Abstract
Genome hypomethylation is a common epigenetic alteration in human tumors, where it often leads to aberrant activation of a group of germline-specific genes, commonly referred to as “cancer-germline” genes. The cellular functions and tumor promoting potential of these genes remain, however, largely uncertain. Here, we report identification of a novel cancer-germline transcript (CT-GABRA3) displaying DNA hypomethylation-dependent activation in various tumors, including melanoma and lung carcinoma. Importantly, CT-GABRA3 harbors a microRNA (miR-105), which has recently been identified as a promoter of cancer metastasis by its ability to weaken vascular endothelial barriers following exosomal secretion. CT-GABRA3 also carries a microRNA (miR-767) with predicted target sites in TET1 and TET3, two members of the ten-eleven-translocation family of tumor suppressor genes, which are involved in the conversion of 5-methylcytosines to 5-hydroxymethylcytosines (5hmC) in DNA. Decreased TET activity is a hallmark of cancer; here, we provide evidence that aberrant activation of miR-767 contributes to this phenomenon. We demonstrate that miR-767 represses TET1/3 mRNA and protein expression and regulates genomic 5hmC levels. Additionally, we show that high CT-GABRA3 transcription correlates with reduced TET1 mRNA levels in vivo in lung tumors. Together, our study identified a cancer-germline gene that produces microRNAs with oncogenic potential. Moreover, our data indicate that DNA hypomethylation in tumors can contribute to reduced 5hmC levels via activation of a TET-targeting microRNA.
Introduction
DNA methylation in mammalian genomes, which occurs mostly at cytosines within CpG dinucleotides, is a potent mechanism of gene repression, and contributes thereby to the establishment and maintenance of cell-type specific gene expression programs.Citation1 Genome methylation patterns often undergo profound alterations in human tumors.Citation2 Both gains (hypermethylation) and losses (hypomethylation) of CpG methylation are observed. DNA hypermethylation often affects CpG-rich promoters of tumor suppressor genes, leading to their irreversible silencing. DNA hypomethylation, on the other hand, has been associated with aberrant activation of a limited group of protein-coding genes, most of which have their transcription normally restricted to the germ line.Citation3,Citation4 Genes in this group, commonly referred to as ‘cancer-germline’ (CG) genes, are indeed characterized by their strict reliance on DNA methylation for repression in normal somatic tissues.Citation5,Citation6 Intriguingly, most CG genes map on the X chromosome.Citation4 It is still unclear, however, if activation of CG genes, which appear to exert a variety of cellular functions, plays a major oncogenic role in hypomethylated tumor cells.Citation3
In the present study, we searched to determine if DNA hypomethylation in tumors also induces aberrant expression of miRNA-producing cancer-germline transcripts. There is indeed mounting evidence that dysregulated expression of miRNAs, which exert important regulatory functions through their ability to induce post-transcriptional inhibition of target mRNAs, contributes to cancer development.Citation7,Citation8
Results
Aberrant activation of GABRA3 and hosted miR-105 and miR-767 in tumors
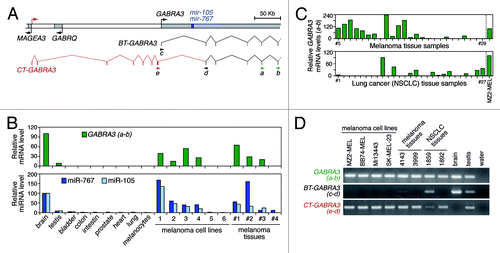
As an initial step in our search for CG-type miRNAs, we performed an in silico screening in miRNA databases (microRNA.org and miRBase.org), using as filtering criteria two characteristics of CG genes: predominant expression in testis and localization on the X chromosome. This led to the selection of 21 X-linked miRNAs with predicted expression in testis and in no more than one normal somatic tissue. Among these, we noticed a pair of miRNAs (miR-105 and miR-767), deriving from the first intron of GABRA3, a gene encoding a receptor subunit for the γ-aminobutyric-acid neurotransmitter. Our interest for these miRNAs was prompted by the fact that, although GABRA3 expression is normally restricted to brain and testis, aberrant transcription of the gene was reported in several tumor types, and was identified as a significant predictor of poor survival in lung cancer patients.Citation9-Citation12 Moreover, GABRA3 is located within a region of the X chromosome (Xq28) that harbors many known CG genes. RT-qPCR experiments with primers located in exons 5 and 6 of GABRA3 confirmed specific expression of this gene in brain and testis, and revealed its activation in melanoma cell lines and tissues (). In parallel, RT-qPCR directed toward miR-105 and miR-767 indicated that expression of these miRNAs strictly mirrors that of their host gene (). Additional analyses in larger sets of tumor samples detected GABRA3 transcripts in 65% of melanoma tissues and in 40% of lung tumors ().
Figure 1. Tumors show aberrant expression of a testis-specific transcript variant of GABRA3, and of the miRNAs it harbors. (A) Schematic representation of the human GABRA3 locus, with broken arrows indicating transcription start sites. The exon/intron structure of the referenced GABRA3 transcript (re-named BT-GABRA3) and of the newly characterized transcript variant (CT-GABRA3) is shown below, with arrowheads indicating the orientation and location of PCR primers used in subsequent expression analyses. (B) Expression analysis of GABRA3, miR-105 and miR-767 in normal human tissues and melanocytes, as well as in melanoma cell lines (1, MI13443-MEL; 2, SK-MEL-23; 3, BB74-MEL; 4, MZ2-MEL; 5, LB2667-MEL; 6, Mi665/2-MEL) and tissue samples (Table S1). Primers a and b amplify both BT-GABRA3 and CT-GABRA3 transcripts. Normalized mRNA (ratio to ACTB) and miRNA (ratio to SNORD44) levels are expressed relative to the brain sample taken as 100% reference. (C) RT-qPCR analysis of GABRA3 with primers a and b, in a larger series of melanoma samples (n = 25), and in non-small-cell lung carcinoma (NSCLC) tissue samples (n = 27). See table S1 for sample descriptions. GABRA3 mRNA levels are expressed relative to the MZ2-MEL melanoma cell line taken as 100% reference. (D) Gel analysis of RT-PCR experiments with primers recognizing either both GABRA3 transcript variants (primers a and b), only BT-GABRA3 transcripts (primers c and d), or only CT-GABRA3 transcripts (primers e and d).
Tumors express a cancer-testis variant of GABRA3: CT-GABRA3
To further characterize the composition of GABRA3 transcripts in tumor cells, RT-PCR experiments with primers located in different exons were performed. Surprisingly, RT-PCR with primers located in exon 1 and 2 of GABRA3 amplified the transcript in brain and testis, but failed to detect it in most tumor cells (). This suggested the existence of an alternative form of GABRA3 transcript in tumors. In order to identify this transcript variant, we performed 5′ RACE experiments in GABRA3-expressing melanoma cell lines. This led to the identification of an alternative transcription start site located 247-kb upstream of the reference GABRA3 start site. We isolated several novel transcript variants originating from this start site, which contained alternatively spliced exons in the 5′ part followed by all exons but exon 1 of GABRA3 (, and Fig. S1). GABRA3 transcripts originating from this alternative start site were named CT-GABRA3 (Cancer-Testis), as opposed to the reference GABRA3 transcript, which, for sake of clarity, we re-named BT-GABRA3 (Brain-Testis). Unlike BT-GABRA3, CT-GABRA3 displayed a typical cancer-germline pattern of expression, as it was expressed in testis but not in brain, and was commonly activated in tumor cells (). CT-GABRA3 transcripts comprise several short upstream open reading frames, which were found to inhibit translation of the GABRA3 protein (Fig. S2). Interestingly, the transcription start site of CT-GABRA3 is located nearby that of a known CG gene, MAGEA3, oriented in the opposite direction (). Both genes appear therefore to share a bidirectional promoter, as supported by their frequent co-activation in melanoma and lung tumor samples (Fig. S3).
CT-GABRA3 activation in tumors is dependent on DNA demethylation
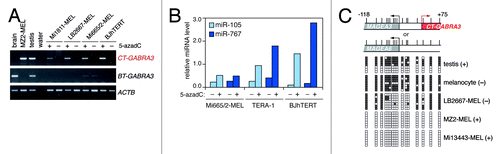
We next investigated whether activation of CT-GABRA3 and its hosted miRNAs in tumors is linked to DNA hypomethylation. Sensitivity of CT-GABRA3 expression to DNA demethylation-dependent activation was demonstrated in an experiment showing induction of this transcript, but not of BT-GABRA3, following treatment of non-expressing cells with the DNA methylation inhibitor, 5-aza-2’-deoxycytidine (5-azadC; ). Not surprisingly, the DNA methylation inhibitor also induced expression of miR-105 and miR-767 (). Moreover, sodium bisulfite sequencing revealed that CT-GABRA3 expression in testis and tumor cells is associated with extensive promoter demethylation (). Consistent with a primary role of genome demethylation in the activation of CT-GABRA3 in tumor cells, we observed a significant trend of co-activation of this gene with other DNA methylation-sensitive CG genes in melanoma cell cultures (Fig. S4). Together, our results indicate that mir-105 and mir-767 are carried by two transcript variants of GABRA3: BT-GABRA3, which is transcribed in brain and testis; and CT-GABRA3, which normally displays specific transcription in testis, but also becomes activated as a result of DNA demethylation in tumor cells. We conclude that DNA hypomethylation in tumors is associated with aberrant activation of miR-105 and miR-767 expression.
Figure 2. Expression of CT-GABRA3, miR-105 and miR-767 is induced by DNA demethylation. (A) Three GABRA3-negative melanoma cell lines (-MEL), and an immortalized fibroblast cell line (BJhTERT) were cultured in the presence (+) or in the absence (-) of 5-azadC. Expression of CT-GABRA3, BT-GABRA3 and ACTB (control) was analyzed by RT-PCR. (B) Expression of miR-105 and miR-767 was analyzed by RT-qPCR in similarly treated cell lines, including the TERA-1 embryonal carcinoma cell line. Relative miRNA levels are expressed as ratio to SNORD44 (x 104). (C) Bisulfite sequencing of the MAGEA3/CT-GABRA3 promoter region. Sequences could not be distinguished from those deriving from the MAGEA6 promoter region, as both loci show 100% sequence identity. Vertical bars indicate location of CpG sites with positions relative to the CT-GABRA3 start site. Open and filled squares represent unmethylated and methylated CpG sites, respectively, and each row represents a single clone. CT-GABRA3 expression status (+) or (-) in samples is indicated (positive samples also express MAGEA3 and MAGEA6). Highly methylated sequences in testis likely derive from somatic cells in the tissue sample.
TET1 and TET3 are targets of miR-767
During the course of our research project, a study was published showing that miR-105 is expressed in metastatic breast cancer cells, and acts as a crucial promoter of cancer metastasis.Citation13 The study revealed indeed that miR-105 undergoes exosome-mediated secretion, and destroys vascular barriers by inhibiting expression of the tight junction protein ZO-1 in endothelial cells. Our data indicate that miR-105 is expressed in other tumor types as well, and reveal that DNA hypomethylation accounts for its tumor-specific activation. In order to get more insight into the function of miR-767 and its potential contribution to tumor development, we decided to search relevant mRNA targets of this miRNA with the help of prediction algorithms. We focused our attention on two potential targets of miR-767, TET1 and TET3, because TET genes were recently shown to exert tumor-suppressive functions.Citation14 The TET family of genes (TET1, TET2, and TET3) encode dioxygenases that are recruited to specific regions of the genome, where they contribute to processes of localized DNA demethylation by converting 5-methylcytosines (5mC) to 5-hydroxymethylcytosines (5hmC).Citation15,Citation16 TET activities are often downregulated in a wide variety of cancers, leading to marked reduction in genomic 5hmC levels.Citation17-Citation19 In a significant proportion of hematopoietic malignancies, this has been associated with mutations in TET2.Citation17 In most solid tumors, however, the origin of reduced TET activities has remained unclear.
Effective targeting of TET1 and TET3 genes by miR-767 was confirmed by transfection experiments showing that synthetic miR-767 molecules, but not control miRNA molecules, induce downregulation of luciferase reporter genes linked to the 3′-UTR of either TET1 or TET3 (). Importantly, we also demonstrated that expressing tumor cells contain sufficient amounts of endogenous miR-767 to inhibit expression of the TET1 3′-UTR luciferase reporter gene, and we confirmed that this inhibition involves a direct interaction, by showing impaired inhibition of a reporter that carries a mutant version of the TET1 3′-UTR lacking miR-767 target sequences (, and Fig. S5). We then assessed whether miR-767 can regulate endogenous TET1 and TET3 mRNA levels. In HEK293T human embryonic kidney cells and in TERA-1 human embryonal carcinoma cells, which both lack constitutive expression of miR-767, transfection of synthetic miR-767 molecules resulted in reduced TET1 and TET3 mRNA levels, although this reduction was not significant for TET3 in TERA-1 cells (). Conversely, inhibition of miR-767 by antisense oligonucleotides in expressing tumor cell lines resulted in a significant elevation in TET1 mRNA levels (). For TET3, we observed a less constant effect of the inhibitor, as only one out of the four treated cell lines showed an increased level of TET3 mRNA (). We also confirmed the ability of miR-767 to inhibit TET1 and TET3 expression at the protein level, by showing decreased amounts of these proteins in HEK293T cells upon transfection with synthetic miR-767 molecules (). Finally, we assessed whether miR-767 can regulate 5hmC levels. Slot blot assays with anti-5hmC antibodies showed that the transfection of synthetic miR-767 molecules in both TERA-1 and HEK293T cells indeed resulted in a significant reduction of global 5hmC levels (). Taken together, these data indicate that miR-767 can function as a regulator of cellular 5hmC levels via targeting of TET genes. In our experiments, miR-767 exhibited a preferential effect on TET1.
Figure 3. miR-767 controls TET1 and TET3 expression levels and regulates cellular 5hmC levels. (A) Red triangles indicate the location of miR-767 seed sequences (7-mer and 8-mer) in TET1 and TET3 3′-UTRs. (B) HEK293T cells were co-transfected with a luciferase reporter linked to the 3′-UTR of either TET1 or TET3, and with the indicated miRNAs (miR-Ctrl and miR-137 served as a negative control). Normalized luciferase activities, which were measured 24h after transfection, are expressed relative to mock cells (no miRNA transfected), and represent means ± SEM (n = 3). *** P < 0.001 for comparison to mock cells (Bonferroni's multiple comparison test). (C) HEK293 cells (miR-767 negative) and Mi13443-MEL cells (miR-767 positive) were stably transfected with a luciferase reporter vector carrying either the wild-type (wt) 3′-UTR of TET1 or a mutant version of the 3′-UTR (mut4), lacking the four miR-767 target sequences. Relative luciferase units represent mean ± SEM (n = 3). *** P < 0.001. (D)TET1 and TET3 mRNA levels were evaluated by RT-qPCR three days after transfection of miR-105 (serving as negative control) or miR-767. Normalized mRNA levels (ratio to ACTB) are expressed relative to mock cells (no miRNA transfected). Values represent mean ± SEM (n ≥ 3). * P < 0.05, for comparison to mock cells (Friedman test with Dunn’s multiple comparison test). (E) miR-767-expressing melanoma cell lines (-MEL) and non-small cell lung carcinoma cell line (GLCP1) were transfected with a LNA inhibitor of miR-767 (miR767-inh) or an irrelevant control LNA inhibitor (Ctrl-inh). TET1 and TET3 mRNA expression levels were assessed by RT-qPCR. Values represent mean ± SEM (n = 3). * P < 0.05 (Wilcoxon signed rank tests). (F) HEK293T cells transfected with the indicated miRNA or siRNA were subjected to western blot analysis for TET1 and TET3 proteins (p80-Ku was assessed to verify equal loading). (G) Genomic 5hmC levels were evaluated by slot blot analysis at day 3 after transfection with the indicated miRNA. Blots were stained with methylene blue to control for loading. Representative blots are shown out of three to five repeats. Quantification of slot blot data was performed. Normalized 5hmC levels are expressed relative to mock-transfected cells, and represent mean ± SEM * P < 0.05 (Friedman test with Dunn’s multiple comparison test). (H) Analysis of microarray data derived from the TCGA collection of lung squamous cell carcinomas (n = 120) revealed significant downregulation of TET1 (but not TET3) in tumor cells that show upregulation of miR-767-harboring GABRA3 transcripts. Positive correlation between GABRA3 and MAGEA3 activation confirmed the validity of the test. Red bars indicate mean mRNA levels. P values were determined by unpaired t tests.
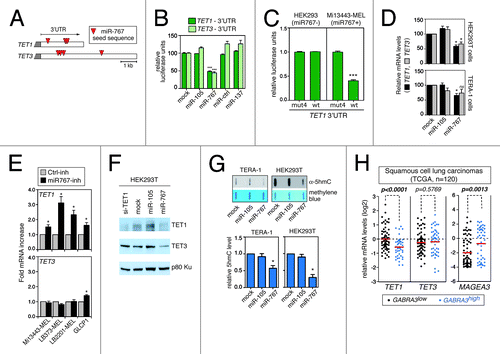
We next searched to determine if miR-767 expression is indeed correlated with reduced expression of TET genes in vivo in tumor tissues. To this end, we analyzed publicly available microarray data, and searched if we could find a negative correlation between the expression of TET1/3 genes and the presence of miR-767-harboring GABRA3 transcripts (probes on microarrays detect both CT- and BT-GABRA3 transcripts). miRNAs usually induce only limited decrease in the level of their target mRNAs. We nevertheless observed a significant correlation between increased GABRA3 expression and reduced TET1 mRNA levels in lung carcinoma tissues (), a tumor type where GABRA3 upregulation was identified as a predictor of poor survival. TET3 did not seem to be affected, suggesting that TET1 is a preferred target of miR-767 in lung carcinoma.
Discussion
One common target of DNA hypomethylation in tumors is the group of CG genes, which normally displays specific expression in germline cells. As a consequence, aberrant transcription of these genes is observed in a large variety of tumor types. Evidence that CG genes exert tumor-promoting roles has however remained scarce. Our identification of CT-GABRA3 provides the first example, to the best of our knowledge, of a CG gene that harbors miRNAs with oncogenic potential. One of these miRNAs, miR-105, was shown very recently to be expressed in breast cancer cells, and to be released in the extracellular environment via exosome secretion.Citation13 Secreted miR-105 targets tight junction protein ZO-1 in endothelial cells, thereby facilitating cancer cell migration to distant locations. Our study establishes therefore a connection between DNA hypomethylation, CG gene activation, and cancer metastasis.
CT-GABRA3 also produces miR-767, and we demonstrate that this miRNA targets genes of the TET family. TET genes encode epigenetic regulators, which were found to exert tumor suppressive functions, notably through their impact on cell differentiation and invasion.Citation18,Citation20,Citation21 A recent study revealed the existence of an extensive network of TET-targeting miRNAs.Citation22 Among these, several miRNAs, including miR-29b and miR-22, were shown to contribute to the development of hematopoietic and breast cancers, in a manner that depended on their effect on TET gene expression.Citation22,Citation23 This demonstrates that TET-targeting miRNAs can have a critical impact on tumor cell functions, even though their effects on TET expression levels are generally very subtle.Citation22,Citation24 In this regard, it is worth noting that monoallelic mutation of Tet2 was sufficient to promote myeloproliferative disorders in mice models, suggesting physiologically relevant gene dosage effects.Citation25,Citation26 The specific contribution of miR-767 to the regulation of TET activities in tumor cells will likely vary according to the level of expression of other TET-targeting miRNAs. Compared with these miRNAs, miR-767 displays a much more contrasted pattern of expression, as it is completely silenced in all tissues, except brain and testis, and becomes activated in a wide variety of tumors. We anticipate therefore that miR-767 can exert a critical tumor-promoting function in several tumor types. Interestingly, a recent study, which conducted a systematic screening for miRNAs involved in cell migration, identified miR-767 as a potential candidate.Citation27 This finding supports a possible pro-metastatic role of miR-767, when expressed in tumor cells.
Intriguingly, our results concerning miR-767 reveal the existence of an unexpected link between DNA methylation and TET genes, whereby genome demethylation can lead to inhibition of TET activities via activation of a TET-targeting miRNA. DNA-hypomethylation-mediated activation of miR-767 in tumors is therefore expected to cause subsequent epigenetic remodeling events. For instance, because TET proteins are required to maintain select genomic sequences in a DNA methylation-free status,Citation28 their downregulation by miR-767 may facilitate DNA hypermethylation at specific gene promoters. This raises the interesting possibility of a link between DNA hypomethylation and DNA hypermethylation in tumor cells.
In a more physiological context, we hypothesize that miR-767 may contribute to an epigenetic regulatory circuit in developing germ line cells, where transient processes of DNA demethylation involving TET enzymes are taking place.Citation29,Citation30 One possibility is that miR-767 becomes activated upon completion of these DNA demethylation processes, and then inhibits TET activities in order to allow subsequent re-methylation of specific DNA sites. The timeframe of activation of CG genes in developing germline cells is compatible with this proposed role of miR-767.Citation31-Citation33 Experimental validation of this hypothesis can be envisaged in the mouse, where miR-767 location in the X-linked Gabra3 gene, and target sites in the 3′-UTR of Tet genes, are conserved.
The other site of constitutive miR-767 expression is brain, where 5hmC levels are generally high.Citation34 Interestingly, TET1 was found to be significantly downregulated by neuronal activity,Citation35 suggesting the existence of intricate regulatory mechanisms, in which miR-767 might be implicated. This may have important implications in memory formation and extinction.Citation35,Citation36
Material and Methods
Cell lines and tumor tissue samples
All human melanoma cell lines, which derive from cutaneous melanoma metastases, and the GLCP1 cell line, which derives from a human NSCLC, were obtained from the Brussels Branch of the Ludwig Institute for Cancer Research, and were cultured as previously described.Citation5 TERA-1 human embryonal carcinoma cells were kindly provided by W. Schultz (Heinrich Heine University, Germany), and their culture conditions are described elsewhere.Citation37 HEK293T cells, which were purchased from Thermo Fisher, HEK293 cells, which were kindly provided by B. Lauwerys (Université catholique de Louvain, Belgium), and BJhTERT cells, which were a gift from F. d’Adda di Fagagna (IFOM foundation, Italy), were maintained in high glucose DMEM (Life Technologies), supplemented with GlutaMAX™ (Life Technologies), 1 x non-essential amino acids (Life Technologies), 1 x Penicillin/Streptavidin (Life Technologies), and 10% fetal bovine serum (Hyclone). Early passage human normal epidermal melanocytes (HNEM) were received from E. De Plaen (Ludwig Institute for Cancer Research, Belgium), and were cultured in Ham’s F10 medium (Life Sciences) supplemented with 6 mM Hepes, 1 x MelanoMax supplement (Gentaur), and 10% fetal bovine serum. A description of human melanoma and NSCLC tissue samples is provided in the table S1. They were obtained from the Brussels Branch of the Ludwig Institute for Cancer Research
RT-PCR and RT-qPCR analyses
Total RNA samples were purchased from Ambion Life Technologies, or prepared from cell lines and surgical specimens (obtained from the Ludwig Institute for Cancer Research, Belgium) using either TriPure Isolation Reagent (Roche Diagnostics GmbH) or the guanidinium-isothiocyanate/cesium chloride procedure.Citation38 Reverse transcription was performed on 2 μg of total RNA using either PrimeScript Reverse transcriptase (Takara) and random hexamers primers, or M-MLV Reverse transcriptase (Invitrogen) and dT18 primers. The transcripts were amplified from 1/40 of the reverse transcription reaction. Conventional PCRs were performed using the DreamTaq polymerase (Thermo Fisher Scientific), in a final reaction volume of 25 μl. Quantitative RT-PCR amplifications were performed using the qPCR Core kit (Eurogentec). All qPCRs where SybrGreen assays, except for the GABRA3 qPCR, which was a Taqman assay. Primers and probe sequences are listed in the table S2.
For miRNA RT-qPCR analyses, 20ng of total RNA was used for the RT with the Universal cDNA Synthesis Kit II (Exiqon). miRNAs were amplified from 1/320 of the reverse transcription reaction, using LNA primers specific for hsa-mir-767–5p (#204238, Exiqon) and hsa-mir-105–5p (#204389, Exiqon). LNA primers specific for SNORD44 (#203902, Exiqon) were used for normalization. qPCR was performed using the qPCR Core Kit (Eurogentec).
Rapid amplification of 5′ cDNA ends (5′-RACE)
Two protocols of 5′-RACE were applied, which were based on the use of either ThermoScript Reverse Transcriptase (Fig. S1B) or PrimeScript Reverse Transcriptase (Fig. S1C).
For Thermoscript 5′-RACE: reverse transcription was performed on 3 μg of total RNA using ThermoScript Reverse Transcriptase (Invitrogen), according to manufacturer’s instructions. Ten pmoles of GABRA3-specific primer (GABRA3b) and 10 pmoles of SMART IV oligo from the SMART™ cDNA Library Construction Kit (Clontech) were used for the reaction. The reaction was incubated during 1 h at 58 °C, and stopped by heating 5 min at 80 °C. The reaction was subsequently incubated for 20 min at 37 °C in the presence of 1 unit of RNaseH (Invitrogen). The 5′-RACE products were amplified from 1/20 of the reverse transcription reaction, with 0.625 units of TaKaRa Taq DNA polymerase (Takara). Three rounds of nested PCR, with primers indicated in Figure S1 (see sequence in accompanying table), were applied. PCR products were run on an agarose gel, and purified with the QIAquick Gel Extraction Kit (Qiagen) before sequencing.
For PrimeScript 5′-RACE: Reverse transcription was performed on 1 μg of total RNA using PrimeScript Reverse Transcriptase (Takara), according to manufacturer’s instructions. Ten pmoles of random hexamers and 10 pmoles of SMART IV oligo were used in the reaction. The reaction was incubated for 1 h at 42 °C, and stopped by heating 7 min at 72 °C. Subsequent steps were as described here above.
DNA fragments generated by 5′-RACE and RT-PCR analyses were sequenced with the BigDye® Terminotor V3.1 Cycle Sequencing kit (Life technologies), according to the manufacturer’s recommendation. Sequencing reactions were purified with the BigDye® XTerminator kit (Life technologies) and were run on an ABI 3130xL Genetic Analyzer (Life technologies). Sequence of the full-length CT-GABRA3 transcript has been submitted to GenBank under the accession number KJ620007.
5-Aza-2′-deoxycytidine treatment
Cells grown to 60–70% confluence were exposed to a single dose of 2 μM 5-aza-2′-deoxycytidine (Sigma-Aldrich), and maintained in culture during 6 d (a period of time corresponding to at least 3 population doublings) before RNA extraction.
Sodium bisulfite genomic sequencing
Sodium bisulfite genomic sequencing of the MAGEA3/CT-GABRA3 promoter region was performed as described previously.Citation39 Primer used for nested PCR amplification of bisulfite treated DNA were respectively TYGATTTTTATTTAGGTAGAATTT and TAAAATAATAACRACCCAACCTAA (1st PCR) and ATTTAGGTAGAATTTAGTTTTAT and CCCTACRAAATAACCCAAA (2nd PCR). These primers sets also amplify the MAGEA6 promoter region, which shows 100% sequence identity.
Construction of luciferase reporter vectors and luciferase assays
TET1 and TET3 3′-UTRs were amplified by PCR using the high fidelity PrimeStar HS DNA polymerase (Takara), with primers carrying a 5′ overhang containing a restriction site for either XhoI (sense primer) or Not1 (antisense primer) (see table S2). PCR fragments were cloned between the corresponding restriction sites into the psiCHECK™-2 vector (Promega). Vector inserts were sequenced to verify the presence of error-free miR-767 target sequences.
The luciferase reporter vector containing the mutant version of TET1 3′-UTR was generated by site directed mutagenesis of the wild-type vector, using the QuickChange multi site-directed mutagenesis kit (Stratagene), and according to the manufacturer’s instructions. Mutagenic primers (TET1-mut1 to -mut4, see Table S1) were designed using the QuickChange primer design program (www.agilent.com/genomics/qcpd).
In co-transfection of miRNA and luciferase reporter experiments, 2.x104 HEK293T cells were seeded in each well of a 96-well plate. After 24h, cells in each well were transfected using the Lipofectamine 2000 reagent (Invitrogen, Life Sciences), with 0.05 ng of the luciferase reporter vector and mirVana miRNA Mimics (Ambion, ThermoScientific) at a final concentration of 10 nM ( or 3 nM (Fig. S3B). Luciferase activities were measured 24h after transfection by using the Dual-Glo® Luciferase Assay System (Promega) and a Glomax® 96 Microplate luminometer (Promega).
In the experiments of stable transfection of luciferase reporters in HEK293 cells, cells were seeded at ~40% confluency in 75 cm2 flasks, and were transfected 24h later, using Lipofectamine 2000, with 20 µg of the reporter vector, 10 µg of genomic DNA, and 1 µg of pCDNA3 (carrying a neomycin resistance gene; Invitrogen). Transfectants were selected in 0.8 mg/ml G418 Geneticin® (Invitrogen, Life Sciences), and were harvested after 19 d of selection for analysis of luciferase activities. For Mi13443-MEL cells, cells were seeded at ~50% confluency in 75 cm2 flasks, and were transfected 24h later, using the Genius DNA transfection reagent (Westburg) according the manufacturer’s instructions, with 10 µg of the reporter vector, and 1 µg of pCDNA3. Transfectants were selected in 0.8 mg/ml G418 Geneticin®, and were harvested at different time points (day 14, 17 and 22) for analysis of luciferase activities.
Transfection of synthetic miRNAs and miRNA inhibitors
For analysis of the effect of miRNA transfection on endogenous TET mRNA and protein levels, and on 5hmC levels, synthetic mirVana miRNA Mimics were transfected at a final concentration of 50 nM using the Lipofectamine 2000 reagent (Invitrogen, Life Sciences) in HEK293T cells; and at a final concentration of 50 nM using the Lipofectin reagent (Invitrogen, Life Sciences) in TERA-1 cells. In both cases, the manufacturer’s instructions were applied.
For analysis of miRNA inhibitors on TET expression levels in tumor cells, 5′-fluorescein-labeled miRCURY LNA™ Power microRNA inhibitors (Exiqon) were transfected at a final concentration of 100 nM, using the ExGen 500 transfection reagent (Thermo Scientific) for Mi13443-MEL cells, the Genius DNA transfection reagent for LB373-MEL cells, and the Lipofectamine 2000 transfection reagent for LB2201-MEL and GLCP1 cells. In all cases, the manufacturer’s instructions were applied, and visual analysis under a fluorescent microscope revealed nearly 100% transfection efficiencies. Cells were harvested at day one after transfection for TET1/3 gene expression analysis.
Western blotting
Whole cell lysates were obtained by harvesting cells in 1 x Laemmli buffer complemented with the cOmplete Mini protease inhibitor cocktail (Roche), PhosphoStop phosphatase inhibitor cocktail (Roche), and 1 mM of phenylmethanesulfonyl fluoride (Sigma-Aldrich). Whole cell lysates were denatured for 10 min at 99 °C, sonicated with a Bioruptor sonicator (Diagenode), and reheated for 5 min at 99 °C before loading and electrophoresis in a 4–15% acrylamide Mini-Protean® TGX gel (Biorad). Proteins were thereafter submitted to an overnight electrotransfer on a polyvinydilene difluoride Immobilon®-P transfer membrane (Millipore) at 4 °C. The membrane was thereafter saturated in a PBS solution containing 4% non-fat milk and 0.05% Tween 20 during 1h at room temperature. Incubation with the primary antibodies was performed in the same solution either overnight at 4 °C for TET1 and TET3, or during 1h at room temperature for p80-Ku. Primary antibodies were: anti-TET1 rabbit polyclonal antibody (1:5000, GT1462, Genetex), anti-TET3 rabbit polyclonal antibody (1:5000, C3, Genetex), and anti-p80-Ku mouse monoclonal antibody (1/500, GE2.9.5, Millipore). Following incubation with the primary antibody, the membrane was washed 3 times in PBS-Tween 0.05% and then incubated at room temperature for 45 min in the presence of either HRP-conjugated goat anti-rabbit IgG antibody (1:10000, Enzo, Life Sciences) or HRP-conjugated goat anti-mouse IgG antibody (1:2000, Santa Cruz). Signals on the membrane were revealed using the SuperSignal West Pico Chemiluminescent Substrate (Pierce, Thermo Scientific), and after exposure to Fuji Medical X-RAY films (Fujifilm). Before applying a new antibody, the membrane was subjected to a 10 min incubation in 0.4 M NaOH at room temperature, three washes in PBS-Tween 0.05%, and incubation in a PBS solution containing 4% non-fat milk and 0.05% Tween 20 during 1h at room temperature.
Slot Blot analysis of 5hmC
Genomic DNA was extracted using the SDS/Proteinase K lysis method as described previously.Citation40 It was then treated with RNase A (1µg/µl final concentration) for 30 min, and purified by phenol/chloroform extractions and ethanol precipitation. The Nanodrop ND-1000 spectrophotometer (Isogen, Life Sciences) was used for quantification. DNA samples were denatured by incubation in a solution of 0.4 mM NaOH and 10 mM EDTA at 99 °C for 10 min, and then chilled on ice. Aliquots (500 ng for HEK293T and 2 µg for TERA-1) were slotted on positively charged nylon Hybond™ membranes (Amesham, GE Healthcare) using a Hybri.Slot 24 blotting apparatus (Core, Life Sciences). Membranes were thereafter washed quickly in 2 x SSC, dried, and cross-linked by UV exposure. Blocking of the membranes was performed by incubation in PBS containing 5% dry milk and 0.05% Tween, during 1h at room temperature. They were then probed with an anti-5hmC rabbit polyconal antibody (1:10000, #39769, Active Motif), during an overnight incubation at 4 °C. After three washes in PBS-Tween 0.05%, the membranes were incubated at room temperature for 45 min in the presence of HRP-conjugated goat anti-rabbit IgG antibody (1:10000, Enzo, Life Sciences). Signals on the membranes were revealed using the SuperSignal West Pico Chemiluminescent Substrate (Pierce, Thermo Scientific), and after exposure to Fuji Medical X-RAY films (Fujifilm). Membranes were thereafter washed quickly in PBS-Tween 0.05%, before staining in a methylene blue solution (0.02% methylene blue, 0.3M NaOH, pH 5.2). Films and stained membranes were scanned, and slot intensities were quantified using ImageJ.
Analysis of lung squamous cell carcinoma mRNA data sets
We used data sets from the TCGA (Nature 2012), which were obtained on Agilent microarrays.Citation41 The gene expression value in each sample was reported to the mean value in all samples. Relative gene expression values were then log2 transformed for subsequent analyses. Samples were sorted according to their relative GABRA3 expression value. GABRA3low and GABRA3high subgroups were defined so as to match the proportion of GABRA3 negative/positive (60% neg. and 40% pos.) samples determined by RT-qPCR experiments in NSCLC lung carcinomas, see ). Two-tailed unpaired t test was used for statistical analyses.
Additional material
Download Zip (4.1 MB)Acknowledgments
The authors wish to acknowledge the excellent technical assistance of Marjorie Mercier. This work was supported by the Fonds special de recherche (FSR), Université catholique de Louvain, Belgium. A.L. was supported by a special grant from the FSR, and by the de Duve Institute, Brussels, Belgium. A.V.T. and J.C. are the recipients of a Télévie grant from the FRS-FNRS, Belgium [#7.4581.13, and #7.4517.13]
References
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002; 16:6 - 21; http://dx.doi.org/10.1101/gad.947102; PMID: 11782440
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3:415 - 28; PMID: 12042769
- De Smet C, Loriot A. DNA hypomethylation and activation of germline-specific genes in cancer. Adv Exp Med Biol 2013; 754:149 - 66; http://dx.doi.org/10.1007/978-1-4419-9967-2_7; PMID: 22956500
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer 2005; 5:615 - 25; http://dx.doi.org/10.1038/nrc1669; PMID: 16034368
- Cannuyer J, Loriot A, Parvizi GK, De Smet C. Epigenetic hierarchy within the MAGEA1 cancer-germline gene: promoter DNA methylation dictates local histone modifications. PLoS One 2013; 8:e58743; http://dx.doi.org/10.1371/journal.pone.0058743; PMID: 23472218
- De Smet C, Lurquin C, Lethé B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol 1999; 19:7327 - 35; PMID: 10523621
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009; 10:704 - 14; http://dx.doi.org/10.1038/nrg2634; PMID: 19763153
- Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett 2011; 585:2087 - 99; http://dx.doi.org/10.1016/j.febslet.2010.08.009; PMID: 20708002
- Liu Y, Guo F, Dai M, Wang D, Tong Y, Huang J, Hu J, Li G. Gammaaminobutyric acid A receptor alpha 3 subunit is overexpressed in lung cancer. Pathol Oncol Res 2009; 15:351 - 8; http://dx.doi.org/10.1007/s12253-008-9128-7; PMID: 19048400
- Liu Y, Li YH, Guo FJ, Wang JJ, Sun RL, Hu JY, Li GC. Gamma-aminobutyric acid promotes human hepatocellular carcinoma growth through overexpressed gamma-aminobutyric acid A receptor alpha 3 subunit. World J Gastroenterol 2008; 14:7175 - 82; http://dx.doi.org/10.3748/wjg.14.7175; PMID: 19084931
- Lu Y, Lemon W, Liu PY, Yi Y, Morrison C, Yang P, Sun Z, Szoke J, Gerald WL, Watson M, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med 2006; 3:e467; http://dx.doi.org/10.1371/journal.pmed.0030467; PMID: 17194181
- Zhang X, Zhang R, Zheng Y, Shen J, Xiao D, Li J, Shi X, Huang L, Tang H, Liu J, et al. Expression of gamma-aminobutyric acid receptors on neoplastic growth and prediction of prognosis in non-small cell lung cancer. J Transl Med 2013; 11:102; http://dx.doi.org/10.1186/1479-5876-11-102; PMID: 23617850
- Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O’Connor ST, Chin AR, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014; 25:501 - 15; http://dx.doi.org/10.1016/j.ccr.2014.03.007; PMID: 24735924
- Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin 2013; 6:10; http://dx.doi.org/10.1186/1756-8935-6-10; PMID: 23634848
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010; 466:1129 - 33; http://dx.doi.org/10.1038/nature09303; PMID: 20639862
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324:930 - 5; http://dx.doi.org/10.1126/science.1170116; PMID: 19372391
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 2010; 468:839 - 43; http://dx.doi.org/10.1038/nature09586; PMID: 21057493
- Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 2012; 150:1135 - 46; http://dx.doi.org/10.1016/j.cell.2012.07.033; PMID: 22980977
- Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 2013; 32:663 - 9; http://dx.doi.org/10.1038/onc.2012.67; PMID: 22391558
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010; 18:553 - 67; http://dx.doi.org/10.1016/j.ccr.2010.11.015; PMID: 21130701
- Hsu CH, Peng KL, Kang ML, Chen YR, Yang YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep 2012; 2:568 - 79; http://dx.doi.org/10.1016/j.celrep.2012.08.030; PMID: 22999938
- Cheng J, Guo S, Chen S, Mastriano SJ, Liu C, D’Alessio AC, Hysolli E, Guo Y, Yao H, Megyola CM, et al. An extensive network of TET2-targeting MicroRNAs regulates malignant hematopoiesis. Cell Rep 2013; 5:471 - 81; http://dx.doi.org/10.1016/j.celrep.2013.08.050; PMID: 24120864
- Song SJ, Ito K, Ala U, Kats L, Webster K, Sun SM, Jongen-Lavrencic M, Manova-Todorova K, Teruya-Feldstein J, Avigan DE, et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 2013; 13:87 - 101; http://dx.doi.org/10.1016/j.stem.2013.06.003; PMID: 23827711
- Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 2013; 154:311 - 24; http://dx.doi.org/10.1016/j.cell.2013.06.026; PMID: 23830207
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011; 20:11 - 24; http://dx.doi.org/10.1016/j.ccr.2011.06.001; PMID: 21723200
- Quivoron C, Couronné L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 2011; 20:25 - 38; http://dx.doi.org/10.1016/j.ccr.2011.06.003; PMID: 21723201
- Zha R, Guo W, Zhang Z, Qiu Z, Wang Q, Ding J, Huang S, Chen T, Gu J, Yao M, et al. Genome-wide screening identified that miR-134 acts as a metastasis suppressor by targeting integrin β1 in hepatocellular carcinoma. PLoS One 2014; 9:e87665; http://dx.doi.org/10.1371/journal.pone.0087665; PMID: 24498348
- Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands?. EMBO Rep 2012; 13:28 - 35; http://dx.doi.org/10.1038/embor.2011.233; PMID: 22157888
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 2013; 339:448 - 52; http://dx.doi.org/10.1126/science.1229277; PMID: 23223451
- Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature 2013; 504:460 - 4; http://dx.doi.org/10.1038/nature12805; PMID: 24291790
- Gjerstorff MF, Kock K, Nielsen O, Ditzel HJ. MAGE-A1, GAGE and NY-ESO-1 cancer/testis antigen expression during human gonadal development. Hum Reprod 2007; 22:953 - 60; http://dx.doi.org/10.1093/humrep/del494; PMID: 17208940
- Koslowski M, Bell C, Seitz G, Lehr HA, Roemer K, Müntefering H, Huber C, Sahin U, Türeci O. Frequent nonrandom activation of germ-line genes in human cancer. Cancer Res 2004; 64:5988 - 93; http://dx.doi.org/10.1158/0008-5472.CAN-04-1187; PMID: 15342378
- Maatouk DM, Kellam LD, Mann MR, Lei H, Li E, Bartolomei MS, Resnick JL. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development 2006; 133:3411 - 8; http://dx.doi.org/10.1242/dev.02500; PMID: 16887828
- Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res 2011; 39:5015 - 24; http://dx.doi.org/10.1093/nar/gkr120; PMID: 21378125
- Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 2013; 79:1086 - 93; http://dx.doi.org/10.1016/j.neuron.2013.08.032; PMID: 24050399
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai LH. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron 2013; 79:1109 - 22; http://dx.doi.org/10.1016/j.neuron.2013.08.003; PMID: 24050401
- Loriot A, Parvizi GK, Reister S, De Smet C. Silencing of cancer-germline genes in human preimplantation embryos: evidence for active de novo DNA methylation in stem cells. Biochem Biophys Res Commun 2012; 417:187 - 91; http://dx.doi.org/10.1016/j.bbrc.2011.11.120; PMID: 22155245
- Davis LG, Dibner MD, Battey JF. Basic methods in molecular biology. New York: Elsevier, 1986.
- De Smet C, Loriot A, Boon T. Promoter-dependent mechanism leading to selective hypomethylation within the 5′ region of gene MAGE-A1 in tumor cells. Mol Cell Biol 2004; 24:4781 - 90; http://dx.doi.org/10.1128/MCB.24.11.4781-4790.2004; PMID: 15143172
- De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci U S A 1996; 93:7149 - 53; http://dx.doi.org/10.1073/pnas.93.14.7149; PMID: 8692960
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489:519 - 25; http://dx.doi.org/10.1038/nature11404; PMID: 22960745
