Abstract
One of the potential mechanisms of imatinib mesylate (IM) resistance in chronic myeloid leukemia (CML) is increased level of P-glycoprotein (Pgp). Pgp is an efflux pump capable of activating the multidrug resistance (MDR) phenotype. The gene encoding Pgp (ABCB1) has several binding sites in its promoter region, along with CpG islands and GC boxes, involved in its epigenetic control. In previous work, we performed a proteomic study to identify proteins involved in IM cross-resistance in acute leukemia. Among these proteins, we identified LRPPRC as a potential regulator of ABCB1 transcription via an invMED1 binding site in ABCB1. Interestingly, this invMED1 binding site overlaps with the GC -100 box. In this work, we investigated the potential role of LRPPRC in the regulation of ABCB1 transcriptional activity in CML resistance. In addition, we evaluated the potential connection between this regulation and the methylation status of the ABCB1 promoter in its GC -100 box. Our results show that LRPPRC binds prominently to the ABCB1 promoter in Lucena cells, an IM-resistant cell line. Luciferase assays showed that ABCB1 transcription is positively regulated by LRPPRC upon its knockdown. Pyrosequencing analysis showed that the ABCB1 promoter is differentially methylated at its GC -100 box in K562 cells compared with Lucena cells, and in CML patients with different response to IM. Chromatin immunoprecipitation and Pgp expression after DNA demethylation treatment showed that LRPPRC binding is affected by the methylation status of ABCB1 GC -100 box. Taken together, our findings indicate that LRPPRC is a transcription factor related to ABCB1 expression and highlight the importance of epigenetic regulation in CML resistance.
Background
Chronic myeloid leukemia (CML) is a myeloproliferative disease characterized by the expression of the BCR-ABL constitutive tyrosine kinase (TK) oncoprotein. Despite the clear progress in therapy with the use of the TK inhibitor imatinib mesylate (IM), resistance is observed, especially among patients in the advanced phases of the disease.Citation1,Citation2 In the past decade, many efforts had been made to elucidate the possible mechanisms surrounding IM resistance.Citation3-Citation7 Among these mechanisms is the rise of multidrug resistance (MDR).Citation8-Citation18 The MDR phenotype is commonly implicated in chemotherapy failure, and it is dependent on the expression of proteins that function as extrusive pumps, such as P-glycoprotein (Pgp), encoded by the ABCB1 gene. In previous work, we performed a set of experiments showing that Lucena cell line (K562/vincristine - VCR) overexpress the ABCB1/Pgp efflux transporter and this protein is involved in IM cross-resistance presented by Lucena cells. K562 cells presented 1 μM as the inhibitory concentration (IC50) to IM treatment, whereas Lucena cells had an IC50 of 5μM. Subsequently, we performed a comparative proteomic study to identify proteins that are potentially involved in IM/MDR-cross resistance in CML, and we reported that Leucine-Rich protein 130 (LRPPRC) is potentially involved in such resistance.Citation19
LRPPRC is a member of the pentatricopeptide repeat protein family and is described as a multifunctional protein involved in homeostasis, microtubule alterations, RNA stability, DNA/RNA binding, transcriptional activity in the mitochondria, metabolic processes, RNA nuclear export and, more recently, in tumorigenesis.Citation20-Citation32 Nuclear transcription activity was described in acute lymphoblastic leukemia (ALL) in an MDR cell model.Citation33 LRPPRC was found to regulate MDR genes (ABCB1 and MVP- major vault protein) through invMED1 binding sites present in their promoters. Nevertheless, that study was the only previous report of LRPPRC as a transcription factor.
The ABCB1 promoter has a complex pattern, which allows it to be regulated by several pathways.Citation34 Moreover, epigenetic regulation, such as histone modification and DNA methylation, add more complexity and has been more extensively studied over the past several years.Citation35-Citation38 Although ABCB1 has been identified to have two different promoters, both of them producing the same protein,Citation39 the downstream promoter is the major promoter in most cells.Citation40-Citation42
The downstream promoter has CpG islands and contains two GC boxes, GC-50 (-56/-45) and GC-100 (-110/-103), for transcriptional regulation by DNA methylation. GC -50 box hypomethylation has been described as a poor prognostic factor in hematological malignancy in patients with or without bone marrow transplantation (BMT), due to ABCB1 upregulation.Citation43 In contrast, the GC-100 box has been poorly studied, but our analysis of this promoter region has revealed that it overlaps the invMED1 binding site, suggesting possible epigenetic control at this site.
Therefore, the aim of this work was to investigate the involvement of LRPPRC in the regulation of ABCB1 transcription in CML. We also investigated whether methylation of ABCB1’s GC -100 box could affect its regulation. Our results support the finding that LRPPRC is a transcription factor and provide information regarding the importance of the GC -100 box in ABCB1 regulation in CML.
Results
LRPPRC binds to the ABCB1 promoter at the invMED1 binding site
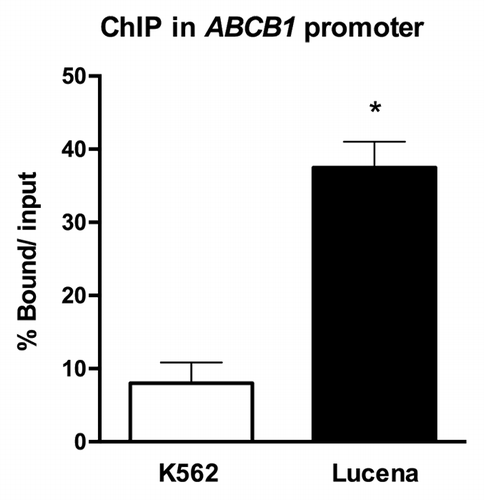
As there has previously been only one report in all of the literature demonstrating that LRPPRC acts as a transcription factor, we first sought to investigate whether LRPPRC could bind to the ABCB1 promoter in CML. Therefore, we used ChIP assays to examine in vivo interactions between nuclear proteins and DNA. Chromatin fractions bound to the LRPPRC antibody in K562 and Lucena cells were quantified by RT-qPCR using primers to amplify the ABCB1 promoter region that contains the invMED1 site. We verified a ≅ 6.0-fold increase in LRPPRC binding in Lucena cells compared with K562 cells after normalization to nonspecific binding of protein A and SMAD8 (). As indicated by the data, LRPPRC binds to the ABCB1 promoter, possibly working as a transcription factor.
Figure 1. ChIP assay for the in vivo quantification of LRPPRC binding to the ABCB1 promoter. RT-qPCR quantification of LRPPRC binding to the ABCB1promoter in K562 and Lucena cells after ChIP assay. DNA amplification was quantified in bound and unbound fractions after normalization with SMAD8 nonspecific amplification. Normalized fractions were used to calculate the bound/input ratio. Results are expressed as mean ± SD for three independent experiments.
LRPPRC regulates ABCB1 expression through the invMED1 binding site
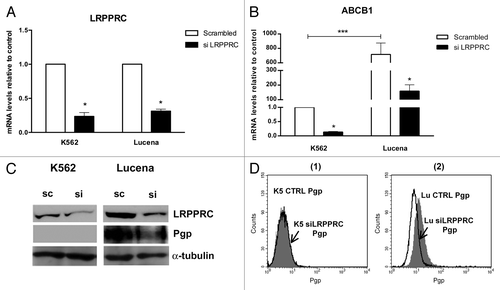
To verify whether the binding of LRPPRC to the ABCB1 promoter could lead to ABCB1 transcriptional activation, we performed functional analyses of LRPPRC depletion in K562 and Lucena cell lines using an siRNA approach. Forty-eight hours after transfection, both mRNA and protein levels of LRPPRC were depleted by more than 70% (-) in both cell lines compared with scrambled controls. We then evaluated ABCB1/Pgp levels in knockdown samples. As shown in --2D, we observed a larger decrease in ABCB1 mRNA (> 75%) and Pgp (≅ 25%) after LRPPRC knockdown in Lucena cells compared with K562 cells. These results demonstrate that the transient knockdown of LRPPRC is involved in the decrease of ABCB1/Pgp levels. Whether its effect is direct or indirect is unclear.
Figure 2. Expression of ABCB1/Pgp levels after LRPPRC depletion in CML cell lines. (A)Analysis of LRPPRC mRNA levels after transient LRPPRC knockdown in K562 and Lucena cells. (B)Analysis of ABCB1 mRNA levels after siLRPPRC. Total RNA was isolated and used in RT-qPCR analysis to determine changes in LRPPRC and ABCB1 mRNA levels after normalization to β-actin expression. All data are presented as fold inductions relative to control group expression (scrambled). (C)Representative western blot analysis of LRPPRC and Pgp expression. Protein extract (50 μg) from both cell lines were separated on a 12% SDS-PAGE gel and probed with anti- LRPPRC and anti-MDR1 antibodies. α-tubulin was used as a loading control. (D) Representative histograms of Pgp expression after 50 nM siLRPPRC (1): K562 ctrl and siLRPPRC (2): Lucena ctrl and siLRPPRC cells. PE-isotype antibody was used as a control. The results are expressed as the mean ± SD for three independent experiments. Ctrl: control; Sc: scrambled; Si: siRNA; K5: K562; LU: Lucena.
To strengthen our observations and to better evaluate the contribution of the invMED1 site in ABCB1 transcription regulation, we transiently transfected the following 3 constructs (): basal promoter sequence without invMED1 site (invMED1_Δ), basal promoter sequence with invMED1 site (invMED1) and basal promoter sequence with mutated invMED1 site (invMED1_mut). After 48 h, we measured Luciferase activity.
Figure 3. Regulation of ABCB1 promoter transcription activity via theinvMED1 binding site.(A) Scheme of invMED1 constructs. (B) Luciferase activity reporter assay in K562 and Lucena cells. All luciferase assay results are expressed as relative light units (RLU). The results are expressed as the mean ± SD for three independent experiments.
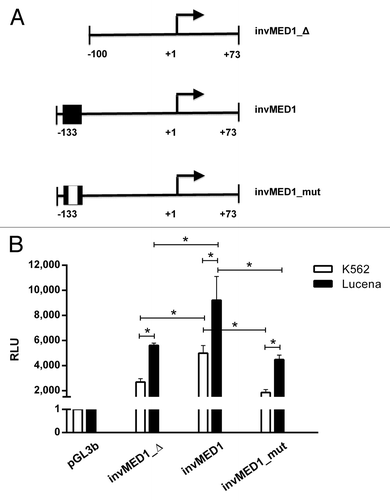
Our results showed that in the presence of the invMED1 binding site, both cell lines exhibited increased Luciferase activity compared with the basal promoter construct. Additionally, the mutated invMED1 construct presented similar Luciferase activity as the basal promoter construct in both cell lines, demonstrating the importance of invMED1 site to activate ABCB1 transcription. Moreover, Luciferase activity was higher in Lucena cells than K562 cells, which confirms the ChIP results that showed increased binding of LRPPRC in Lucena cells. These data suggest the direct involvement of LRPPRC in ABCB1 transcription activity through the invMED1 site.
Role of GC -100 box methylation in ABCB1 transcription through LRPPRC
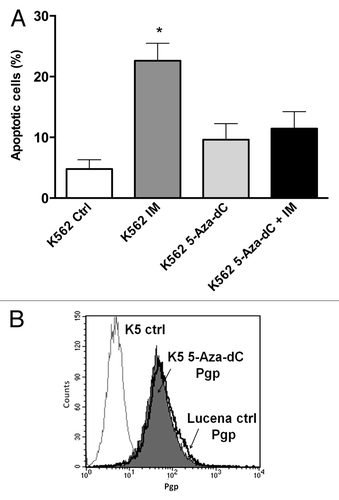
Because we showed that there was a direct relationship between LRPPRC expression and ABCB1 regulation, we next wanted to determine the potential role of the GC -100 box methylation in this regulation. Both cell lines were treated for 24 h with 0.5 μM or 1 μM 5-Aza-dC, a demethylating agent, and cell viability was evaluated. As shown in , K562 and Lucena cell viability decreased by ≅ 20% after 1 μM treatment, which led us to use a 0.5 μM dose in subsequent experiments. To correlate DNA methylation and ABCB1 regulation in the GC -100 box, we evaluated ABCB1/Pgp levels and performed ChIP assays in 5-Aza-dC-treated cells. Our results showed that ABCB1 mRNA levels increased 100% in K562 cells after 5-Aza-dC treatment (). This increase was accompanied by a 3.0-fold increase in Pgp levels (). ChIP assays with 5-Aza-dC-treated cells revealed a 2.0-fold increase in LRPPRC binding in K562 cells (). In order to investigate if ABCB1/Pgp upregulation observed in K562 cells after demethylation treatment could interfere in the cells response to IM treatment, we pre-treated K562 cells with 5 μM 5-Aza-dC for 3 wk, as previously resported,Citation43 and then performed apoptosis assay with 1 μM IM. This IM dose was previously reported by our group as the IC50 for K562 cells, corroborating with literature data.Citation19 We verified reduced apoptosis induction in K562 cells treated with IM after the 5-Aza-dC treatment (), compared with K562 cells treated with IM alone. As we observed that demethylating treatment caused a significant downregulation in apoptosis levels, we investigated if this protective effect could be due to upregulation of Pgp levels. Indeed, as shown in , K562 cells treated with 5 μM 5-Aza-dC showed upregulated Pgp levels, similar to the expression level observed in Lucena cells. Together, these results suggest that the ABCB1 promoter is differentially methylated between the two cell lines, as no difference in LRPPRC binding was observed in Lucena cells.
Figure 4. 5-Aza-dC treatment alters cell viability. Viable K562 and Lucena cells were assessed by Trypan blue staining after 24 h culture in the absence (ctrl) or presence of 0.5μM and 1 μM 5-Aza-dC. Results are expressed as mean ± SD for three independent experiments. Ctrl: control.
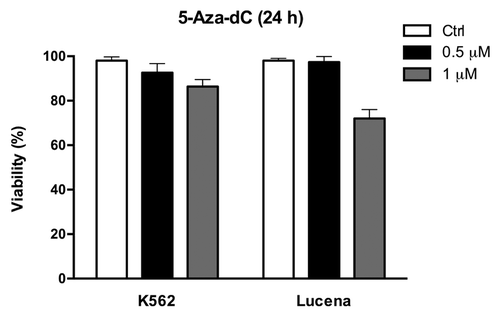
Figure 5. Effect of 5-Aza-dC treatment on ABCB1/Pgp levels and LRPPRC binding to the invMED1 site. (A)ABCB1 mRNA levels were evaluated after 24 h of 0.5 μM 5-Aza-dC treatment. Total RNA was isolated and used in RT-qPCR to determine changes in ABCB1 mRNA levels after normalization to β-actin expression. (B) Representative histograms of Pgp expression after 0.5 μM 5-Aza-dC treatment (1): K562 ctrl and 5-Aza-dC-treated K562 cells (2): Lucena ctrl and Lucena cells treated with 5-Aza-dC. PE-isotype antibody was used as a control. (C) RT-qPCR quantification of LRPPRC binding to the ABCB1 promoter in 5-Aza-dC-treated K562 and Lucena cells. DNA amplification was quantified in bound and unbound fractions after normalization with SMAD8 nonspecific amplification. Normalized fractions were used to calculate the bound/input ratio. The results are expressed as the mean ± SD for three independent experiments. Ctrl: control; K5: K562; LU: Lucena.
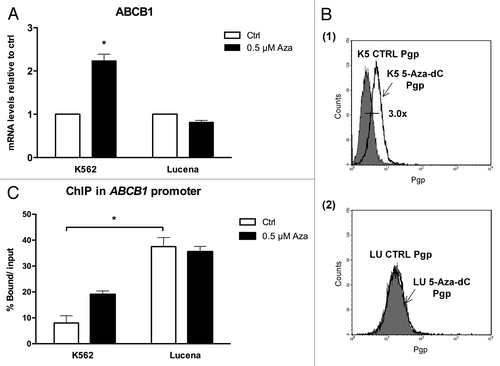
Figure 6. 5-Aza-dC treatment alters response to IM treatment in K562 cells. (A) Apoptosis level was assessed in K562 cells under the following conditions: 1 μM IM, 5μM 5-Aza-dC (3 wk treatment), co-treatment, and at the absence (ctrl) of both drugs. (B) Representative histograms of Pgp expression after 5 μM 5-Aza-dC treatment after 3 wk: K562 ctrl, 5-Aza-dC –treated K562 cells and Lucena cells (used for positive control of Pgp expression). PE-isotype antibody was used as a control. The results are expressed as the mean ± SD for three independent experiments. Ctrl: control.
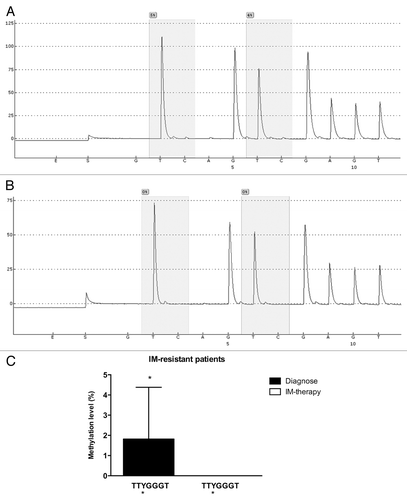
Once our results indicated a difference in the methylation status of the ABCB1 promoter between K562 and Lucena cells, we performed pyrosequencing analysis to evaluate GC -100 box methylation. As expected, pyrosequencing showed that the GC -100 box in the ABCB1 promoter was more methylated in K562 cells (15%) compared with Lucena cells (4%) (). In order to deepen our investigation regarding the relevance of GC -100 box methylation in CML, we also analyzed a total of 20 CML patients with different responses to IM (9 IM-responsive and 11 IM-resistant). Pyrosequencing showed that ≅ 45% of IM-resistant patients (5 patients) showed a methylation percentage of 3–5% at the GC -100 box and completely lost this methylation status after treatment (, ), maintaining the same fold-change as the one observed in K562 and Lucena cell lines. No difference was found in responsive patients (data not shown). Taken together, these results demonstrate that methylation in ABCB1GC -100 box is involved in its transcriptional regulation through LRPPRC binding and could be an important approach to improve prediction of IM response in CML.
Figure 7. Pyrosequencing analysis of the GC -100 box in CML cell lines. Representative pyrosequencing analysis of (A) K562 and (B) Lucena cells for the methylation level (%) at GC -100 box. (C) Results are expressed as the mean ± SD for three independent experiments. * Position of potential 5mC (Y).
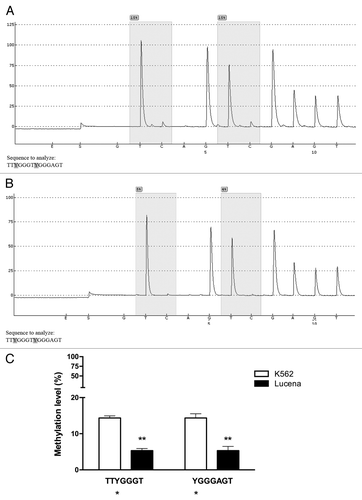
Figure 8. Pyrosequencing analysis of the GC -100 box in CML patients. Representative pyrosequencing analysis for the methylation level (%) at GC -100 box. (A) IM-resistant patient at diagnoses and (B) the same IM-resistant patient at relapse (C) Results are expressed as the mean ± SD for three independent experiments. * Position of potential 5mC (Y).
Discussion
Although our understanding of cancer biology has drastically increased in the past decades, and has sometimes been transferred into therapy in some particular cancer types, the MDR phenotype is still an emergent problem.Citation44,Citation45 MDR is multifactorial, but the number of different proteins and other molecules involved in the rise of the phenotype was underestimated. Indeed, several pathways play direct and indirect roles in ABC transport regulation, and there is redundancy, but it may also be that some particular members play specific functions, suggesting that MDR rise is not as nonspecific as it was once believed. It is still unclear how typical MDR (ABC transporters) and atypical MDR (non ABC transporters) are regulated and how they fit into the new molecular target therapy.Citation46-Citation48
Our previous work demonstrated that LRPPRC was potentially involved in IM and MDR cross-resistance. LRPPRC is a 130 kDa protein that was first described in hepatocarcinoma cells.Citation49 This protein has been associated with several cellular processes. Among these processes, it has a well-defined function in metabolic pathways due to its mitochondria association; nevertheless, recent studies have focused on the role of LRPPRC in tumorigenesis. This particular interest developed as it became known that accelerated energy consumption and enhanced function of mitochondria in tumor cells compared with normal cells is a prerequisite for tumor development.
Tian and colleagues have found that LRPPRC is abundantly expressed in various tumor types, such as lung, stomach, colon, mammary and endometrial adenocarcinomas, esophageal squamous cell carcinoma and lymphoma, compared with its expression in surrounding non-neoplastic cells (for a review see ref. Citation31). It has also been shown that LRPPRC is overexpressed in gastric cancer.Citation50 In hepatocarcinoma cells, LRPPRC was proven to contribute to apoptosis resistance (for a review see ref. Citation32). Additionally, LRPPRC was found highly expressed in the side population of lung adenocarcinoma cell lines, a fraction of cells in which cancer stem cells are enriched.Citation51 This finding is of great interest since it became accepted that cancer stem cells play an important role in tumor resistance.Citation52-Citation54 LRPPRC was reported to be a transcriptional regulator of ABCB1 and MVP genes in ALL. This regulation was mediated through its invMED1 binding site located in the -105/-100 promoter region (for a review see ref. Citation33). The GC -100 box (-108/-103) is located in the same region, which could potentially allow ABCB1 to be regulated through DNA methylation. Because we previously found that ABCB1 and LRPPRC were overexpressed in CML IM-resistant patients, we sought to investigate whether ABCB1 could be regulated by LRPPRC in CML resistance, and whether this regulation was influenced by GC -100 box methylation. Together, our data supported the hypothesis that the ABCB1 promoter was differentially methylated between K562 and Lucena cells.
In fact, it is known that MDR partially arises from promoter hypomethylation. This relationship has already been investigated in several types of cancer, including breast cancer, gastric, prostate, acute myeloid leukemia, and ALL, among others.Citation55-Citation61 However, one outstanding question is whether the decreased LRPPRC binding to ABCB1 invMED1 site in K562 cells was due to a lower availability of LRPPRC or because the ABCB1 promoter is methylated in K562 cells, or a combination of both.
To address this question, we treated both cell lines with a specific DNA demethylating agent and evaluated ABCB1/Pgp levels and analyzed in vivo binding of LRPPRC to ABCB1 invMED1 site. As expected, we observed an increase in ABCB1/Pgp levels only in K562 cells, which supports the previously reported data. Furthermore, LRPPRC binding to the ABCB1 promoter in K562 cells increased compared with untreated cells, suggesting that the invMED1 site is more methylated in non-resistant cells, which was confirmed by pyrosequencing. Moreover, it has been demonstrated that a 3-wk treatment of 5-Aza-dC at 5 μM dose induced upregulation of ABCB1 mRNA levels in K562 cells, comparable to the MDR cell line in this study (see reference Citation43). Using the same approach, we demonstrated that overexpression of Pgp in K562 cells was induced resistance to 1 μM IM treatment. Pyrosequencing analyses of CML patients showed a difference of methylation status among IM resistant patients at the time of diagnoses compared with in resistance/relapse samples. Although we understand this is a low methylation status, the fold-change detected in IM-resistant patients was similar to the fold-change found in K562 and Lucena cells. It is clear that other methylation sites in ABCB1 promoter are contributing to ABCB1 gene expression control, as we observe significant differences in global DNA demethylation after 5-aza-dC treatment. A large cohort of patients would be necessary in order to demonstrate the potential importance of GC -100 box analysis as a support approach in IM-resistance prediction.
Although it is well recognized that epigenetic changes result in an increase on MDR phenotype, this is the first report on the contribution of ABCB1 GC -100 box in its regulation. Nevertheless, our results do not exclude the importance of LRPPRC nuclear localization/availability. Further studies are needed to uncover which signaling pathways are involved in LRPPRC activation/nuclear translocation. Moreover, studies to date have been addressing LRPPRC as a potential prognostic marker in different types of cancer due to its overexpression, but its importance in tumor biology/development/progression remains clearly unexplored.
Taken together, our data indicate that LRPPRC is a transcription factor involved in ABCB1 regulation with considerable implications in MDR/IM resistance. This regulation is dependent on the methylation status of ABCB1 promoter GC -100 box, suggesting the importance of this epigenetic signature in CML resistance.
Materials and Methods
Culture conditions
Lucena [K562 multidrug-resistant cell line induced by vincristine (VCR)] cells overexpressing ABCB1 were kindly provided by Dra. Vivian Rumjanek (Departamento de Bioquímica Médica, Universidade Federal do Rio de Janeiro, Brazil).Citation62 The human myelogenous leukemia cell line (K562) and its vincristine-resistant derivative, the Lucena cell line, were grown in RPMI 1640 medium (Sigma-Aldrich, cat. n. R5886) supplemented with 10% FBS (GIBCO, cat n. 10437–028), 50 units/mL penicillin G / 50 µg/L streptomycin (Invitrogen, cat n. 15140–122) and 2 mM l-glutamine (Invitrogen, cat n. 25030–081) at 37 °C in a humidified atmosphere containing 5% CO2. Lucena medium was supplemented with 60 nM VCR (Sigma, cat n. V8879).
Patients samples
All bone marrow samples were obtained from CML patients in all disease phases (chronic, accelerated and blastic phases) at the time of diagnose and follow up: IM-responsive patients (3 to 6 mo follow up) and IM-resistant or relapse after initial response (3 to 24 mo follow up). Patients were admitted or registered at the Instituto Nacional de Câncer (Rio de Janeiro, Brazil), according to the guidelines of the local Ethics Committee and the Helsinki declaration. We selected 9 IM-responsive patients (mean age = 41.5, range = 28–68, male:female ratio = 2:7) and 11 IM-resistant patients (mean age = 49, range = 17–59, male:female ratio = 7:4). Diagnoses and follow-ups were based on hematologic, cytogenetic and molecular assays. IM-responsive patients exhibited a major molecular response and complete hematologic and cytogenetic response, whereas IM-resistant patients lacked hematologic, cytogenetic and molecular responses. The inclusion criterion was to investigate CML patients that received IM as a first-line therapy. The exclusion criterion was CML patients with BCR-ABL mutations. Marrow aspirates were collected in heparinized tubes and processed on the day they were collected. Bone marrow mononuclear cells were isolated from 2–5 mL of aspirate in a Ficoll-Hypaque density gradient (Ficoll 1.077 g/mL; GE, Sweden) according to manufacturer’s protocol. Cells were washed 3 times in PBS and subsequently used for RNA extraction.
5-Aza-2'-deoxycytidine (5-Aza-dC) treatment
Treatment with the demethylation agent 5-Aza-2'-deoxycytidine (5-Aza-dC) (Sigma-Aldrich, cat n. A3656) at 0.5 μM and 1 μM doses was performed in 12-well culture plates for a period of 24 h with a cell density of 8.0 x 104 cells/mL in both cell lines. Also, a 3 wk treatment at 5 μM dose was performed in 12-well culture plates with a cell density of 8.0 x 104 cells/mL in both cell lines. K562 and Lucena cells exposed to 5-Aza-dC were further used for cell viability assays, real-time quantitative PCR, Pgp expression assays and chromatin immunoprecipitation assays. All experiments were performed in triplicate.
Real-time Quantitative PCR (RT-qPCR)
Analysis of ABCB1, LRPPRC, and β-ACTIN mRNA levels was performed by RT-qPCR. Two micrograms of Trizol extracted RNA from cell lines using Trizol (Invitrogen, cat n. 15596–026) were treated with DNase Amplification Grade I (Invitrogen, cat n. 18068–015) and reverse-transcribed with Superscript III Reverse Transcriptase® (Invitrogen, cat n. 18080044). Diluted cDNAs (1:100) were mixed with SYBR Green PCR Master Mix® (Applied Biosystems, cat n. 4309155) and the following primers: ABCB1: (F) 5′-CCC ATC ATT GCA ATA GCA GG-3′ and (R) 5′-GTT CAA ACT TCT GCT CCT GA-3′; LRPPRC: (F) 5′- GAG AGA TGC CGG AAT TGA GC -3′, and (R) 5′- CTC GGA CTT CTC CAC CTT CT -3′; β-ACTIN: (F) 5′-ACC TGA GAA CTC CAC TAC CCT-3′ and (R) 5′-GGT CCC ACC CAT GTT CCA G-3′. RT-qPCR was performed in a Rotor Gene 6000 thermocycler (Cobertt) with 50 cycles of 20 s at 95 °C, 30 s at 60 °C and 30 s at 72 °C. For each sample, the expression of target genes was normalized to β-actin mRNA levels. Changes in the mRNA levels of genes were evaluated.Citation63
Western blot analysis
Cell lysates from K562 and Lucena cells (that underwent scrambled RNA and siLRPPRC RNA treatments) were run on 7% sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGE), transferred to nitrocellulose membranes (Bioead) and incubated with LRPPRC, MDR1(Santa Cruz Technologies) and α-tubulin (Sigma) antibodies. Antibody binding was detected using enhanced chemiluminescence ECL Plus Western Blotting detection Reagents (GE).
Pgp expression assay
To determine Pgp expression, we analyzed both cell lines with anti-Pgp-PE (phycoerythrin) antibody (Beckman Coulter, cat n. IM2370U) according to the manufacturer’s instructions. Briefly, after treatment, 5.0 x 105 cells were harvested, washed twice with cold PBS, resuspended in 1 mL PBS/BSA (0.2% Azide, 1% BSA) and incubated for 15 min. After incubation, cells were harvested, and anti-Pgp-PE (5 µL) was added, and the sample was incubated for 30 min in the dark. After incubation, 2 mL of PBS/BSA was added to each sample. Cells were harvested and resuspended in PBS/1% Formol. For each condition, 20.000 events were acquired using a FACSCalibur Flow Cytometer (Becton Dickinson) and analyzed using CellQuest v.3.1 Software (Becton Dickinson). The results are expressed as the mean relative fluorescence intensity (MRFI), which was calculated by subtracting the mean fluorescence intensity (MFI) for each specific antibody from the MFI of the respective isotype control.All experiments were performed in triplicate.
Chromatin immunoprecipitation (ChIP) assays
Chromatin immunoprecipitation from K562 and Lucena cells was assessed using a SimpleChIPTM Enzymatic Chromatin IP kit (Cell Signaling Technology®, cat n. #9003) according to the manufacturer’s instructions. Immunoprecipitation was performed with an anti-LRPPRC antibody (Santa Cruz Biotech, cat n. sc-66844). DNA extractions from bound fractions were performed following the Abcam (www.abcam.com) protocol. The immunoprecipitated DNA was amplified for sequences containing the invMED1 binding site by using the following ABCB1 promoter sequence primers: ABCB1p (F) 5′- GCT GAT GCG CGT TTC TCT ACT -3′ and ABCB1p (R) 5′- CCG GGC CGG GAG CAG TCA TC -3′. Quantification was evaluated by RT-qPCR analysis.
RNAi knockdown (siRNA) and transfection
RNAi knockdown (siRNA) and transfections were performed with Trifectin reagent (Integrated DNA Technologies) following the manufacturer’s protocol. LRPPRC siRNA duplex was purchased from Qiagen – cat n. SI03243940 (F) 5′-GCCUGCCGAUUGAACCAAATT-3` (R) 5′-UUUGGUU CAAUCGGCAGGCAA-3′. Universal negative control siRNA duplex (scrambled) (Santa Cruz Biotech, cat n. sc-37007) was used at the same concentration as the experimental siRNA. K562 and Lucena cells (5.5 × 104 cells per well) were split in 24-well plates at 60% confluence in RPMI medium 1 d prior to transfection. A FITC-labeled transfection control duplex was used to monitor the transfection efficiency according to the manufacturer’s recommendations. Only experiments in which transfection efficiencies were ≥ 80% were evaluated. mRNA levels were measured 48 h after transfection. Duplexes were evaluated at 50 nM. All transfections were minimally performed in duplicate, and the data were averaged. LRPPRC depletion was evaluated by RT-qPCR as described above.
Apoptosis assay
To determine the percentage of apoptotic cells, we analyzed phosphatidyl serine externalization and membrane integrity by double staining with Annexin V PE and 7-AAD (PE Annexin V Apoptosis Detection Kit I, BD PharMingen, USA) according to manufacturer's instructions. Briefly, after treatment, 1.0 × 105 cells were harvested, washed twice with cold PBS and resuspended in 100 μL of 1 × binding buffer. Annexin V PE (5 μL) and 7-AAD (5 μL) were added, and samples were incubated for 15 min in the dark. After incubation, 400 μL of 1X binding buffer was added to each sample. Cells positive for Annexin V PE and 7-AAD were considered apoptotic. For every condition, 20.000 events were acquired using a FACSCalibur Flow Cytometer (Becton Dickinson, USA) and analyzed using CellQuest v.3.1 Software (Becton Dickinson, USA). All experiments were performed in triplicate.
Reporter vector design
The ABCB1 firefly luciferase reporter constructs were synthesized through PCR amplification and cloned into the firefly PGL3-Basic vector (Promega, cat n. E1751) upstream of the luciferase reporter gene. The constructs named invMED1_ Δ (-100pb/+73 pb), invMED1 (-133pb/+73 pb) and invMED1_mut (-133pb/+73 bp) were inserted into the KpnI and BglII restriction sites of pGL3-basic.
Transient transfection and luciferase reporter assay
K562 and Lucena cells (1X10Citation5 cells/mL) were plated into 12-well plates and transfected with 1.0 μg of empty pGL3 reporter vectors, constructions containing ABCB1 promoter inserts or pGL3-basic vector. To normalize transfection efficiency, we co-transfected 0.01 μg of Renilla luciferase pRL-TK (Promega, cat n. E2241) plasmid into each well. Transfection was performed using Lipofectamine 2000 (Invitrogen, cat n. 11668027) according to the manufacturer’s protocol. Firefly and Renilla luciferase activities were measured in cell lysates 48 h after transfection using the Dual-Glo Luciferase Assay System (Promega, cat n.E1910) on a Veritas TM Microplate Luminometer (Turner Biosystems) following the manufacturer’s protocol. All experiments were performed in triplicate. Ratios of Renilla luciferase readings to firefly luciferase readings were taken for each experiment, and triplicates were averaged. The average values of the tested constructs were normalized to the activity of the empty pGL3-basic vector, which was arbitrarily set at a value of 1.
Pyrosequencing analysis
Genomic DNA was obtained from RNeasy kit (Qiagen, cat n. 74104) extraction, according to the manufacturer’s recommendations. A total of 500 ng of genomic DNA were treated with sodium bisulfite using the EZ DNA Methylation-Gold Kit (Zymo Research, cat n. D5005). The DNA was eluted to reach a final concentration of 25 ng/μL. To quantify the percentage of methylated cytosine in individual CpG sites, bisulfite-converted DNA was sequenced using a pyrosequencing system (PyroMark Q96, Qiagen).Citation64 This method treats each individual CpG site as a C/T polymorphism and generates quantitative data for the relative proportion of the methylated vs. the unmethylated allele. The assay for the analysis of ABCB1 regulatory region in downstream promoter was designed using the software PyroMark Assay Design version 2.0.1.15 (Qiagen). PCR reaction was performed with 25 ng of bisulfite-treated DNA, primers forward: 5′ GAGAGGTGTA AAGGAAGTTA GAATATTTT and reverse: 5′ biotin-ATTCCTACCC CACCAATCAA CCTCA), and Platinum Taq DNA Polymerase (Invitrogen, cat n. 10966026), according to the manufacturer’s protocol. After initial denaturation at 95 °C for 15 min, amplification was performed for 50 cycles at 95 °C for 30 s, 50 °C for 30 s, 72 °C for 30 s, followed by a final elongation at 72 °C for 10 min. Pyrosequencing was performed with the sequencing primer 5′ GAGGAATTAG TATTTAGTTA A in accordance with the manufacturer’s protocol (Qiagen). The target CpGs were evaluated by converting the resulting pyrograms into numerical values for peak heights.
Statistical Analysis
Cell viability and mRNA levels between K562 and Lucena cells from different assays were compared by a paired t test. These statistical analyses were performed using GraphPad Prism® v.5 software (GraphPad).
| Abbreviations: | ||
| CML | = | chronic myeloid leukemia |
| TK | = | tyrosine kinase |
| IM | = | imatinib mesylate |
| MDR | = | multidrug resistance |
| ABC | = | ATP-binding cassette |
| ALL | = | acute lymphoblastic leukemia |
| VCR | = | vincristine |
| Cat n. | = | catalog number |
| FBS | = | fetal bovine serum |
| MRFI | = | mean relative fluorescence intensity |
| MFI | = | mean fluorescence intensity |
| PE | = | phycoerythrin |
| PI | = | propidium iodide |
| PBS | = | phosphate buffered saline |
| BSA | = | bovine serum albumin |
| FITC | = | fluorescein isothiocyanate |
| RT-qPCR | = | real-time quantitative PCR |
| F | = | forward |
| R | = | reverse |
| siRNA | = | small interfering RNA |
| ChIP | = | chromatin immunoprecipitation |
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgments
We thank Dra. Vivian Rumjanek (Departamento de Bioquímica Médica, Universidade Federal do Rio de Janeiro, Brazil) for providing the Lucena cell line. This work was supported by FINEP, FAPERJ, CNPQ and Ministério da Saúde (MS).
Authors' contributions
SC performed the experiments, statistical analysis and drafted the manuscript. RB assisted the Luciferase constructions design and assisted the experiment. BDR participated in flow cytometry experiments. GF: assisted cloning experiments. PC: performed the caracterization, monitoring and follow up analyses of CML patients. SSL and LFP: provided the Pyrosequencing Facility Core, participated/assisted the pyrosequencing design and experiment and critically revised the manuscript for intellectual content. AM contributed in study conception and intellectual content. EA made substantial contributions to the study conception and design and critically revised the manuscript for intellectual content. All authors read and approved the final manuscript.
References
- Jiang X, Zhao Y, Smith C, Gasparetto M, Turhan A, Eaves A, Eaves C. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia 2007; 21:926 - 35; PMID: 17330101
- Jørgensen HG, Holyoake TL. Characterization of cancer stem cells in chronic myeloid leukaemia. Biochem Soc Trans 2007; 35:1347 - 51; http://dx.doi.org/10.1042/BST0351347; PMID: 17956348
- Mahon FX, Deininger M, Schultheis B, Chabrol J, Reiffers J, Goldman JM, Melo JV. Selection and characterization of BCR-ABL positive cell lines with differential sensitivity to the tyrosine kinase inhibitor STI571: diverse mechanisms of resistance. Blood 2000; 96:1070 - 9; PMID: 10910924
- Hochhaus A. Chronic myelogenous leukemia (CML): resistance to tyrosine kinase inhibitors. Ann Oncol 2006; 17:Suppl 10 x274 - 9; http://dx.doi.org/10.1093/annonc/mdl273; PMID: 17018738
- Mughal TI, Goldman JM. Emerging strategies for the treatment of mutant Bcr-Abl T315I myeloid leukemia. Clin Lymphoma Myeloma 2007; 7:Suppl 2 S81 - 4; http://dx.doi.org/10.3816/CLM.2007.s.006; PMID: 17382017
- Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer 2007; 7:441 - 53; http://dx.doi.org/10.1038/nrc2147; PMID: 17522713
- Quintás-Cardama A, Kantarjian HM, Cortes JE. Mechanisms of primary and secondary resistance to imatinib in chronic myeloid leukemia. Cancer Control 2009; 16:122 - 31; PMID: 19337198
- List AF, Kopecky KJ, Willman CL, Head DR, Slovak ML, Douer D, Dakhil SR, Appelbaum FR. Cyclosporine inhibition of P-glycoprotein in chronic myeloid leukemia blast phase. Blood 2002; 100:1910 - 2; PMID: 12176916
- Hamada A, Miyano H, Watanabe H, Saito H. Interaction of imatinib mesilate with human P-glycoprotein. J Pharmacol Exp Ther 2003; 307:824 - 8; http://dx.doi.org/10.1124/jpet.103.055574; PMID: 12975485
- Mahon FX, Belloc F, Lagarde V, Chollet C, Moreau-Gaudry F, Reiffers J, Goldman JM, Melo JV. MDR1 gene overexpression confers resistance to imatinib mesylate in leukemia cell line models. Blood 2003; 101:2368 - 73; http://dx.doi.org/10.1182/blood.V101.6.2368; PMID: 12609962
- Gurney H, Wong M, Balleine RL, Rivory LP, McLachlan AJ, Hoskins JM, Wilcken N, Clarke CL, Mann GJ, Collins M, et al. Imatinib disposition and ABCB1 (MDR1, P-glycoprotein) genotype. Clin Pharmacol Ther 2007; 82:33 - 40; http://dx.doi.org/10.1038/sj.clpt.6100201; PMID: 17495881
- Dulucq S, Bouchet S, Turcq B, Lippert E, Etienne G, Reiffers J, Molimard M, Krajinovic M, Mahon FX. Multidrug resistance gene (MDR1) polymorphisms are associated with major molecular responses to standard-dose imatinib in chronic myeloid leukemia. Blood 2008; 112:2024 - 7; http://dx.doi.org/10.1182/blood-2008-03-147744; PMID: 18524988
- Stromskaya TP, Rybalkina EY, Kruglov SS, Zabotina TN, Mechetner EB, Turkina AG, Stavrovskaya AA. Role of P-glycoprotein in evolution of populations of chronic myeloid leukemia cells treated with imatinib. Biochemistry (Mosc) 2008; 73:29 - 37; http://dx.doi.org/10.1134/S0006297908010045; PMID: 18294126
- Kim DH, Sriharsha L, Xu W, Kamel-Reid S, Liu X, Siminovitch K, Messner HA, Lipton JH. Clinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemia. Clin Cancer Res 2009; 15:4750 - 8; http://dx.doi.org/10.1158/1078-0432.CCR-09-0145; PMID: 19584153
- Ni LN, Li JY, Miao KR, Qiao C, Zhang SJ, Qiu HR, Qian SX. Multidrug resistance gene (MDR1) polymorphisms correlate with imatinib response in chronic myeloid leukemia. Med Oncol 2011; 28:265 - 9; http://dx.doi.org/10.1007/s12032-010-9456-9; PMID: 20204543
- Maffioli M, Camós M, Gaya A, Hernández-Boluda JC, Alvarez-Larrán A, Domingo A, Granell M, Guillem V, Vallansot R, Costa D, et al. Correlation between genetic polymorphisms of the hOCT1 and MDR1 genes and the response to imatinib in patients newly diagnosed with chronic-phase chronic myeloid leukemia. Leuk Res 2011; 35:1014 - 9; http://dx.doi.org/10.1016/j.leukres.2010.12.004; PMID: 21185600
- Yamakawa Y, Hamada A, Nakashima R, Yuki M, Hirayama C, Kawaguchi T, Saito H. Association of genetic polymorphisms in the influx transporter SLCO1B3 and the efflux transporter ABCB1 with imatinib pharmacokinetics in patients with chronic myeloid leukemia. Ther Drug Monit 2011; 33:244 - 50; PMID: 21311410
- Racil Z, Razga F, Polakova KM, Buresova L, Polivkova V, Dvorakova D, Zackova D, Klamova H, Cetkovsky P, Mayer J. Assessment of adenosine triphosphate-binding cassette subfamily B member 1 (ABCB1) mRNA expression in patients with de novo chronic myelogenous leukemia: the role of different cell types. Leuk Lymphoma 2011; 52:331 - 4; http://dx.doi.org/10.3109/10428194.2010.533220; PMID: 21133723
- Corrêa S, Pizzatti L, Du Rocher B, Mencalha A, Pinto D, Abdelhay E. A comparative proteomic study identified LRPPRC and MCM7 as putative actors in imatinib mesylate cross-resistance in Lucena cell line. Proteome Sci 2012; 10:23; http://dx.doi.org/10.1186/1477-5956-10-23; PMID: 22458888
- Ghiso NS, Lennon GG. lrp130 gene assigned to chromosome 2. In Vitro Cell Dev Biol Anim 1994; 30A:744; http://dx.doi.org/10.1007/BF02631295; PMID: 7881626
- Small ID, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 2000; 25:46 - 7; http://dx.doi.org/10.1016/S0968-0004(99)01520-0; PMID: 10664580
- Mili S, Shu HJ, Zhao Y, Piñol-Roma S. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol Cell Biol 2001; 21:7307 - 19; http://dx.doi.org/10.1128/MCB.21.21.7307-7319.2001; PMID: 11585913
- Liu L, McKeehan WL. Sequence analysis of LRPPRC and its SEC1 domain interaction partners suggests roles in cytoskeletal organization, vesicular trafficking, nucleocytosolic shuttling, and chromosome activity. Genomics 2002; 79:124 - 36; http://dx.doi.org/10.1006/geno.2001.6679; PMID: 11827465
- Tsuchiya N, Fukuda H, Sugimura T, Nagao M, Nakagama H. LRP130, a protein containing nine pentatricopeptide repeat motifs, interacts with a single-stranded cytosine-rich sequence of mouse hypervariable minisatellite Pc-1. Eur J Biochem 2002; 269:2927 - 33; http://dx.doi.org/10.1046/j.1432-1033.2002.02966.x; PMID: 12071956
- Liu L, Amy V, Liu G, McKeehan WL. Novel complex integrating mitochondria and the microtubular cytoskeleton with chromosome remodeling and tumor suppressor RASSF1 deduced by in silico homology analysis, interaction cloning in yeast, and colocalization in cultured cells. In Vitro Cell Dev Biol Anim 2002; 38:582 - 94; http://dx.doi.org/10.1290/1543-706X(2002)38<582:NCIMAT>2.0.CO;2; PMID: 12762840
- Mili S, Piñol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol Cell Biol 2003; 23:4972 - 82; http://dx.doi.org/10.1128/MCB.23.14.4972-4982.2003; PMID: 12832482
- Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci U S A 2003; 100:605 - 10; http://dx.doi.org/10.1073/pnas.242716699; PMID: 12529507
- Tsuchiya N, Fukuda H, Nakashima K, Nagao M, Sugimura T, Nakagama H. LRP130, a single-stranded DNA/RNA-binding protein, localizes at the outer nuclear and endoplasmic reticulum membrane, and interacts with mRNA in vivo. Biochem Biophys Res Commun 2004; 317:736 - 43; http://dx.doi.org/10.1016/j.bbrc.2004.03.103; PMID: 15081402
- Liu L, Vo A, Liu G, McKeehan WL. Putative tumor suppressor RASSF1 interactive protein and cell death inducer C19ORF5 is a DNA binding protein. Biochem Biophys Res Commun 2005; 332:670 - 6; http://dx.doi.org/10.1016/j.bbrc.2005.05.006; PMID: 15907802
- Cooper MP, Qu L, Rohas LM, Lin J, Yang W, Erdjument-Bromage H, Tempst P, Spiegelman BM. Defects in energy homeostasis in Leigh syndrome French Canadian variant through PGC-1alpha/LRP130 complex. Genes Dev 2006; 20:2996 - 3009; http://dx.doi.org/10.1101/gad.1483906; PMID: 17050673
- Tian T, Ikeda J, Wang Y, Mamat S, Luo W, Aozasa K, Morii E. Role of leucine-rich pentatricopeptide repeat motif-containing protein (LRPPRC) for anti-apoptosis and tumourigenesis in cancers. Eur J Cancer 2012; 48:2462 - 73; http://dx.doi.org/10.1016/j.ejca.2012.01.018; PMID: 22326293
- Michaud M, Barakat S, Magnard S, Rigal D, Baggetto LG. Leucine-rich protein 130 contributes to apoptosis resistance of human hepatocarcinoma cells. Int J Oncol 2011; 38:169 - 78; PMID: 21109938
- Labialle S, Dayan G, Gayet L, Rigal D, Gambrelle J, Baggetto LG. New invMED1 element cis-activates human multidrug-related MDR1 and MVP genes, involving the LRP130 protein. Nucleic Acids Res 2004; 32:3864 - 76; http://dx.doi.org/10.1093/nar/gkh722; PMID: 15272088
- Labialle S, Gayet L, Marthinet E, Rigal D, Baggetto LG. Transcriptional regulators of the human multidrug resistance 1 gene: recent views. Biochem Pharmacol 2002; 64:943 - 8; http://dx.doi.org/10.1016/S0006-2952(02)01156-5; PMID: 12213590
- Baker EK, Johnstone RW, Zalcberg JR, El-Osta A. Epigenetic changes to the MDR1 locus in response to chemotherapeutic drugs. Oncogene 2005; 24:8061 - 75; http://dx.doi.org/10.1038/sj.onc.1208955; PMID: 16091741
- Reed K, Hembruff SL, Laberge ML, Villeneuve DJ, Côté GB, Parissenti AM. Hypermethylation of the ABCB1 downstream gene promoter accompanies ABCB1 gene amplification and increased expression in docetaxel-resistant MCF-7 breast tumor cells. Epigenetics 2008; 3:270 - 80; http://dx.doi.org/10.4161/epi.3.5.6868; PMID: 19001875
- Baker EK, El-Osta A. The rise of DNA methylation and the importance of chromatin on multidrug resistance in cancer. Exp Cell Res 2003; 290:177 - 94; http://dx.doi.org/10.1016/S0014-4827(03)00342-2; PMID: 14567978
- Jain S, Wojdacz TK, Su YH. Challenges for the application of DNA methylation biomarkers in molecular diagnostic testing for cancer. Expert Rev Mol Diagn 2013; 13:283 - 94; http://dx.doi.org/10.1586/erm.13.9; PMID: 23570406
- Ueda K, Pastan I, Gottesman MM. Isolation and sequence of the promoter region of the human multidrug-resistance (P-glycoprotein) gene. J Biol Chem 1987; 262:17432 - 6; PMID: 2891692
- Ueda K, Clark DP, Chen CJ, Roninson IB, Gottesman MM, Pastan I. The human multidrug resistance (mdr1) gene. cDNA cloning and transcription initiation. J Biol Chem 1987; 262:505 - 8; PMID: 3027054
- Raguz S, Randle RA, Sharpe ER, Foekens JA, Sieuwerts AM, Meijer-van Gelder ME, Melo JV, Higgins CF, Yagüe E. Production of P-glycoprotein from the MDR1 upstream promoter is insufficient to affect the response to first-line chemotherapy in advanced breast cancer. Int J Cancer 2008; 122:1058 - 67; http://dx.doi.org/10.1002/ijc.23149; PMID: 17955490
- Raguz S, Tamburo De Bella M, Tripuraneni G, Slade MJ, Higgins CF, Coombes RC, Yagüe E. Activation of the MDR1 upstream promoter in breast carcinoma as a surrogate for metastatic invasion. Clin Cancer Res 2004; 10:2776 - 83; http://dx.doi.org/10.1158/1078-0432.CCR-03-0517; PMID: 15102684
- Shi CJ, Wang F, Ren MF, Mi YJ, Yan YY, To KK, Dai CL, Wang YS, Chen LM, Tong XZ, et al. Up-regulation of ABCB1/P-glycoprotein by escaping promoter hypermethylation indicates poor prognosis in hematologic malignancy patients with and without bone marrow transplantation. Leuk Res 2011; 35:73 - 9; http://dx.doi.org/10.1016/j.leukres.2010.04.019; PMID: 20488541
- Shtil AA, Azare J. Redundancy of biological regulation as the basis of emergence of multidrug resistance. Int Rev Cytol 2005; 246:1 - 29; http://dx.doi.org/10.1016/S0074-7696(05)46001-5; PMID: 16164965
- Lage H. An overview of cancer multidrug resistance: a still unsolved problem. Cell Mol Life Sci 2008; 65:3145 - 67; http://dx.doi.org/10.1007/s00018-008-8111-5; PMID: 18581055
- Hochhaus A. Chronic myelogenous leukemia (CML): resistance to tyrosine kinase inhibitors. Ann Oncol 2006; 17:Suppl 10 x274 - 9; http://dx.doi.org/10.1093/annonc/mdl273; PMID: 17018738
- Tang C, Schafranek L, Watkins DB, Parker WT, Moore S, Prime JA, White DL, Hughes TP. Tyrosine kinase inhibitor resistance in chronic myeloid leukemia cell lines: investigating resistance pathways. Leuk Lymphoma 2011; 52:2139 - 47; http://dx.doi.org/10.3109/10428194.2011.591013; PMID: 21718141
- Chomel JC, Turhan AG. Chronic myeloid leukemia stem cells in the era of targeted therapies: resistance, persistence and long-term dormancy. Oncotarget 2011; 2:713 - 27; PMID: 21946665
- Hou J, Wang F, McKeehan WL. Molecular cloning and expression of the gene for a major leucine-rich protein from human hepatoblastoma cells (HepG2). In Vitro Cell Dev Biol Anim 1994; 30A:111 - 4; http://dx.doi.org/10.1007/BF02631402; PMID: 8012652
- Li X, Lv L, Zheng J, Zhou J, Liu B, Chen H, Liang C, Wang R, Su L, Li X, et al. The significance of LRPPRC overexpression in gastric cancer. Med Oncol 2014; 31:818; http://dx.doi.org/10.1007/s12032-013-0818-y; PMID: 24375316
- Seo DC, Sung JM, Cho HJ, Yi H, Seo KH, Choi IS, Kim DK, Kim JS, El-Aty AM A, Shin HC. Gene expression profiling of cancer stem cell in human lung adenocarcinoma A549 cells. Mol Cancer 2007; 6:75; http://dx.doi.org/10.1186/1476-4598-6-75; PMID: 18034892
- Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell 2012; 10:717 - 28; http://dx.doi.org/10.1016/j.stem.2012.05.007; PMID: 22704512
- Baccelli I, Trumpp A. The evolving concept of cancer and metastasis stem cells. J Cell Biol 2012; 198:281 - 93; http://dx.doi.org/10.1083/jcb.201202014; PMID: 22869594
- Bibber B, Sinha G, Lobba AR, Greco SJ, Rameshwar P. A review of stem cell translation and potential confounds by cancer stem cells. Stem Cells Int 2013; 2013:241048; http://dx.doi.org/10.1155/2013/241048; PMID: 24385986
- Enokida H, Shiina H, Igawa M, Ogishima T, Kawakami T, Bassett WW, Anast JW, Li LC, Urakami S, Terashima M, et al. CpG hypermethylation of MDR1 gene contributes to the pathogenesis and progression of human prostate cancer. Cancer Res 2004; 64:5956 - 62; http://dx.doi.org/10.1158/0008-5472.CAN-04-0081; PMID: 15342374
- Taylor KH, Pena-Hernandez KE, Davis JW, Arthur GL, Duff DJ, Shi H, Rahmatpanah FB, Sjahputera O, Caldwell CW. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Res 2007; 67:2617 - 25; http://dx.doi.org/10.1158/0008-5472.CAN-06-3993; PMID: 17363581
- Klajic J, Fleischer T, Dejeux E, Edvardsen H, Warnberg F, Bukholm I, Lønning PE, Solvang H, Børresen-Dale AL, Tost J, et al. Quantitative DNA methylation analyses reveal stage dependent DNA methylation and association to clinico-pathological factors in breast tumors. BMC Cancer 2013; 13:456; http://dx.doi.org/10.1186/1471-2407-13-456; PMID: 24093668
- Lee TB, Park JH, Min YD, Kim KJ, Choi CH. Epigenetic mechanisms involved in differential MDR1 mRNA expression between gastric and colon cancer cell lines and rationales for clinical chemotherapy. BMC Gastroenterol 2008; 8:33; http://dx.doi.org/10.1186/1471-230X-8-33; PMID: 18673531
- Tahara T, Arisawa T, Shibata T, Yamashita H, Yoshioka D, Hirata I. Effect of promoter methylation of multidrug resistance 1 (MDR1) gene in gastric carcinogenesis. Anticancer Res 2009; 29:337 - 41; PMID: 19331170
- Sharma G, Mirza S, Parshad R, Srivastava A, Datta Gupta S, Pandya P, Ralhan R. CpG hypomethylation of MDR1 gene in tumor and serum of invasive ductal breast carcinoma patients. Clin Biochem 2010; 43:373 - 9; http://dx.doi.org/10.1016/j.clinbiochem.2009.10.009; PMID: 19879256
- Nakayama M, Wada M, Harada T, Nagayama J, Kusaba H, Ohshima K, Kozuru M, Komatsu H, Ueda R, Kuwano M. Hypomethylation status of CpG sites at the promoter region and overexpression of the human MDR1 gene in acute myeloid leukemias. Blood 1998; 92:4296 - 307; PMID: 9834236
- Rumjanek VM, Trindade GS, Wagner-Souza K, de-Oliveira MC, Marques-Santos LF, Maia RC, Capella MA. Multidrug resistance in tumour cells: characterization of the multidrug resistant cell line K562-Lucena 1. An Acad Bras Cienc 2001; 73:57 - 69; http://dx.doi.org/10.1590/S0001-37652001000100007; PMID: 11246270
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402 - 8; http://dx.doi.org/10.1006/meth.2001.1262; PMID: 11846609
- Vaissière T, Cuenin C, Paliwal A, Vineis P, Hoek G, Krzyzanowski M, Airoldi L, Dunning A, Garte S, Hainaut P, et al. Quantitative analysis of DNA methylation after whole bisulfitome amplification of a minute amount of DNA from body fluids. Epigenetics 2009; 4:221 - 30; PMID: 19458486
