Abstract
Silencing of tumor suppressor genes (TSGs) by DNA promoter hypermethylation is an early event in carcinogenesis and a potential target for personalized cancer treatment. In head and neck cancer, little is known about the role of promoter hypermethylation in survival. Using methylation specific multiplex ligation-dependent probe amplification (MS-MLPA) we investigated the role of promoter hypermethylation of 24 well-described genes (some of which are classic TSGs), which are frequently methylated in different cancer types, in 166 HPV-negative early oral squamous cell carcinomas (OSCC), and 51 HPV-negative early oropharyngeal squamous cell carcinomas (OPSCC) in relation to clinicopathological features and survival. Early OSCC showed frequent promoter hypermethylation in RARB (31% of cases), CHFR (20%), CDH13 (13%), DAPK1 (12%), and APC (10%). More hypermethylation (≥ 2 genes) independently correlated with improved disease specific survival (hazard ratio 0.17, P = 0.014) in early OSCC and could therefore be used as prognostic biomarker. Early OPSCCs showed more hypermethylation of CDH13 (58%), TP73 (14%), and total hypermethylated genes. Hypermethylation of two or more genes has a significantly different effect on survival in OPSCC compared with OSCC, with a trend toward worse instead of better survival. This could have a biological explanation, which deserves further investigation and could possibly lead to more stratified treatment in the future.
Introduction
Head and neck cancer is the sixth most common malignancy worldwide and appears to be a highly heterogeneous group of malignant diseases, of which approximately one third consists of oral squamous cell carcinoma (OSCC). Despite improvements in therapy, the five-year survival rate has not significantly changed over the past decades and remains approximately 50%.Citation1,Citation2 To improve the outcome of patients with OSCC, it is pivotal to understand the molecular biology of distinctive tumors and find predictive biomarkers for targeted therapy.Citation3
Besides genetic changes, epigenetic alterations may lead to changes in gene expression as well; these include modifications of the genome without changes in the underlying DNA sequence.Citation4 Epigenetic regulation plays a central role in both embryogenesis and cell type differentiation of normal cells. DNA promoter hypermethylation of tumor suppressor genes (TSGs) is the best characterized epigenetic event in carcinogenesis.Citation5,Citation6 Aberrant methylation of cytosine occurs at CpG dinucleotide (or CpG islands) rich promoter regions of TSGs and is catalyzed by DNA methyltransferases (DNMTs). This promoter hypermethylation results in a closed chromatin configuration and therefore blocks the access of transcription factors to the promoter, leading to the transcriptional silencing of these TSGs.Citation7,Citation8 In contrast to genetic events, DNA methylation is reversible and could therefore serve as an attractive target for new therapeutic strategies using DNMT inhibitors to reactivate methylation-silenced TSGs.Citation4,Citation7 In many cancers, gene silencing by promoter methylation seems to be an early event in carcinogenesis and may occur even more frequently than structural inactivation of genes by mutations and deletions.Citation5,Citation9
Promoter hypermethylation profiles of head and neck squamous cell carcinoma have been explored widely, though most promoter hypermethylation studies evaluated only a limited number of genes or combined a mixture of different tumor stages of OSCC and oropharyngeal squamous cell carcinomas (OPSCC) and did not take high-risk types of human papillomavirus (HPV) status into account.Citation10,Citation11 This is important because prevalence of HPV is higher in OPSCC than in OSCC and HPV-driven squamous cell carcinomas are known to show more promoter hypermethylation than HPV-negative tumors.Citation3,Citation10 Moreover, most studies did not correlate promoter hypermethylation with clinical outcome, such as survival.
In this study we correlated promoter hypermethylation of 24 common methylated genes in cancer with clinicopathological features and survival in a large group of early HPV-negative OSCC. In addition, we compared these results with a group of early HPV-negative OPSCC to see if these clinically similar groups of head and neck cancers are epigenetically different.
Results
Methylation status of early OSCC and HPV-status
In 30 tumors, not enough DNA could be extracted. In another 15 cases, the amount and/or quality of the extracted DNA was insufficient for analysis. Therefore, 166 OSCC samples and 24 normal oral mucosa samples were available for further analysis. No HPV-positive tumors were found in this cohort of 166 early OSCC. The extent of promoter methylation of these samples and normal oral tissue is shown inFigure S1.
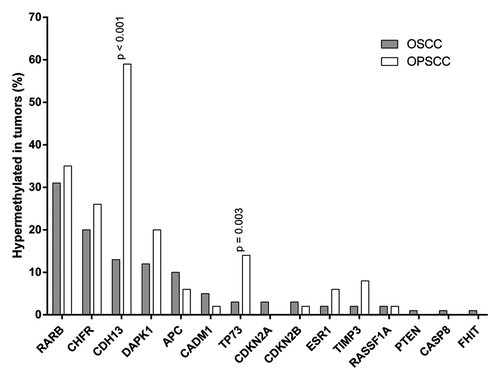
Fifty-eight percent of the OSCC samples showed hypermethylation of at least one gene, with a maximum of six hypermethylated genes. Hypermethylation frequencies of these 24 genes in OSCC are presented in . Hypermethylation was most frequent for RARB (31%), CHFR (20%), CDH13 (13%), DAPK1 (12%), and APC (10%). There was no hypermethylation in GSTP1, CD44, HIC1, VHL, ATM, CDKN1B, MLH1, BRCA1, and BRCA2.
Table 1. Gene promoter hypermethylation early oral (166) and oropharyngeal (51) cancers
Methylation status of early OSCC and clinicopathological features
Compared with OSCC of the tongue, OSCC of the floor of the mouth showed a higher level of hypermethylation of RARB (46% vs 21%, P = 0.001), DAPK1 (19% vs 8%, P = 0.030), and CHFR (30% vs 14%, P = 0.009). The total number of hypermethylated genes compared with OSCC of the tongue was also higher in univariate analysis. After correction for differences in TNM-classification, smoking and alcohol consumption, hypermethylation of RARB (P = 0.011) and CHFR (P = 0.023), and a higher total number of hypermethylated genes correlated with early OSCC of the floor of the mouth. Promoter hypermethylation of none of the 24 genes correlated with age (in continuum), nodal metastasis, or aggressive growth patterns (a non-cohesive tumor front, vascular invasive or perineural growth).
Survival analysis of early OSCC
Although promoter hypermethylation of individual genes did not correlate with survival, hypermethylation of two or more genes (P = 0.007) correlated significantly with improved disease specific survival in early OSCC (hazard ratio 0.18, P = 0.002), see . From the baseline characteristics, age (hazard ratio 1.03, P = 0.044) and nodal metastases (hazard ratio 3.93, P < 0.001) also correlated with decreased disease specific survival. None of the baseline characteristics turned out to be a confounder or effect modifier. Multivariate analysis revealed the presence of nodal metastases (hazard ratio 4.13, P < 0.001) and hypermethylation of two or more genes (hazard ratio 0.17, P = 0.014) as independent prognostic factors for disease specific survival in early OSCC (see .)
Figure 1. Promoter hypermethylation and disease specific survival in early OSCC samples. Log Rank test: P = 0.007; Cox regression analysis: hazard ratio 0.18 (95% confidence interval: 0.04 - 0.75), P = 0.002. *(patients/events): < 2 meth. genes 29 events in 123 patients; ≥ 2 meth. genes 2 events in 43 patients.
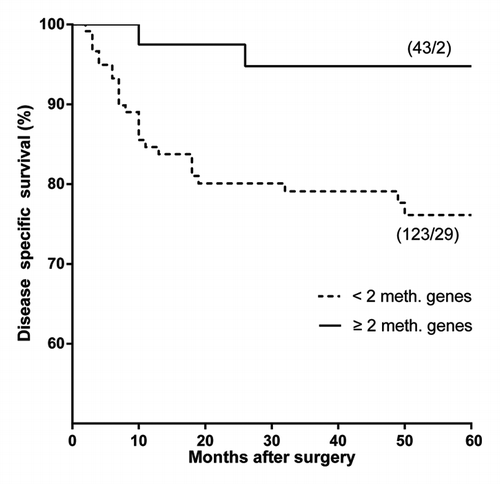
Table 2. Cox regression analysis of disease specific survival in early OSCC
Comparison of methylation status between early OSCC and OPSCC
HPV-positive OPSCC has a distinct molecular biology and clinical behavior. As none of the OSCC samples was HPV-positive, only HPV-negative early OPSCC were compared with early OSCC. Besides the known differences in HPV status between these sub-sites of head and neck cancer, there were several significant clinical differences between these cohorts: in the OPSCC group the cigarette and alcohol consumption was higher and tumor size was larger. Patients with OPSCC were clinically suspected to have lymph node metastases and their primary treatment consisted of radiotherapy instead of surgery (see .)
Table 3. Characteristics of early OSCC and OPSCC
The differences between promoter hypermethylation of OSCC and OPSCC are illustrated in and . In univariate analysis, CDH13 (P < 0.001) and TP73 (P = 0.003) showed different hypermethylation levels. After correction for baseline sub-site differences, multivariate analysis revealed that promoter regions of TP73 and CDH13 were significantly less frequently hypermethylated in OSCC. In multivariate analysis, the percentage of tumors with 2 or more hypermethylated genes was significantly lower in OSCC compared with OPSCC (P < 0.001).
Figure 2. Promoter hypermethylation in early OSCC and OPSCC samples. Only genes with hypermethylation in at least one sample are illustrated.
Although there is no significant correlation between hypermethylation and overall survival in OSCC (P = 0.055) or OPSCC (P = 0.080), the location (oral cavity or oropharynx) appeared to be a significant effect modifier (P = 0.012) for the correlation between 2 or more methylated genes and overall survival, see . In OSCC, the 5-year overall survival in less methylated tumors was 70% and in more methylated tumors 85%. In contrast, the 5-year overall survival in less methylated OPSCCs was 76% and in more methylated tumors only 43%. Due to limited events we could not construct a robust multivariate model; however, there were no significant differences in TMA-classification, cigarette or alcohol consumption, and treatment of primary tumor between subgroups with OSCC or OPSCC (see .)
Figure 3. Promoter hypermethylation and overall survival in patients with early OSCC and OPSCC. ≥ 2 hypermethylated genes is an effect modifier in oral and oropharyngeal SCC, P = 0.012. Log Rank test: OSCC (P = 0.054); OPSCC (P = 0.080). *(patients/events): OSCC < 2 meth. genes 36 events in 122 patients; ≥ 2 meth. genes 6 events in 43 patients; OPSCC < 2 meth. genes 4 events in 22 patients; ≥ 2 meth. genes 11 events in 25 patients. One OSCC patient out of analysis due to missing data.
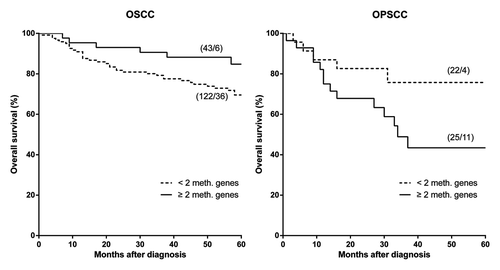
Table 4. Baseline characteristics in OSCC and OPSCC in methylated subgroups
Discussion
Promoter hypermethylation of TSGs disrupts the tumor suppressor function by gene-silencing and is thought to be an early event in carcinogenesis.Citation8,Citation12 Understanding the epigenetic role of promoter hypermethylation in early oral cancer is important to gain new insight into oral carcinogenesis, and to identify diagnostic and prognostic biomarkers, as well as potential therapeutic targets.Citation11 Although multiple studies have evaluated the role of promoter hypermethylation in head and neck cancer, wide ranges of hypermethylation frequencies have been reported. Differences in methylation testing methodology, variations in sample processing, differences in composition of patient cohorts (mixing different sub-sites and tumor stages) and the lack HPV-status determination probably account for this wide range of reported methylation data.Citation10,Citation11 Therefore, MS-MLPA was used to investigate promoter hypermethylation of multiple frequently methylated genes in a large homogeneous group of early oral (tongue and floor of mouth) squamous cell carcinomas to evaluate its prognostic value.
From our 24-gene panel, five genes showed promoter hypermethylation in at least 10% of early OSCC samples and may be involved in oral carcinogenesis: RARB (31%), CHFR (20%), CDH13 (13%), DAPK1 (12%), and APC (10%). This is in accordance with earlier studies evaluating promoter hypermethylation in head and neck cancer using MS-MLPA or methylation-specific PCR (MSP), although published methylation rates vary widely: RARB (15–80%), CHFR (19–61%), CDH13 (10–90%), DAPK1 (7–77%), APC (9–34%).Citation11,Citation13-Citation15 Two genes, MLH1 and RASSF1A, are rarely methylated in our cohorts of OSCC and OPSCC, although they have been described to be methylated in a wide range (2–84%) in earlier head and neck cancer studies.Citation11 These apparently inconsistent results could be explained by differences in methodology, investigated CpGs, and cohort composition. Many previous studies evaluated promoter hypermethylation using MS-PCR with bisulfite-modified templates, a method which is prone to overestimate the number of methylated samples due to incomplete bisulfite conversion.Citation16,Citation17 Comparison of multiple techniques showed this overestimation for RARB and RASSF1A in head and neck cancer.Citation14 Together with differences in thresholds to define hypermethylation, this could explain the wide range of results in the literature. Advantages of MS-MLPA over MSP are the possibility to use genomic DNA instead of bisulfite-modified templates, the quantitative nature of MLPA, and the analysis of multiple genes in one reaction. MS-MLPA is restricted to methylation sites containing a restriction site (GCGC) for the methylation-sensitive HhaI enzyme, while MSP targets a specific CpG with the CpG island, which could also explain differences between MS-MLPA and MSP. However, multiple studies show a good correlation between MS-MLPA and pyrosequencing or (Q)MSP. For example, Furlan et al. showed a correlation of 95% between MS-MLPA and MSP (n = 102) and a correlation of 96% between MS-MLPA and bisulfite pyrosequencing (n = 96). We believe these high correlation percentages justify the reliability of MS-MLPA.Citation18-Citation25
The presence of two or more hypermethylated genes proved to be an independent predictor for better disease specific survival in our cohort of early OSCC, which seems paradoxical since promoter hypermethylation leads to gene silencing, as mentioned above. However, multiple studies found correlations between promoter hypermethylation of TSGs and increased survival in lung, oral and gastric cancer.Citation26-Citation28 A possible explanation for this seemingly contradictory phenomenon may be that hypermethylated carcinomas have fewer genetic alterations, i.e., mutations and/or deletions, and are, therefore, less aggressive.Citation26,Citation28 In gastric cancer, patients with promoter hypermethylation of E-cadherin have a better survival rate than patients with somatic mutations of E-cadherin.Citation26 This supports the theory of a less aggressive phenotype in hypermethylated tumors.
OPSCC samples showed higher promoter hypermethylation than OSCC samples in this study. Although HPV-driven OPSCC are known to have higher level of promoter hypermethylation, HPV-positive tumors were excluded in our analysis.Citation10 Differences in smoking habits may partially account for the higher promoter hypermethylation in OPSCC samples, which is in line with reports on lung cancer in which smoking habits correlated with promoter methylation patterns.Citation28-Citation30 However, even after correction for smoking habits, TP73 and CDH13 still showed significantly different promoter hypermethylation levels in OSCC vs. OPSCC. This indicates a different biological behavior of tumors between these sub-sites in head and neck cancer, which is in line with earlier studies.Citation31,Citation32
Although we found no significant correlation between hypermethylation and overall survival in both OSCC and OPSCC samples, the effect of hypermethylation on survival in these sub-sites is significantly different. This difference may be explained by the treatment modality of the primary tumor, which was radiotherapy instead of surgery. The study by Huang et al. reported a similar phenomenon with worse outcome after radiotherapy, but not surgery, in oral cancer in tumors with promoter methylation of RASSF1A, RASSF2A, or HIN-1. These genes are involved in the Ras/P13K/AKT pathway and known to be associated with radio-resistance, which could explain the differences in outcome.Citation33 Frequently hypermethylated genes (> 10% of patients) associated with radiosensitivity in our OPSCC group are: CDH13, RARB, CHFR, and PT73. T-cadherin (CDH13) is an intercellular adhesion molecule, which plays a role in the regulation of cell proliferation, invasion, and intracellular signaling during cancer progression.Citation34 It is an inhibitor of the Ras/P13K/AKT pathway and promoter hypermethylation could therefore be involved in the development of radioresistance.Citation35 Retinoic acid receptor β (RARB) is associated with both cell growth and differentiation.Citation7 Loss of RARB in cancer is mostly the result of promoter hypermethylation instead of genetic aberrations.Citation16 Decrease of both gene and protein expression are correlated with late response to radiotherapy in cervical cancer.Citation36 Checkpoint with forkhead and ring finger domains (CHFR) has recently been identified as a checkpoint protein, which safeguards mitotic entry and, therefore, affects cell cycle progression.Citation37 CHFR silencing leads to upregulation of PARP1, a DNA repair enzyme associated with resistance to radiotherapy.Citation38,Citation39 TP73 encodes for p73, family of p53, which is involved in cell cycle regulation and induction of apoptosis. In contrast to p53, p73 is rarely mutated in human cancer but seems to be silenced by promoter hypermethylation. Overexpression of p73 is associated with cellular radiosensitivity in cervical cancers, which indicates an important role for p73 in response to radiotherapy.Citation40 The mutual function of these frequently methylated genes in development of radio-resistance could possibly explain these differences in survival between OSCC and OPSCC samples; however, further research is needed to confirm this hypothesis. In the future, this may have implications for further stratified treatment, such as combination of radiotherapy with DNMT inhibitors or surgery as treatment modality of first choice in early OPSCC with more hypermethylated genes.Citation4
In conclusion, promoter methylation analysis of a large cohort of early OSCC using MS-MLPA identified RARB, CHFR, CDH13, APC, and DAPK1 as frequently hypermethylated genes and, therefore, potential therapeutic targets in oral cancer, given the reversible nature of epigenetic gene silencing. In addition, hypermethylation of 2 or more genes in this 24-gene panel could be used as prognosticator in early OSCC. Compared with HPV-negative early OPSCC, early OSCC show distinct methylation patterns, which illustrates that epigenetic changes observed in head and neck cancer are sub-site dependent. Furthermore, hypermethylation in this 24-gene panel shows a different effect on survival in OPSCCs compared with OSCCs, with a trend toward worse instead of better survival. This might have a biological explanation that deserves further research and might indicate more stratified treatments in the future.
Materials and Methods
Patients and clinicopathological information
All patients with a histologically confirmed early OSCC (cT1–2 N0, stage I-II), primarily treated by surgery between January 2004 and December 2010 at the University Medical Center Utrecht were included. Exclusion criteria were: a previous history of head and neck squamous cell carcinoma or synchronous primary tumor. These criteria resulted in a total of 211 OSCCs. Demographical, clinical and survival data were retrieved from electronic medical records. Fifty-one HPV-negative early oropharyngeal cancers from a consecutive cohort of 200 patients with OPSCC were used for comparison.
A dedicated head and neck pathologist (SMW) assessed margin status, tumor diameter, tumor thickness, and the histological features of the tumor front, i.e., invasive pattern, perineural and vascular invasive growth. All this information was handled in a coded fashion, according to Dutch national ethical guidelines. The standard treatment agreement with patients in our hospital includes anonymous use of redundant tissue for research purposes (Code for proper secondary use of human tissue, Dutch Federation of Medical Scientific Societies).Citation41 HPV status was determined for all tumors by a combination of p16 immunohistochemistry and molecular analysis as described below. In addition, normal oral cavity mucosa of 24 patients treated for an oral fibroma (due to chronic irritation by dentures or dental prosthesis) with no history of head and neck cancer was used as control tissue.
DNA isolation
A dedicated head and neck pathologist (SMW) identified tumor areas with at least 30% tumor cells on HE slides for DNA extraction. For the control tissue, normal oral mucosa was identified on HE slides as well. Corresponding areas were dissected from deparaffinized 5μm slides and suspended in direct lysis buffer (50 mM Tris-HCL, pH 8.0; 0.5% Tween 20). After overnight incubation with proteinase K (10mg/ml; Roche) at 56 degrees, followed by boiling for 10 min, the supernatant was extracted after centrifugation. DNA was stored at -20 degrees until use.
HPV DNA detection
HPV-16 status was determined according to a well-established algorithm for HPV determination in paraffin embedded head and neck cancer tissue.Citation42 Immunohistochemistry for p16 (p16INK4A-specific primary mouse monoclonal antibody, clone 16P07, Neomarkers) and the Linear Array HPV Genotyping Test (Linear Array HPV Genotyping Kit: 03378179 190, Linear Array HPV Detection Kit: 208693; Roche) were performed as described earlier.Citation43 For p16 expression both intensity (0, 1+, 2+ or 3+) and percentage of stained tumor cells were scored by a dedicated head and neck pathologist (SMW). In case of p16 overexpression, defined as strong (2+/3+) nuclear and/or cytoplasmic staining of over 70% of tumor cells, the Linear Array HPV Genotyping Test was performed to identify HPV-16 positive cases.
Methylation-Specific Multiplex Ligation-dependent Probe Amplification (MS-MLPA)
For promoter methylation analysis, MS-MLPA was performed according to the manufacturer’s instructions using the SALSA MS-MLPA probemix ME001-C2 (MRC Holland), which contains 15 control probes and 26 HhaI-sensitive probes of 24 TSGs or genes with a tumor suppressor like function. Multiple genes of this kit have been associated with hypermethylation in head and neck cancer in earlier studies.Citation11,Citation44 Table S1 shows an overview of the functions of these genes and their associated hallmark of cancer.Citation45
All runs were performed on a Veriti 96-well thermal cycler (Applied Biosystems, Foster city, CA, USA). Positive (100% methylated, CpG methylase treated) and negative controls, both derived from human blood were taken along in each MS-MLPA run in duplicate. PCR fragments were separated by electrophoresis (ABI 3730 capillary sequencer, Applied Biosystems, Foster city, CA, USA). Genemapper software version 4.1 (Applied Biosystems) and Coffalyser.NET analysis software (MRC Holland) were used for methylation status analysis, see Table S2 for representative examples of the MS-MLPA assay including controls (negative and positive). The MS-MLPA platform for promoter methylation analysis has been described in more detail in existing literature.Citation46 As MLH1 and RASSF1A both contained two probes for different CpG sites, the mean value of these two probes was used for further analysis. Promoter methylation analysis of 24 normal oral mucosa samples was performed using the same method and tumor suppressor kit to define a cutoff point for promoter hypermethylation. None of the normal oral mucosa tissues exceeded 15% methylation in our panel of genes. Therefore we defined 15% promoter methylation as cutoff for promoter hypermethylation as before, see Figure S1.
Statistics
Pearson X2 test (or Fisher’s exact when appropriate) for categorical variables and ANOVA for continuous variables were used to compare baseline characteristics and frequency of hypermethylation of individual genes between OSCC and OPSCC. To adjust for baseline differences, backward logistic regression was performed to compare methylation in OSCC and OPSCC, entering significant univariate features. Overall and disease specific survival was examined using Kaplan-Meier survival curves, and differences between strata were tested using LogRank test. Cox regression analysis (backward logistic regression, probability for stepwise entry 0.05 and removal 0.10) was used for multivariate analysis in case of sufficient events. Prior to multivariate analysis, baseline characteristics were screened for effect modifiers by cox regression effect modification analysis. Baseline characteristics with a significant correlation with survival and baseline characteristics revealed as possible confounders by cox regression analysis were included in the multivariate model. All p-values were based on two-tailed statistical analysis and p-values < 0.05 were considered to be statistically significant. Statistical analyses were performed using IBM SPSS 20.0 statistical software.
| Abbreviations: | ||
| OSCC | = | oral squamous cell carcinoma |
| TSG | = | tumor suppressor gene |
| DNMT | = | DNA methyltransferase |
| OPSCC | = | oropharyngeal squamous cell carcinoma |
| HPV | = | human papillomavirus |
| MS-MLPA | = | Methylation-Specific Multiplex Ligation-dependent Probe Amplification |
| MSP | = | methylation-specific PCR |
| HR | = | hazard ratio |
| CI | = | confidence interval |
| NA | = | not available |
Additional material
Download Zip (708.8 KB)Conflict of Interest
No conflicts to disclose
Acknowledgments
RN is funded by the Dutch Cancer Society (research grant: 2014–6620).
SMW is funded by the Dutch Cancer Society (clinical fellowship: 2011–4964).
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61:69 - 90; http://dx.doi.org/10.3322/caac.20107; PMID: 21296855
- Crowe DL, Hacia JG, Hsieh CL, Sinha UK, Rice H. Molecular pathology of head and neck cancer. Histol Histopathol 2002; 17:909 - 14; PMID: 12168802
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011; 11:9 - 22; http://dx.doi.org/10.1038/nrc2982; PMID: 21160525
- Tsai HC, Baylin SB. Cancer epigenetics: linking basic biology to clinical medicine. Cell Res 2011; 21:502 - 17; http://dx.doi.org/10.1038/cr.2011.24; PMID: 21321605
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3:415 - 28; PMID: 12042769
- Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005; 6:597 - 610; http://dx.doi.org/10.1038/nrg1655; PMID: 16136652
- Gasche JA, Goel A. Epigenetic mechanisms in oral carcinogenesis. Future Oncol 2012; 8:1407 - 25; http://dx.doi.org/10.2217/fon.12.138; PMID: 23148615
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet 2000; 16:168 - 74; http://dx.doi.org/10.1016/S0168-9525(99)01971-X; PMID: 10729832
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003; 349:2042 - 54; http://dx.doi.org/10.1056/NEJMra023075; PMID: 14627790
- van Kempen PM, Noorlag R, Braunius WW, Stegeman I, Willems SM, Grolman W. Differences in methylation profiles between HPV-positive and HPV-negative oropharynx squamous cell carcinoma: a systematic review. Epigenetics 2014; 9:194 - 203; http://dx.doi.org/10.4161/epi.26881; PMID: 24169583
- Demokan S, Dalay N. Role of DNA methylation in head and neck cancer. Clin Epigenetics 2011; 2:123 - 50; http://dx.doi.org/10.1007/s13148-011-0045-3; PMID: 22704334
- Verschuur-Maes AH, de Bruin PC, van Diest PJ. Epigenetic progression of columnar cell lesions of the breast to invasive breast cancer. Breast Cancer Res Treat 2012; 136:705 - 15; http://dx.doi.org/10.1007/s10549-012-2301-4; PMID: 23104224
- Chen K, Sawhney R, Khan M, Benninger MS, Hou Z, Sethi S, Stephen JK, Worsham MJ. Methylation of multiple genes as diagnostic and therapeutic markers in primary head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg 2007; 133:1131 - 8; http://dx.doi.org/10.1001/archotol.133.11.1131; PMID: 18025318
- Yalniz Z, Demokan S, Suoglu Y, Ulusan M, Dalay N. Simultaneous methylation profiling of tumor suppressor genes in head and neck cancer. DNA Cell Biol 2011; 30:17 - 24; http://dx.doi.org/10.1089/dna.2010.1090; PMID: 20860434
- Sun D, Zhang Z, Van N, Huang G, Ernberg I, Hu L. Aberrant methylation of CDH13 gene in nasopharyngeal carcinoma could serve as a potential diagnostic biomarker. Oral Oncol 2007; 43:82 - 7; http://dx.doi.org/10.1016/j.oraloncology.2006.01.007; PMID: 16807071
- Shaw RJ, Hall GL, Lowe D, Liloglou T, Field JK, Sloan P, Risk JM. The role of pyrosequencing in head and neck cancer epigenetics: correlation of quantitative methylation data with gene expression. Arch Otolaryngol Head Neck Surg 2008; 134:251 - 6; http://dx.doi.org/10.1001/archoto.2007.50; PMID: 18347248
- Claus R, Wilop S, Hielscher T, Sonnet M, Dahl E, Galm O, Jost E, Plass C. A systematic comparison of quantitative high-resolution DNA methylation analysis and methylation-specific PCR. Epigenetics 2012; 7:772 - 80; http://dx.doi.org/10.4161/epi.20299; PMID: 22647397
- Hömig-Hölzel C, Savola S. Multiplex ligation-dependent probe amplification (MLPA) in tumor diagnostics and prognostics. Diagn Mol Pathol 2012; 21:189 - 206; http://dx.doi.org/10.1097/PDM.0b013e3182595516; PMID: 23111197
- Stuppia L, Antonucci I, Palka G, Gatta V. Use of the MLPA Assay in the Molecular Diagnosis of Gene Copy Number Alterations in Human Genetic Diseases. Int J Mol Sci 2012; 13:3245 - 76; http://dx.doi.org/10.3390/ijms13033245; PMID: 22489151
- Suijkerbuijk KP, Pan X, van der Wall E, van Diest PJ, Vooijs M. Comparison of different promoter methylation assays in breast cancer. Anal Cell Pathol (Amst) 2010; 33:133 - 41; PMID: 20978321
- Leong KJ, Wei W, Tannahill LA, Caldwell GM, Jones CE, Morton DG, Matthews GM, Bach SP. Methylation profiling of rectal cancer identifies novel markers of early-stage disease. Br J Surg 2011; 98:724 - 34; http://dx.doi.org/10.1002/bjs.7422; PMID: 21360524
- Cardoso LC, Tenorio Castaño JA, Pereira HS, Lima MA, Dos Santos AC, de Faria PS, Ferman S, Seuánez HN, Nevado JB, de Almeida JC, et al. Constitutional and somatic methylation status of DMRH19 and KvDMR in Wilms tumor patients. Genet Mol Biol 2012; 35:714 - 24; http://dx.doi.org/10.1590/S1415-47572012005000073; PMID: 23271929
- López F, Sampedro T, Llorente JL, Domínguez F, Hermsen M, Suárez C, Alvarez-Marcos C. Utility of MS-MLPA in DNA methylation profiling in primary laryngeal squamous cell carcinoma. Oral Oncol 2014; 50:291 - 7; http://dx.doi.org/10.1016/j.oraloncology.2014.01.003; PMID: 24444674
- Furlan D, Sahnane N, Mazzoni M, Pastorino R, Carnevali I, Stefanoli M, Ferretti A, Chiaravalli AM, La Rosa S, Capella C. Diagnostic utility of MS-MLPA in DNA methylation profiling of adenocarcinomas and neuroendocrine carcinomas of the colon-rectum. Virchows Arch 2013; 462:47 - 56; http://dx.doi.org/10.1007/s00428-012-1348-2; PMID: 23224118
- Pineda M, Mur P, Iniesta MD, Borràs E, Campos O, Vargas G, Iglesias S, Fernández A, Gruber SB, Lázaro C, et al. MLH1 methylation screening is effective in identifying epimutation carriers. Eur J Hum Genet 2012; 20:1256 - 64; http://dx.doi.org/10.1038/ejhg.2012.136; PMID: 22763379
- Corso G, Carvalho J, Marrelli D, Vindigni C, Carvalho B, Seruca R, Roviello F, Oliveira C. Somatic mutations and deletions of the E-cadherin gene predict poor survival of patients with gastric cancer. J Clin Oncol 2013; 31:868 - 75; http://dx.doi.org/10.1200/JCO.2012.44.4612; PMID: 23341533
- Marsit CJ, Posner MR, McClean MD, Kelsey KT. Hypermethylation of E-cadherin is an independent predictor of improved survival in head and neck squamous cell carcinoma. Cancer 2008; 113:1566 - 71; http://dx.doi.org/10.1002/cncr.23770; PMID: 18711702
- Castro M, Grau L, Puerta P, Gimenez L, Venditti J, Quadrelli S, Sánchez-Carbayo M. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med 2010; 8:86; http://dx.doi.org/10.1186/1479-5876-8-86; PMID: 20849603
- Kim DH, Nelson HH, Wiencke JK, Zheng S, Christiani DC, Wain JC, Mark EJ, Kelsey KT. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res 2001; 61:3419 - 24; PMID: 11309302
- Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y, Virmani AK, Zochbauer-Muller S, Tsukuda K, Sugio K, et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer 2003; 103:153 - 60; http://dx.doi.org/10.1002/ijc.10787; PMID: 12455028
- Lleras RA, Smith RV, Adrien LR, Schlecht NF, Burk RD, Harris TM, Childs G, Prystowsky MB, Belbin TJ. Unique DNA methylation loci distinguish anatomic site and HPV status in head and neck squamous cell carcinoma. Clin Cancer Res 2013; 19:5444 - 55; http://dx.doi.org/10.1158/1078-0432.CCR-12-3280; PMID: 23894057
- Colacino JA, Dolinoy DC, Duffy SA, Sartor MA, Chepeha DB, Bradford CR, McHugh JB, Patel DA, Virani S, Walline HM, et al. Comprehensive analysis of DNA methylation in head and neck squamous cell carcinoma indicates differences by survival and clinicopathologic characteristics. PLoS One 2013; 8:e54742; http://dx.doi.org/10.1371/journal.pone.0054742; PMID: 23358896
- Huang KH, Huang SF, Chen IH, Liao CT, Wang HM, Hsieh LL. Methylation of RASSF1A, RASSF2A, and HIN-1 is associated with poor outcome after radiotherapy, but not surgery, in oral squamous cell carcinoma. Clin Cancer Res 2009; 15:4174 - 80; http://dx.doi.org/10.1158/1078-0432.CCR-08-2929; PMID: 19509163
- Conacci-Sorrell M, Zhurinsky J, Ben-Ze’ev A. The cadherin-catenin adhesion system in signaling and cancer. J Clin Invest 2002; 109:987 - 91; http://dx.doi.org/10.1172/JCI0215429; PMID: 11956233
- Adachi Y, Takeuchi T, Nagayama T, Furihata M. T-cadherin modulates tumor-associated molecules in gallbladder cancer cells. Cancer Invest 2010; 28:120 - 6; http://dx.doi.org/10.3109/07357900903124472; PMID: 20121546
- Kim WY, Lee JW, Park YA, Choi JJ, Sung CO, Song SY, Choi CH, Kim TJ, Huh SJ, Kim BG, et al. RAR-beta expression is associated with early volumetric changes to radiation therapy in cervical cancer. Gynecol Obstet Invest 2011; 71:11 - 8; http://dx.doi.org/10.1159/000320719; PMID: 21160189
- Sanbhnani S, Yeong FM. CHFR: a key checkpoint component implicated in a wide range of cancers. Cell Mol Life Sci 2012; 69:1669 - 87; http://dx.doi.org/10.1007/s00018-011-0892-2; PMID: 22159584
- Chow JP, Man WY, Mao M, Chen H, Cheung F, Nicholls J, Tsao SW, Li Lung M, Poon RY. PARP1 is overexpressed in nasopharyngeal carcinoma and its inhibition enhances radiotherapy. Mol Cancer Ther 2013; 12:2517 - 28; http://dx.doi.org/10.1158/1535-7163.MCT-13-0010; PMID: 23979918
- Khan K, Araki K, Wang D, Li G, Li X, Zhang J, Xu W, Hoover RK, Lauter S, O’Malley B Jr., et al. Head and neck cancer radiosensitization by the novel poly(ADP-ribose) polymerase inhibitor GPI-15427. Head Neck 2010; 32:381 - 91; PMID: 19672867
- Liu SS, Leung RC, Chan KY, Chiu PM, Cheung AN, Tam KF, Ng TY, Wong LC, Ngan HY. p73 expression is associated with the cellular radiosensitivity in cervical cancer after radiotherapy. Clin Cancer Res 2004; 10:3309 - 16; http://dx.doi.org/10.1158/1078-0432.CCR-03-0119; PMID: 15161684
- van Diest PJ. No consent should be needed for using leftover body material for scientific purposes. For. BMJ 2002; 325:648 - 51; http://dx.doi.org/10.1136/bmj.325.7365.648; PMID: 12242180
- Smeets SJ, Hesselink AT, Speel EJ, Haesevoets A, Snijders PJ, Pawlita M, Meijer CJ, Braakhuis BJ, Leemans CR, Brakenhoff RH. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer 2007; 121:2465 - 72; http://dx.doi.org/10.1002/ijc.22980; PMID: 17680565
- Farshadpour F, Konings S, Speel EJ, Hordijk GJ, Koole R, van Blokland M, Slootweg PJ, Kummer JA. Human Papillomavirus and Oropharyngeal Squamous Cell Carcinoma: A Case-Control Study regarding Tobacco and Alcohol Consumption. Patholog Res Int. 2011;2011:806345
- Shaw R. The epigenetics of oral cancer. Int J Oral Maxillofac Surg 2006; 35:101 - 8; http://dx.doi.org/10.1016/j.ijom.2005.06.014; PMID: 16154320
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646 - 74; http://dx.doi.org/10.1016/j.cell.2011.02.013; PMID: 21376230
- Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, Schouten JP, Errami A. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res 2005; 33:e128; http://dx.doi.org/10.1093/nar/gni127; PMID: 16106041
