379; PMID: 15211354
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 2002; 99:15524 - 9; http://dx.doi.org/10.1073/pnas.242606799; PMID: 12434020
- Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 2003; 1:882 - 91; PMID: 14573789
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 2004; 64:3753 - 6; http://dx.doi.org/10.1158/0008-5472.CAN-04-0637; PMID: 15172979
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65:7065 - 70; http://dx.doi.org/10.1158/0008-5472.CAN-05-1783; PMID: 16103053
- Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol 2009; 24:652 - 7; http://dx.doi.org/10.1111/j.1440-1746.2008.05666.x; PMID: 19175831
- Yoon SO, Chun SM, Han EH, Choi J, Jang SJ, Koh SA, Hwang S, Yu E. Deregulated expression of microRNA-221 with the potential for prognostic biomarkers in surgically resected hepatocellular carcinoma. Hum Pathol 2011; 42:1391 - 400; http://dx.doi.org/10.1016/j.humpath.2010.12.010; PMID: 21458843
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005; 353:1793 - 801; http://dx.doi.org/10.1056/NEJMoa050995; PMID: 16251535
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 2006; 9:435 - 43; http://dx.doi.org/10.1016/j.ccr.2006.04.020; PMID: 16766263
- Nakamura T, Canaani E, Croce CM. Oncogenic All1 fusion proteins target Drosha-mediated microRNA processing. Proc Natl Acad Sci U S A 2007; 104:10980 - 5; http://dx.doi.org/10.1073/pnas.0704559104; PMID: 17581865
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 2005; 55:74 - 108; http://dx.doi.org/10.3322/canjclin.55.2.74; PMID: 15761078
- Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol 2006; 12:354 - 62; PMID: 16489633
- Ushijima T, Nakajima T, Maekita T. DNA methylation as a marker for the past and future. J Gastroenterol 2006; 41:401 - 7; http://dx.doi.org/10.1007/s00535-006-1846-6; PMID: 16799880
- Shin SH, Park SY, Kang GH. Down-regulation of dual-specificity phosphatase 5 in gastric cancer by promoter CpG island hypermethylation and its potential role in carcinogenesis. Am J Pathol 2013; 182:1275 - 85; http://dx.doi.org/10.1016/j.ajpath.2013.01.004; PMID: 23402999
- Wang X, Kang GH, Campan M, Weisenberger DJ, Long TI, Cozen W, Bernstein L, Wu AH, Siegmund KD, Shibata D, et al. Epigenetic subgroups of esophageal and gastric adenocarcinoma with differential GATA5 DNA methylation associated with clinical and lifestyle factors. PLoS One 2011; 6:e25985; http://dx.doi.org/10.1371/journal.pone.0025985; PMID: 22028801
- Suzuki H, Tokino T, Shinomura Y, Imai K, Toyota M. DNA methylation and cancer pathways in gastrointestinal tumors. Pharmacogenomics 2008; 9:1917 - 28; http://dx.doi.org/10.2217/14622416.9.12.1917; PMID: 19072648
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 2007; 67:1424 - 9; http://dx.doi.org/10.1158/0008-5472.CAN-06-4218; PMID: 17308079
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res 2008; 68:4123 - 32; http://dx.doi.org/10.1158/0008-5472.CAN-08-0325; PMID: 18519671
- Yan H, Choi AJ, Lee BH, Ting AH. Identification and functional analysis of epigenetically silenced microRNAs in colorectal cancer cells. PLoS One 2011; 6:e20628; http://dx.doi.org/10.1371/journal.pone.0020628; PMID: 21698188
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 2002; 416:552 - 6; http://dx.doi.org/10.1038/416552a; PMID: 11932749
- Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res 2010; 38:391 - 9; http://dx.doi.org/10.1093/nar/gkp992; PMID: 19906696
- Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood 2006; 108:3646 - 53; http://dx.doi.org/10.1182/blood-2006-01-030015; PMID: 16882713
- Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev 2006; 16:203 - 8; http://dx.doi.org/10.1016/j.gde.2006.02.012; PMID: 16503132
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 2003; 115:787 - 98; http://dx.doi.org/10.1016/S0092-8674(03)01018-3; PMID: 14697198
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A 2005; 102:13944 - 9; http://dx.doi.org/10.1073/pnas.0506654102; PMID: 16166262
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A 2008; 105:13556 - 61; http://dx.doi.org/10.1073/pnas.0803055105; PMID: 18768788
- Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernández-Piqueras J, Malumbres M. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell 2008; 13:496 - 506; http://dx.doi.org/10.1016/j.ccr.2008.04.018; PMID: 18538733
- Suzuki H, Yamamoto E, Nojima M, Kai M, Yamano HO, Yoshikawa K, Kimura T, Kudo T, Harada E, Sugai T, et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis 2010; 31:2066 - 73; http://dx.doi.org/10.1093/carcin/bgq203; PMID: 20924086
- Nadal E, Chen G, Gallegos M, Lin L, Ferrer-Torres D, Truini A, Wang Z, Lin J, Reddy RM, Llatjos R, et al. Epigenetic inactivation of microRNA-34b/c predicts poor disease-free survival in early-stage lung adenocarcinoma. Clin Cancer Res 2013; 19:6842 - 52; http://dx.doi.org/10.1158/1078-0432.CCR-13-0736; PMID: 24130071
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol 2007; 17:1298 - 307; http://dx.doi.org/10.1016/j.cub.2007.06.068; PMID: 17656095
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435:834 - 8; http://dx.doi.org/10.1038/nature03702; PMID: 15944708
- Heinzelmann J, Henning B, Sanjmyatav J, Posorski N, Steiner T, Wunderlich H, Gajda MR, Junker K. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J Urol 2011; 29:367 - 73; http://dx.doi.org/10.1007/s00345-010-0633-4; PMID: 21229250
- Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010; 101:2087 - 92; http://dx.doi.org/10.1111/j.1349-7006.2010.01650.x; PMID: 20624164
- Kalimutho M, Di Cecilia S, Del Vecchio Blanco G, Roviello F, Sileri P, Cretella M, Formosa A, Corso G, Marrelli D, Pallone F, et al. Epigenetically silenced miR-34b/c as a novel faecal-based screening marker for colorectal cancer. Br J Cancer 2011; 104:1770 - 8; http://dx.doi.org/10.1038/bjc.2011.82; PMID: 21610744
- Kamimae S, Yamamoto E, Yamano HO, Nojima M, Suzuki H, Ashida M, Hatahira T, Sato A, Kimura T, Yoshikawa K, et al. Epigenetic alteration of DNA in mucosal wash fluid predicts invasiveness of colorectal tumors. Cancer Prev Res (Phila) 2011; 4:674 - 83; http://dx.doi.org/10.1158/1940-6207.CAPR-10-0214; PMID: 21543345
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12:861 - 74; http://dx.doi.org/10.1038/nrg3074; PMID: 22094949
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 2007; 449:689 - 94; http://dx.doi.org/10.1038/nature06192; PMID: 17851529
- Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res 2007; 17:850 - 7; http://dx.doi.org/10.1038/cr.2007.83; PMID: 17923864
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 2008; 22:1865 - 70; http://dx.doi.org/10.1101/gad.1673508; PMID: 18628393
- Ramadoss S, Chen X, Wang CY. Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J Biol Chem 2012; 287:44508 - 17; http://dx.doi.org/10.1074/jbc.M112.424903; PMID: 23152497
- Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J 1996; 10:940 - 54; PMID: 8801176
- Kakizuka A, Miller WH Jr., Umesono K, Warrell RP Jr., Frankel SR, Murty VV, Dmitrovsky E, Evans RM. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell 1991; 66:663 - 74; http://dx.doi.org/10.1016/0092-8674(91)90112-C; PMID: 1652368
- He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi PP. Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential responses to RA in APL. Nat Genet 1998; 18:126 - 35; http://dx.doi.org/10.1038/ng0298-126; PMID: 9462740
- Pandolfi PP. Oncogenes and tumor suppressors in the molecular pathogenesis of acute promyelocytic leukemia. Hum Mol Genet 2001; 10:769 - 75; http://dx.doi.org/10.1093/hmg/10.7.769; PMID: 11257111
- Brown NJ, Ramalho M, Pedersen EW, Moravcsik E, Solomon E, Grimwade D. PML nuclear bodies in the pathogenesis of acute promyelocytic leukemia: active players or innocent bystanders?. Front Biosci (Landmark Ed) 2009; 14:1684 - 707; http://dx.doi.org/10.2741/3333; PMID: 19273155
- Zhu J, Gianni M, Kopf E, Honoré N, Chelbi-Alix M, Koken M, Quignon F, Rochette-Egly C, de Thé H. Retinoic acid induces proteasome-dependent degradation of retinoic acid receptor alpha (RARalpha) and oncogenic RARalpha fusion proteins. Proc Natl Acad Sci U S A 1999; 96:14807 - 12; http://dx.doi.org/10.1073/pnas.96.26.14807; PMID: 10611294
- Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996; 93:9821 - 6; http://dx.doi.org/10.1073/pnas.93.18.9821; PMID: 8790415
