Abstract
Chromatin insulators affect interactions between promoters and enhancers/silencers and function as barriers for the spreading of repressive chromatin. Drosophila insulator protein dCTCF marks active promoters and boundaries of many histone H3K27 trimethylation domains associated with repressed chromatin. In particular, dCTCF binds to such boundaries between the parasegment-specific regulatory domains of the Bithorax complex. Here we demonstrate that the evolutionarily conserved protein ENY2 is recruited to the zinc-finger domain of dCTCF and is required for the barrier activity of dCTCF-dependent insulators in transgenic lines. Inactivation of ENY2 by RNAi in BG3 cells leads to the spreading of H3K27 trimethylation and Pc protein at several dCTCF boundaries. The results suggest that evolutionarily conserved ENY2 is responsible for barrier activity mediated by the dCTCF protein.
Introduction
Eukaryotic genomes are organized into functional units containing individual genes together with the corresponding regulatory elements, clusters of co-expressed genes, or group of genes that share regulatory elements.Citation1-Citation3 The current model suggests that these functional units need to be isolated from each other in order to prevent illegitimate interactions of enhancers with inappropriate promoters. This is apparently achieved due to a special class of regulatory elements, known as insulators, which can block enhancer–promoter communication.Citation4-Citation12 The ability of insulators to disrupt the communication between an enhancer and a promoter when inserted between them has allowed their identification in the Drosophila and vertebrate genomes.Citation13-Citation19 In addition, a number of insulators display barrier activity and can block the spreading of heterochromatization and consequent gene silencing.Citation6,Citation9,Citation12,Citation20,Citation21
Both activities of insulators are required for blocking the repressive effect of Polycomb response elements (PREs).Citation22-Citation25 PREs are bound by proteins of the Polycomb group (PcG), inducing silencing of both endogenous target genes and reporter genes.Citation26 It appears that PREs can interact with promoters and enhancers by looping out the intervening sequences.Citation27-Citation29 PREs also participate in the formation of transcriptionally inactive chromatin domains enriched in H3K27me3 modification.Citation26,Citation30 Loci repressed by PcG proteins are embedded in broad H3K27me3 domains that are regulated by PREs bound by two main multiprotein PcG complexes: the Polycomb repressive complex 1 (PRC1) and the E(Z) complex (PRC2).Citation29-Citation31 The canonical PRC2 complex has histone H3 lysine 27 methyltransferase activity and is responsible for di- and tri-methylation of H3K27. PRC1 contains the Pc protein, whose chromodomain preferentially binds histone H3 with di- or tri-methylated lysine 27, and the dRing module that ubiquitinates H2A at lysine 119. These properties of the PcG complexes have given rise to the model that H3K27 trimethylation by PRC2 leads to the spreading of PRC1 complex that maintains the transcriptionally repressed state of chromatin by ubiquitinating histone H2AK119. In accordance with the ability of insulator proteins to be a barrier for repression, they have been frequently found at the boundaries of domains with the repressive H3K27me3 mark.Citation32-Citation35
One of the best model systems for studying the role of insulators in gene regulation is the regulatory region of the homeotic Abdominal-B (Abd-B) gene of the bithorax complex.Citation36,Citation37 The large cis-regulatory region of the bithorax complex (BX-C) is divided into nine parasegment-specific chromatin domains that control the expression of the three homeotic genes, Ultrabithorax (Ubx), Abdominal-A (Abd-A), and Abdominal-B (Abd-B).Citation38 These genes are responsible for specifying the identity of parasegments 5 to 14 (PS5−PS14), which form the posterior half of the thorax and all abdominal segments of an adult fly.Citation39,Citation40 The 300-kb regulatory region can be divided into nine discrete segment-specific iab domains, which are aligned on the chromosome in the same order as the body segments in which they operate. Each iab domain appears to contain at least one enhancer that initiates Abd-A or Abd-B expression in the early embryo, as well as a PRE silencer element that maintains the expression pattern throughout development.Citation40-Citation48 A specific class of boundary elements, or insulators, are proposed to exist between the iab domains to allow their autonomy in properly specifying segmental identity.Citation16,Citation40,Citation41,Citation45 Insulators named Miscadastral (Mcp), Frontadominal-6 (Fab-6), Frontadominal-7 (Fab-7), and Frontadominal-8 (Fab-8), have been functionally identified by deletion analysis within the bithorax complex and in experiments with transgenic enhancer-blocking assay.Citation16,Citation41,Citation45,Citation49-Citation56
Binding sites for dCTCF, the Drosophila homolog of vertebrate insulator protein CTCF, have been identified in the bithorax complexCitation56-Citation59 as well as in the Mcp, Fab-6 and Fab-8 insulators.Citation56,Citation57,Citation59 It has been suggested that dCTCF is the key protein involved in organization of chromatin domains in the bithorax complex.Citation57,Citation58 Domains enriched in the H3K27me3 mark are frequently bordered by dCTCF.Citation33,Citation35
In this study, we have found that dCTCF interacts with the ENY2/Sus1 protein in vivo and in vitro. ENY2 is a small (101 amino acids) evolutionarily conserved and ubiquitously expressed protein that was first found in Drosophila and shown to be chromatin-associated and capable of co-activating transcription on a chromatin template in vitro.Citation37,Citation60,Citation61 ENY2 homologs have subsequently been identified in different organisms as components of SAGA, TREX-2, and THO complexes.Citation62-Citation66
The ENY2 protein was also described as the component of Su(Hw)-dependent insulators that is required for their barrier activity.Citation23 The results presented here show that dCTCF recruits ENY2 to chromatin and that ENY2 interferes with the spreading of repressive H3K27me3 histone mark at most of tested dCTCF binding sites. ENY2 is essential for dCTCF-mediated blocking of repression in transgenic constructs containing PRE. Thus, ENY2 is a common co-factor for at least two different Drosophila insulator proteins.
Results
ENY2 interacts with zinc finger domain of dCTCF in Drosophila
To test whether dCTCF can interact with ENY2, as does Su(Hw),Citation23 we used the yeast two-hybrid assay (Y2H). dCTCF has 11 zinc fingers (aa 287 to 610) that share strong homology with the similar domain of mammalian CTCF ().
Figure 1. Interaction of dCTCF and ENY2 proteins. (A) ENY2 interacts with zinc-finger domain of dCTCF in the yeast two-hybrid assay. The scheme shows the structure of the full-length dCTCF protein and the polypeptides tested. The plus and minus signs indicate relatively strong interaction and the absence of interaction, respectively. Different fragments of dCTCF were individually fused to the C terminus of the GAL4 activating domain and analyzed for the interaction with E(y)2 fused to the DNA-binding domain of GAL4. All dCTCF fragments were tested for the absence of interaction with GAL4. (B) Co-immunoprecipitation of dCTCF and ENY2 proteins from S2 cell extract. The immunoprecipitated complexes were washed with 300 mM, 500 mM and 150 mM NaCl-containing buffers before resolving them by SDS-PAGE for western blot analysis with the indicated antibodies.
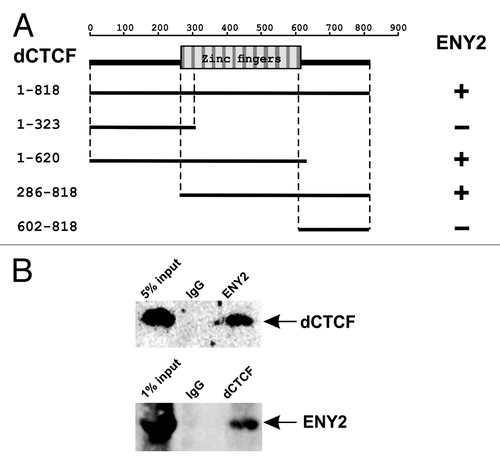
Different dCTCF fragments were individually fused to the GAL4 activating domain and assayed for the interaction with ENY2 fused to GAL4 DNA-binding domain (). As a result, we found that the zinc finger domain of dCTCF was essential for the interaction with ENY2.
To confirm the results obtained in Y2H assay, we checked whether dCTCF and ENY2 are physically associated in vivo by co-immunoprecipitation of the nuclear extract from Drosophila S2 cells. Anti-dCTCF antibodies proved to co-precipitate part of ENY2, and vice versa (), indicating that dCTCF and ENY2 interact in vivo.
To further confirm the interaction between dCTCF and ENY2, we used an alternative method, MBP pull-down assay with bacterially expressed proteins. Different parts of dCTCF were fused to MBP, the fusion protein was immobilized on amylose resin beads, and then recombinant His-tagged ENY2 was incubated with the beads. However, no consistent interaction was observed between ENY2 and dCTCF–MBP (data not shown). Since this contradicted the results of our Y2H and co-IP experiments, we considered that bacterially expressed ENY2 failed to interact with dCTCF in the pull-down assay.
Careful examination of X-ray structures (Fig. S1A) of the yeast homolog of ENY2, named Sus1,Citation67-Citation70 suggested that it could form homodimers in solution. Molecular docking of two Sus1 molecules (Fig. S1B) also revealed a probable dimerization interface within the inner cleft of the molecule, which is essential for interactions between Sus1 and its partners, Sgf11 (Fig. S1A) or Sac3 protein.Citation67,Citation68 This interface is preferable in molecular docking with zinc fingers of CTCF (Fig. S1C). Therefore, dimerization may block this interface, thereby preventing the interaction of ENY2 with other proteins (Fig. S1B).
Using crosslinking with glutaraldehyde, we confirmed the presence of ENY2 dimers in solution (). In this experiment, we used GST as a positive control for dimerization and thioredoxin as a monomer molecule. In the yeast two-hybrid assay, we detected no signs of dimerization between full-length ENY2 molecules, probably because of some steric problems. At the same time, the ENY2 protein displayed the ability to interact with its parts ().
Figure 2. Tests for the interaction between ENY2 and dCTCF in vitro. (A) Cross-linking of ENY2 by incubation with increasing concentration of glutaraldehyde (GA). Proteins were separated in 5–12% gradient polyacrylamide gels and visualized by silver staining (for experimental details, see Materials and Methods). GST was used as a positive control of dimerization. Thioredoxin used as a negative control is shown presented as a monomer molecule. (B) ENY2 interacts with parts of ENY2 in the yeast two-hybrid assay. (C) ENY2 interacts with zinc fingers 7–11 of dCTCF in co-expression assay. Indicated MBP-fused zinc-finger domains were co-expressed with 6 × His-ENY2. The results of 6 × His (stained with Coomassie) and MBP (stained with Coomassie/anti-ENY2 antibodies) pull-down assays are shown. “Input” refers to bacterial lysate. For Sgf11–ENY2, the results of GST and 6 × His pull down assays stained with Coomassie are shown. Arrows indicate positions of MBP-fused zinc fingers of dCTCF or Zw5, GST-fused N-terminus of Sgf11, and 6 × His-ENY2. The zinc-finger domain of Zw5 protein (negative control) displayed no direct interaction with ENY2. The Sgf11 protein (positive control) interacted with ENY2.
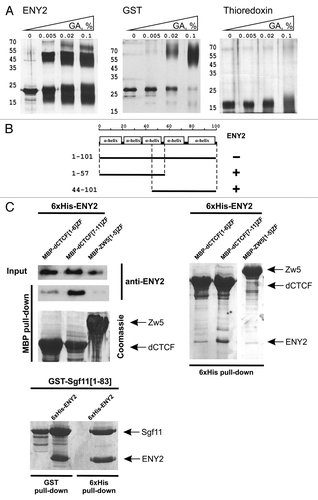
Thus, it was dimerization of ENY2 that could suppress its interaction with dCTCF in vitro. To overcome this obstacle, we used compatible vectors that allowed co-expression of dCTCF287–610 zinc-finger domain and ENY2 protein in bacteria. Pull-down assays from bacterial lysates () revealed a strong, specific interaction of ENY2 with the dCTCF fragment containing zinc fingers 7–11 but not 1–6, and no interaction with unrelated zinc fingers of Zw5 used as negative control. Taken together, these results suggest that ENY2 efficiently interacts with zinc fingers in vivo and in vitro.
Genetic interactions demonstrate functional cooperation between ENY2 and dCTCF
To assess the functional role of association between dCTCF and ENY2 in gene regulation, we studied the interaction between mutations in the dCTCF and e(y)2 genes. The GE24185 mutation leads to strong inactivation of the dCTCF gene.Citation58 Flies homozygous for the GE24185 mutation showed a highly penetrant mild held-out wing phenotype and lethality of embryos derived from F2 females homozygous for the GE24185 mutation (Fig. S2A). A weak e(y)2u1 mutation was generated by inserting the Stalker mobile element in the promoter-proximal region of the gene, which reduced the mRNA content to one-third of normal.Citation60,Citation61 The e(y)2u1 mutation has diverse weak effects on fly morphology (Fig. S2A), which are manifested in a short stocky body, abnormal morphology of tergites 9 and 10, separated wings, eyes with altered facets, and low fertility.Citation60 The phenotypic expression of the e(y)2u1 mutation at 18оС is very weak, but it becomes much stronger after the flies are transferred to 25 °C . We compared the amounts of e(y)2 transcripts in 2- to 3-d-old e(y)2u1 and e(y)2+ males at 18 °C and 25 °C using RT-PCR. The results showed that the amounts of these transcripts in both mutant and wild-type flies were approximately the same at 18 °C, but the transcription level in e(y)2u1 mutants dropped at 25 °C (Fig. S2B).
While e(y)2u1 and GE24185/GE24185 (F1) males showed only a moderate decrease in viability, males with the combination of mutations e(y)2u1/Y; GE24185/GE24185 or even e(y)2u1/Y; GE24185/TM6,Tb failed to survive at 25 °C, which was indicative of a functional interaction between proteins encoded by these genes.
Testing the role of ENY2 in the barrier activity of dCTCF-dependent insulators
Recent studies suggest multiple functions for insulator proteins in organizing boundaries between active and repressed chromatin.Citation33,Citation35 In BG3 cells, 54% of domain boundaries between active and repressive histone marks coincide with robust insulator protein binding sites.Citation35 However, dCTCF knockdown in BG3 cells has shown that the insulator protein is required for the formation of boundaries of about 15–20 H3K27me3 domains.Citation35 Using these results, we selected nine dCTCF-dependent boundaries to test the role of dCTCF in recruiting ENY2 and organizing the functional barrier (Fig. S3). In one case, two dCTCF-dependent boundaries (57B4 and 57B4R) flanked the H3K27me3 domain. Inactivation of dCTCF resulted in H3K27me3 spreading only at 57B4, suggesting that additional proteins are responsible for the barrier activity at 57B4R.
We tested for interdependence of dCTCF and ENY2 binding to selected boundaries and the Fab-8 region in BG3 cells. ENY2 was found to strongly bind to six sites and Fab-8, and weakly bind to three sites (). Next, we induced knockdown of ENY2 or dCTCF (Fig. S4). The results showed that inactivation of ENY2 had no effect on dCTCF binding, whereas knockdown of dCTCF strongly reduced ENY2 binding at all “strong” sites, confirming the role of dCTCF in recruiting of ENY2 to chromatin ().
Figure 3. dCTCF and ENY2 determine some boundaries of H3K27me3 domains in BG3 cells. Histograms show the results of ChIP for the relative amounts of dCTCF or ENY2 or Pc or H3K27me3 in previously defined dCTCF binding regions on chromatin isolated from BG3 cells treated with specific dsRNA from dCTCF (dCTCF_Ri) or e(y)2 (ENY2_Ri) coding regions and incubated with corresponding antibodies. “C” is a mock RNAi control obtained with BG3 cells treated with GFP dsRNA. The bxd region (PRE) of bithorax complexCitation25 was used as dCTCF/ENY2-independent region of Pc binding. Error bars show standard deviations of triplicate PCR measurements. The results are presented as a percentage of input DNA. The results for ChIP with histone modifications are presented as a percentage of input DNA normalized relative to the amount of the H3 histone at the tested regions. To test for relative amount of H3K27me3, we used the pairs of primers inside (in), at the boundary (cts), and outside (out) of the H3K27me3 domains. The distribution of dCTCF and H3K27me3 is shown at the bottom. Relative locations of primers (in, cts, out) for ChIP on the cytological map are indicated in Figure S3.
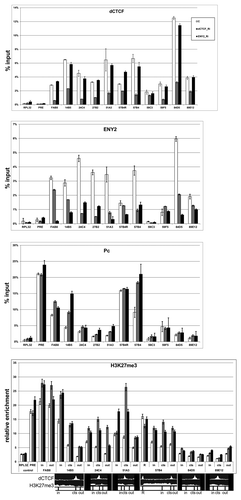
We also examined binding of the Polycomb (Pc) protein to selected sites (, S5). Only Fab-8, 14B5, 59F5 boundaries and 57B4–57B4R domain were significantly enriched with Pc. Inactivation of either dCTCF or ENY2 resulted in considerable enhancement of Pc binding to 14B5 and 57B4 regions. Once again, inactivation of either dCTCF or ENY2 promoted Pc binding to 14B5 and 57B4 but not to 57B4R.
To further test whether dCTCF and ENY2 are involved in the function of boundary between active and repressed chromatin, we examined seven sites strongly enriched with dCTCF and ENY2 for the distribution of chromatin marks (H3K27me3 and H3K27ac) inside (in), at the boundary (cts), and outside (out) of repressed domains (). As a result, we found that inactivation of either dCTCF or ENY2 did not affect H3K27ac modification of histones (Fig. S5); at the same time, inactivation of either protein caused a strong increase in the H3K27me3 mark at all tested regions except 57B4R. Taken together, these results suggest that ENY2 is required for the barrier activity mediated by the dCTCF protein, but the activity of ENY2 and dCTCF at certain boundaries, such as 57B4R, appears to be redundant.
ENY2 is essential for the barrier activity of dCTCF-dependent Mcp and Fab-8 insulators in transgenic flies
To test the role of ENY2 in the barrier activity of dCTCF-dependent insulators Mcp and Fab-8, we used the yellow regulatory system as a model. The yellow gene is required for dark pigmentation of larval and adult cuticle and bristles. The enhancer driving yellow expression in bristles is located in the intron.Citation71 In this model¸ we used the 660-bp PRE from the bxd region of the Ultrabithorax gene.Citation22,Citation25,Citation72
In the (PRE)(M)YW and (PRE)(F8)YW constructs, PRE was inserted at –1603 relative to the yellow transcription start site, and either 340-bp Mcp (M) or 550-bp Fab-8 (F8) insulator was inserted at –893 between PRE and the yellow promoter (). The insulator and PRE were flanked by lox and frt sites, respectively. This allowed us to compare the effects of the absence or presence of either the PRE or the insulator at a given transgene insertion site (elements that could be excised are parenthesized in all constructs).
Figure 4. ENY2 is essential for the barrier activity of dCTCF-dependent insulators Fab-8 and Mcp in transgenic lines. (A) The Fab-8 and Mcp insulators are indicated by gray rectangle. In schemes of the constructs (drawn not to scale), the yellow coding region and the white gene (marker) are shown as white rectangles. The yellow coding region is separated by the intron. Arrows indicate the direction of yellow and white transcription. The enhancer controlling yellow expression in bristles is represented by white oval. The 660-bp PRE is indicated by black pentagon. Downward arrows mark the lox and frt sites. In construct names, the corresponding excisable element is parenthesized. Ey is the abbreviation for “enhancers of yellow gene.” The yellow expression in bristles is shown in the selected transgenic lines before and after deletion of either the insulator or PRE in the e(y)2u1 background. The degree of yellow expression in bristles of the thorax and head was scored using a five-point scale: (1) loss of pigmentation, (ev) extreme variegation, (mv) moderate variegation, (wv) weak variegation, and (5) wild-type pigmentation (for details, see the text). The “degree” column shows the numbers of transgenic lines with different bristle pigmentation levels. N is the number of lines in which flies acquired a new yellow phenotype relative to the initial lines. T is the total number of lines examined for each particular construct. (B) The Fab-8 and Mcp insulators with mutated dCTCF binding sites shown as white rectangles.
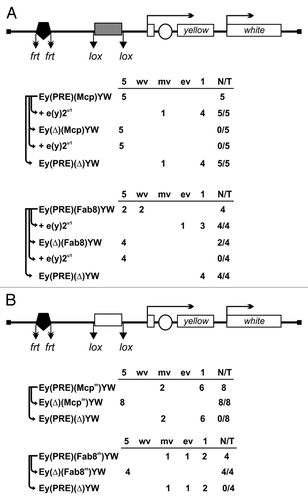
Thus, we obtained five (PRE)(M)YW and four (PRE)(F8)YW independent transgenic lines in which PRE completely repressed yellow expression in bristles when the insulator was deleted (). In most of transgenic lines, the Mcp and Fab-8 insulators completely prevented gene repression by PRE. At the same time, transgenic lines with the e(y)2u1 mutation demonstrated almost complete repression of bristle pigmentation, as after the deletion of the corresponding insulator at this position. When PRE was deleted, the e(y)2u1 mutation did not affect yellow pigmentation. These results support the role of ENY2 in barrier activity mediated by Fab-8 and Mcp insulators.
Two representative lines with either Mcp or Fab-8 insulator were analyzed in more detail. ChIP experiments were performed in 2- to 3-d pupae, because adult bristle pigmentation required yellow expression during the second stage of pupation. Strong dCTCF and ENY2 binding to the insulator regions was observed in both transgenic lines (). The Pc protein showed strong binding to PRE and its flanking region (including insulator) from the proximal to PRE side, while the yellow promoter region was free of Pc. Deletion of the insulator resulted in complete elimination of the ENY2 and dCTCF proteins, while the amount of the Pc protein was increased ().
Figure 5. dCTCF-dependent Mcp and Fab-8 insulators block the spreading of Pc. (A) Binding of dCTCF, ENY2, and Pc to the constructs from homozygous transgenic lines. The scale bar shows distances in kilobases. (B) Binding of the same proteins to the construct from the derivative homozygous line obtained by deletion of the Mcp insulator. Protein binding to the constructs from homozygous lines was analyzed by ChIP followed by real-time PCR quantification. Each ChIP experiment with 2- to 3-d pupae was performed in at least two independent replications. The results of ChIP are presented as a percentage of input DNA normalized relative to the endogenous positive binding site for dCTCF and ENY2 proteins from the CG1354 region and for Pc protein from the endogenous region PRE of the Ubx gene. Error bars show standard deviations of triplicate PCR measurements. C1–C7 are regions flanked by primers for ChIP analysis. Other designations are as in .
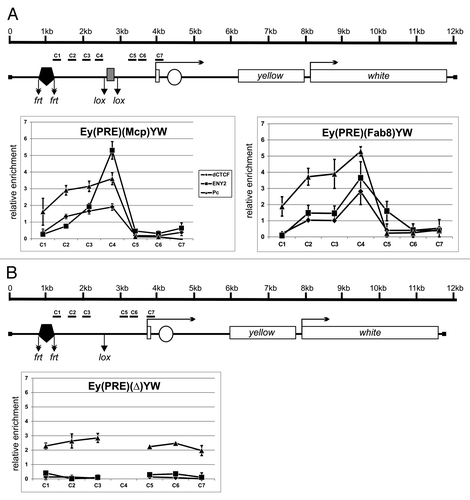
To test the role of dCTCF binding sites in the barrier activity of the insulators, we used previously described Mcp and Fab-8 insulators with the mutated binding sites for dCTCF.Citation73 We made a similar construct with the mutated Mcp (Mm) and Fab-8 (F8m) insulators and obtained a total of 12 transgenic lines with both mutations (). In all these lines, the mutated insulators failed to protect yellow expression from PRE-mediated repression. The dCTCF and ENY2 proteins failed to bind to the mutant insulators in pupae (Fig. S6), and the mutant insulators did not block the spreading of Pc. This is additional evidence that dCTCF binding sites are essential for the barrier activity of the Mcp and Fab-8 insulators.
Discussion
One of predicted activities of boundaries in the regulatory regions of the bithorax complex is to restrict PcG-mediated repression.Citation36,Citation38 Here we have found that ENY2 interacts with dCTCF and is required for blocking PRE-mediated repression by the Fab-8 and Mcp boundaries in transgenic lines. Thus, our results confirm that dCTCF-dependent insulators can protect gene expression from PcG repression and that the recruitment of ENY2 by dCTCF is essential for this process.
We have previously found that ENY2 directly interacts with several C2H2 zinc fingers of Su(Hw) and colocalizes with many Su(Hw) binding sites.Citation23 In transgenic lines, partial ENY2 inactivation in the e(y)2u1 mutation affect only the barrier activity of Su(Hw) in protecting the yellow reporter from PcG repression. Thus, ENY2 binds to zinc fingers of two different insulator proteins, suggesting that ENY2 recruitment by C2H2 zinc fingers might be a general mechanism for restricting PcG silencing.
As recently shown in several independent studies, domains enriched with the H3K27me3 mark are bordered by actively transcribed genes or insulators.Citation33,Citation35 Recognition of histone H3 by the PRC2 complex is suppressed by H3K4me3 and H3K36me2/3 histone marks that are associated with active transcription.Citation74 The insulator proteins are dispensable for the maintenance of boundaries formed by actively transcribed genes.Citation35 At the same time, the barrier activity of insulator proteins is partially redundant when domain boundaries contain insulator protein binding sites but not active genes. Here we have found that dCTCF-dependent boundaries in BG3 cells are also dependent on the recruitment of ENY2. The redundant activity of dCTCF might be explained by the ability of other, unknown zinc-finger proteins to bind to the boundaries of H3K27me3 domains and recruit ENY2.
In conclusion, our current and previous resultsCitation23 show that ENY2 is responsible for the barrier activity of the best studied Drosophila insulators, Su(Hw) and dCTCF, and suggest that ENY2 may mediate the barrier activity of other zinc-finger proteins.
Materials and Methods
Protein expression and purification
Recombinant protein was expressed in E. coli BL21 cells and purified using standard procedures. Full-length ENY2-coding region was cloned in the modified (without thioredoxin, which was digested with NdeI) pET32a(+) vector in frame with 6 × His tag. Recombinant protein was expressed in E. coli BL21 cells and purified using standard procedures. Briefly, the cells expressing ENY2 were disrupted by sonication in buffer A (40 mM HEPES-KOH, pH 7.7; 400 mM NaCl, 5 mM β-mercaptoethanol, 20 mM imidazole, 1 mM PMSF, 1:1000 Complete Protease Inhibitor Cocktail VII (Calbiochem catalog # 539138)). The lysate was cleared by centrifugation and applied onto a Ni-NTA column. After washing, the bound proteins were eluted with buffer A containing 300 mM imidazole and dialyzed against appropriate buffer.
Chemical crosslinking was performed for 10 min at room temperature in buffer containing 20 mM HEPES-KOH, pH 7.7; 150 mM NaCl, 20 mM imidazole, 1 mM β-mercaptoethanol. Crosslinking was quenched with 50 mM glycine and samples were resolved using SDS-PAGE followed by silver staining.
Pull-down assays
For pull-down assays, we performed co-expression of full-length ENY2 protein fused with 6 × His and zinc-finger domains of dCTCF protein fused with MBP in E. coli BL21 cells. ENY2-coding region fused with 6 × His tag was cloned into the vector derived from pACYC and pET28a(+) (Novagen) bearing p15A replication origin, Kanamycin-resistance gene and pET28a(+) MCS. cDNAs coding “zinc-finger” domains were cloned in the vector pMAL-C5X (New England Biolabs) in frame with MBP: 286–462 aa (1–6 ZFs) and 460–631 aa (7–11 ZFs) from dCTCF protein and 188–591 aa from Zw5 protein. BL21 cells were grown in LB medium at 37 °C to an A600 of 1.0 and then induced with 1 mM IPTG at 18 °C overnight. Before induction, ZnCl2 was added to a final concentration of 200 μM. MBP pull-down was performed with Amylose resin (New England Biolabs) in buffer B (20 mM HEPES-KOH, pH 7.7; 150 mM NaCl, 10mM MgCl2, 0.1 mM ZnCl2, 0.1% NP40, 10% (w/w) glycerol). Cells were disrupted by sonication, centrifuged, and applied onto the resin for 10 min at room temperature. After binding, the resin was washed four times with buffer B containing 500 mM NaCl and eluted with buffer 20 mM HEPES-KOH, pH 7.7; 200 mM NaCl, 10 mM maltose, 10 mM MgCl2, 0.1 mM ZnCl2 for 15 min. The mixture was then centrifuged at 2000 rpm for 1 min, and the supernatant was analyzed by SDS-PAGE with Coomassie staining.
Molecular docking
Structural data were taken from 3FWB, 4DHX and 3KIK pdb data files. Molecular docking was performed using ZDOCKCitation75 with an angular step size of 6, estimating the electrostatic and desolvatation energy. The generated protein poses were re-ranked with ZRANK.Citation76 One hundred top-scored structures were analyzed manually, and more than 70% of protein poses demonstrated the formation of an intertwined dimer with the involvement of Sus1 inner cleft and one of its α-helices. Protein structural alignment and homology model building were performed with MODELER.Citation77
S2 cell nuclear lysate preparation and immunoprecipitation
An aliquot of 1 × 108 S2 cells was washed twice in 10 mL of ice-cold PBS, resuspended in 10 mL of ice-cold IP-Sucrose buffer (10 mM Tris, pH 7.5; 10 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 250 mM sucrose, 0.5 mM PMSF) with Complete (EDTA-free) Protease Inhibitor Cocktail V (Calbiochem catalog # 539137), incubated on ice for 10 min, and homogenized with a Dounce loose pestle (20 strokes). The nuclei were then pelleted by centrifugation at 3000 g, 4°C for 10 min. The pellet was resuspended in 500 mL of ice-cold IP-10 buffer (10 mM Tris, pH 7.5; 10 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.1% NP-40, 10% glycerol, 0.5 mM PMSF, and Complete Protease Inhibitor Cocktail V), homogenized with a Dounce tight pestle (20 strokes), and mixed with an equal volume of IP-850 buffer (10 mM Tris, pH 7.5, 850 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.1% NP-40, 10% glycerol, 0.5 mM PMSF, and Complete Protease Inhibitor Cocktail V). The suspension was incubated on ice for 10 min and then centrifuged at 20 000 rpm, 4°C, for 10 min. The supernatant fluid (the nuclear fraction) was collected for immunoprecipitation experiments. Rabbit antibodies against dCTCF (1:400) and ENY2 (1:100) were conjugated with Protein A agarose beads (Pierce); in respective control experiments, rabbit pre-immune serum was used. An aliquot of an antibody was mixed with 30 μL of agarose beads equilibrated in IP buffer with 150 mM NaCl (IP-150) and incubated on a rotator at 4°C for 3 h. The beads were then washed with IP-150, blocked with 1% BSA for 30 min under the same conditions, and washed with two portions of IP-150. The nuclear extract was adjusted to 150 mM NaCl, and its 1-mL aliquot containing approximately 1 mg of total protein was mixed with 30 μL of “fresh” agarose beads equilibrated in IP- 150 and incubated at 4°C for 1 h to pre-clear the sample. The beads were pelleted, and the supernatant fluid was transferred to a new tube and mixed with antibody-conjugated beads. The samples were incubated on a rotator at 4°C for 3 h, and the beads were washed with three portions of IP buffer with 300 mM NaCl, one portion of IP buffer with 500 mM NaCl, and one portion of IP buffer with 150 mM NaCl. After the last washing step, the beads were resuspended in SDS–PAGE loading buffer, boiled, and analyzed by western blotting. Proteins were detected using the SuperSignal West Fempto substrate (Pierce).
Antibodies
Antibodies against dCTCF [606–818 aa] fragment and ENY2 were raised in rabbits and rats and purified from the sera by ammonium sulfate fractionation followed by affinity purification on CNBr-activated Sepharose (GE Healthcare) according to standard protocols. Antibodies against lamin ADL67.10 were from the Hybridoma Bank at the University of Iowa. Antibodies against Polycomb protein were a kind gift from G. Cavalli. Antibodies against histone H3 (catalog #39163) and chromatin modifications H3K27me3 (catalog #39155) and H3acK27 (catalog #39135) were from Active Motif.
RNA interference in Drosophila BG3 cells
The BG3 cells were cultured in Schneider medium in 60-mm petri dishes at 25 °C. RNAi experiments were performed when the culture reached a density of about 0.2 × 106 cells/mL. dsRNA were synthesized with MEGAScript kit (Pierce) according to manufacturer’s recommendations. The cells were treated with 100 μg of corresponding dsRNA, and the treatment was repeated after 3 d. On day 7 after the first dsRNA treatment, the cells were used for subsequent experiments (preparation of total protein, total RNA, and chromatin). The sequences of PCR primers used to produce DNA templates for dsRNA synthesis are listed in Table S1.
RNA isolation and real-time PCR
Total RNA was isolated using the TRI reagent (Molecular Research Center) according to the manufacturer’s protocol. RNA was treated with two units of Turbo DNase I (Ambion) for 30 min at 37 °C to eliminate genomic DNA. The synthesis of cDNA was performed using 2 μg of RNA, ArrayScript reverse transcriptase (Ambion), and oligo(dT) as a primer. The amounts of specific cDNA fragments were quantified by real-time PCR. At least three independent measurements were made for each RNA sample. Relative levels of mRNA expression were calculated in the linear amplification range by calibration to a standard genomic DNA curve to account for differences in primer efficiencies. Individual expression values were normalized with reference to rpl32 and tub mRNAs. The primers used for qPCR are shown in Table S1.
Chromatin Immunoprecipitation
Chromatin was prepared from BG3 cells and mid-late pupae. Detailed information is presented in the Supplemental Materials online. The enrichment of specific DNA fragments was analyzed by real-time PCR, using a StepOne Plus Thermal Cycler (Applied Biosystems). The primers used for PCR in ChIP experiments for genome fragments are shown in Table S1.
Yeast two-hybrid assay
Yeast two-hybrid assay was performed using yeast strain pJ69–4A, with plasmids and protocols from Clontech. For growth assays, plasmids were transformed into yeast strain pJ69–4A by the lithium acetate method, according to the manufacturer's protocol, and plated on media without tryptophan and leucine. After 2 d of growth at 30 °C, the cells were plated on selective media without tryptophan, leucine, histidine, and adenine, and their growth was compared after 2–3 d. Each assay was repeated three times.
Genetic crosses
The transgenic construct and P25.7wc plasmid were injected into yacw1118 pre-blastoderm embryos.Citation78 The resultant flies were crossed with yacw1118 flies, and the transgenic progeny were identified by their eye color under a Stemi 2000 stereomicroscope (Carl Zeiss). The transformed lines were tested for transposon integrity and copy number by RT-PCR. Only single-copy transformants were included in analysis. Details of the crosses used for genetic analysis and the excision of functional elements and determination the levels of yellow expression in bristles are available in the Supplemental Materials online.
Additional material
Download Zip (11 MB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to N.A. Gorgolyuk for his help in preparing the manuscript. The authors would like to acknowledge support from the Ministry of Education and Science of the Russian Federation (project no. 14.B25.31.0022), the Molecular and Cellular Biology Program of the Russian Academy of Sciences (to P.G.), the Russian Foundation for Basic Research (project no. 12–04–00845-а to O.M.), RF Presidential Stipend no. SP-1960.2012.4 (to O.M.). This study was performed using the equipment of the IGB RAS facilities supported by the Ministry of Science and Education of the Russian Federation.
References
- Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell 2013; 49:773 - 82; http://dx.doi.org/10.1016/j.molcel.2013.02.011; PMID: 23473598
- Krijger PH, de Laat W. Identical cells with different 3D genomes; cause and consequences?. Curr Opin Genet Dev 2013; 23:191 - 6; http://dx.doi.org/10.1016/j.gde.2012.12.010; PMID: 23415810
- Tanay A, Cavalli G. Chromosomal domains: epigenetic contexts and functional implications of genomic compartmentalization. Curr Opin Genet Dev 2013; 23:197 - 203; http://dx.doi.org/10.1016/j.gde.2012.12.009; PMID: 23414654
- Brasset E, Vaury C. Insulators are fundamental components of the eukaryotic genomes. Heredity (Edinb) 2005; 94:571 - 6; http://dx.doi.org/10.1038/sj.hdy.6800669; PMID: 15815711
- Chetverina D, Aoki T, Erokhin M, Georgiev P, Schedl P. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. Bioessays 2014; 36:163 - 72; http://dx.doi.org/10.1002/bies.201300125; PMID: 24277632
- Herold M, Bartkuhn M, Renkawitz R. CTCF: insights into insulator function during development. Development 2012; 139:1045 - 57; http://dx.doi.org/10.1242/dev.065268; PMID: 22354838
- Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Curr Opin Cell Biol 2003; 15:259 - 65; http://dx.doi.org/10.1016/S0955-0674(03)00039-5; PMID: 12787766
- Kyrchanova O, Georgiev P. Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett 2014; 588:8 - 14; http://dx.doi.org/10.1016/j.febslet.2013.10.039; PMID: 24211836
- Matzat LH, Lei EP. Surviving an identity crisis: A revised view of chromatin insulators in the genomics era. Biochim Biophys Acta 2014; 1839:203 - 14; PMID: 24189492
- Mukhopadhyay S, Schedl P, Studitsky VM, Sengupta AM. Theoretical analysis of the role of chromatin interactions in long-range action of enhancers and insulators. Proc Natl Acad Sci U S A 2011; 108:19919 - 24; http://dx.doi.org/10.1073/pnas.1103845108; PMID: 22123989
- Nikolaev LG, Akopov SB, Didych DA, Sverdlov ED. Vertebrate Protein CTCF and its Multiple Roles in a Large-Scale Regulation of Genome Activity. Curr Genomics 2009; 10:294 - 302; http://dx.doi.org/10.2174/138920209788921038; PMID: 20119526
- Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev 2007; 17:400 - 7; http://dx.doi.org/10.1016/j.gde.2007.08.005; PMID: 17913488
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 1999; 98:387 - 96; http://dx.doi.org/10.1016/S0092-8674(00)81967-4; PMID: 10458613
- Cai H, Levine M. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature 1995; 376:533 - 6; http://dx.doi.org/10.1038/376533a0; PMID: 7637789
- Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 1992; 6:1865 - 73; http://dx.doi.org/10.1101/gad.6.10.1865; PMID: 1327958
- Gyurkovics H, Gausz J, Kummer J, Karch F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J 1990; 9:2579 - 85; PMID: 1973385
- Holdridge C, Dorsett D. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol Cell Biol 1991; 11:1894 - 900; PMID: 1900919
- Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol 1992; 12:2424 - 31; PMID: 1569958
- Scott KS, Geyer PK. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J 1995; 14:6258 - 67; PMID: 8557045
- Barkess G, West AG. Chromatin insulator elements: establishing barriers to set heterochromatin boundaries. Epigenomics 2012; 4:67 - 80; http://dx.doi.org/10.2217/epi.11.112; PMID: 22332659
- Ghirlando R, Giles K, Gowher H, Xiao T, Xu Z, Yao H, Felsenfeld G. Chromatin domains, insulators, and the regulation of gene expression. Biochim Biophys Acta 2012; 1819:644 - 51; http://dx.doi.org/10.1016/j.bbagrm.2012.01.016; PMID: 22326678
- Comet I, Savitskaya E, Schuettengruber B, Nègre N, Lavrov S, Parshikov A, Juge F, Gracheva E, Georgiev P, Cavalli G. PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev Cell 2006; 11:117 - 24; http://dx.doi.org/10.1016/j.devcel.2006.05.009; PMID: 16824958
- Kurshakova M, Maksimenko O, Golovnin A, Pulina M, Georgieva S, Georgiev P, Krasnov A. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol Cell 2007; 27:332 - 8; http://dx.doi.org/10.1016/j.molcel.2007.05.035; PMID: 17643381
- Mallin DR, Myung JS, Patton JS, Geyer PK. Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics 1998; 148:331 - 9; PMID: 9475743
- Sigrist CJ, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 1997; 147:209 - 21; PMID: 9286681
- Kassis JA, Brown JL. Polycomb group response elements in Drosophila and vertebrates. Adv Genet 2013; 81:83 - 118; http://dx.doi.org/10.1016/B978-0-12-407677-8.00003-8; PMID: 23419717
- Delest A, Sexton T, Cavalli G. Polycomb: a paradigm for genome organization from one to three dimensions. Curr Opin Cell Biol 2012; 24:405 - 14; http://dx.doi.org/10.1016/j.ceb.2012.01.008; PMID: 22336329
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 2007; 134:223 - 32; http://dx.doi.org/10.1242/dev.02723; PMID: 17185323
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol 2008; 20:266 - 73; http://dx.doi.org/10.1016/j.ceb.2008.03.002; PMID: 18439810
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 2011; 471:480 - 5; http://dx.doi.org/10.1038/nature09725; PMID: 21179089
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell 2007; 128:735 - 45; http://dx.doi.org/10.1016/j.cell.2007.02.009; PMID: 17320510
- Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J 2009; 28:877 - 88; http://dx.doi.org/10.1038/emboj.2009.34; PMID: 19229299
- Nègre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. A cis-regulatory map of the Drosophila genome. Nature 2011; 471:527 - 31; http://dx.doi.org/10.1038/nature09990; PMID: 21430782
- Nègre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet 2010; 6:e1000814; http://dx.doi.org/10.1371/journal.pgen.1000814; PMID: 20084099
- Schwartz YB, Linder-Basso D, Kharchenko PV, Tolstorukov MY, Kim M, Li HB, Gorchakov AA, Minoda A, Shanower G, Alekseyenko AA, et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res 2012; 22:2188 - 98; http://dx.doi.org/10.1101/gr.138156.112; PMID: 22767387
- Maeda RK, Karch F. Gene expression in time and space: additive vs hierarchical organization of cis-regulatory regions. Curr Opin Genet Dev 2011; 21:187 - 93; http://dx.doi.org/10.1016/j.gde.2011.01.021; PMID: 21349696
- Georgiev PG. Identification of mutations in three genes that interact with zeste in the control of white gene expression in Drosophila melanogaster. Genetics 1994; 138:733 - 9; PMID: 7851770
- Maeda RK, Karch F. The bithorax complex of Drosophila an exceptional Hox cluster. Curr Top Dev Biol 2009; 88:1 - 33; http://dx.doi.org/10.1016/S0070-2153(09)88001-0; PMID: 19651300
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature 1978; 276:565 - 70; http://dx.doi.org/10.1038/276565a0; PMID: 103000
- Mihaly J, Hogga I, Barges S, Galloni M, Mishra RK, Hagstrom K, Müller M, Schedl P, Sipos L, Gausz J, et al. Chromatin domain boundaries in the Bithorax complex. Cell Mol Life Sci 1998; 54:60 - 70; http://dx.doi.org/10.1007/s000180050125; PMID: 9487387
- Barges S, Mihaly J, Galloni M, Hagstrom K, Müller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 2000; 127:779 - 90; PMID: 10648236
- Busturia A, Wightman CD, Sakonju S. A silencer is required for maintenance of transcriptional repression throughout Drosophila development. Development 1997; 124:4343 - 50; PMID: 9334282
- Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev 1996; 10:3202 - 15; http://dx.doi.org/10.1101/gad.10.24.3202; PMID: 8985188
- Hagstrom K, Muller M, Schedl P. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics 1997; 146:1365 - 80; PMID: 9258680
- Mihaly J, Barges S, Sipos L, Maeda R, Cléard F, Hogga I, Bender W, Gyurkovics H, Karch F. Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development 2006; 133:2983 - 93; http://dx.doi.org/10.1242/dev.02451; PMID: 16818450
- Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 1997; 124:1809 - 20; PMID: 9165128
- Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P. The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 1999; 153:1333 - 56; PMID: 10545463
- Zhou J, Barolo S, Szymanski P, Levine M. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev 1996; 10:3195 - 201; http://dx.doi.org/10.1101/gad.10.24.3195; PMID: 8985187
- Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol Cell Biol 2005; 25:3682 - 9; http://dx.doi.org/10.1128/MCB.25.9.3682-3689.2005; PMID: 15831473
- Iampietro C, Cléard F, Gyurkovics H, Maeda RK, Karch F. Boundary swapping in the Drosophila Bithorax complex. Development 2008; 135:3983 - 7; http://dx.doi.org/10.1242/dev.025700; PMID: 18987027
- Iampietro C, Gummalla M, Mutero A, Karch F, Maeda RK. Initiator elements function to determine the activity state of BX-C enhancers. PLoS Genet 2010; 6:e1001260; http://dx.doi.org/10.1371/journal.pgen.1001260; PMID: 21203501
- Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Schedl P. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res 1994; 22:3138 - 46; http://dx.doi.org/10.1093/nar/22.15.3138; PMID: 7915032
- Rodin S, Kyrchanova O, Pomerantseva E, Parshikov A, Georgiev P. New properties of Drosophila fab-7 insulator. Genetics 2007; 177:113 - 21; http://dx.doi.org/10.1534/genetics.107.075887; PMID: 17890362
- Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 2004; 168:1371 - 84; http://dx.doi.org/10.1534/genetics.104.029561; PMID: 15579691
- Schweinsberg SE, Schedl P. Developmental modulation of Fab-7 boundary function. Development 2004; 131:4743 - 9; http://dx.doi.org/10.1242/dev.01343; PMID: 15329342
- Smith ST, Wickramasinghe P, Olson A, Loukinov D, Lin L, Deng J, Xiong Y, Rux J, Sachidanandam R, Sun H, et al. Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev Biol 2009; 328:518 - 28; http://dx.doi.org/10.1016/j.ydbio.2008.12.039; PMID: 19210964
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet 2007; 3:e112; http://dx.doi.org/10.1371/journal.pgen.0030112; PMID: 17616980
- Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J 2007; 26:4203 - 14; http://dx.doi.org/10.1038/sj.emboj.7601851; PMID: 17805343
- Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep 2005; 6:165 - 70; http://dx.doi.org/10.1038/sj.embor.7400334; PMID: 15678159
- Georgieva S, Nabirochkina E, Dilworth FJ, Eickhoff H, Becker P, Tora L, Georgiev P, Soldatov A. The novel transcription factor e(y)2 interacts with TAF(II)40 and potentiates transcription activation on chromatin templates. Mol Cell Biol 2001; 21:5223 - 31; http://dx.doi.org/10.1128/MCB.21.15.5223-5231.2001; PMID: 11438676
- Krasnov AN, Kurshakova MM, Ramensky VE, Mardanov PV, Nabirochkina EN, Georgieva SG. A retrocopy of a gene can functionally displace the source gene in evolution. Nucleic Acids Res 2005; 33:6654 - 61; http://dx.doi.org/10.1093/nar/gki969; PMID: 16314324
- Galán A, Rodríguez-Navarro S. Sus1/ENY2: a multitasking protein in eukaryotic gene expression. Crit Rev Biochem Mol Biol 2012; 47:556 - 68; http://dx.doi.org/10.3109/10409238.2012.730498; PMID: 23057668
- Jani D, Lutz S, Hurt E, Laskey RA, Stewart M, Wickramasinghe VO. Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export. Nucleic Acids Res 2012; 40:4562 - 73; http://dx.doi.org/10.1093/nar/gks059; PMID: 22307388
- Kopytova DV, Orlova AV, Krasnov AN, Gurskiy DY, Nikolenko JV, Nabirochkina EN, Shidlovskii YV, Georgieva SG. Multifunctional factor ENY2 is associated with the THO complex and promotes its recruitment onto nascent mRNA. Genes Dev 2010; 24:86 - 96; http://dx.doi.org/10.1101/gad.550010; PMID: 20048002
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 2008; 29:92 - 101; http://dx.doi.org/10.1016/j.molcel.2007.12.011; PMID: 18206972
- Pascual-García P, Govind CK, Queralt E, Cuenca-Bono B, Llopis A, Chavez S, Hinnebusch AG, Rodríguez-Navarro S. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev 2008; 22:2811 - 22; http://dx.doi.org/10.1101/gad.483308; PMID: 18923079
- Ellisdon AM, Jani D, Köhler A, Hurt E, Stewart M. Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1. J Biol Chem 2010; 285:3850 - 6; http://dx.doi.org/10.1074/jbc.M109.070839; PMID: 20007317
- Jani D, Lutz S, Marshall NJ, Fischer T, Köhler A, Ellisdon AM, Hurt E, Stewart M. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol Cell 2009; 33:727 - 37; http://dx.doi.org/10.1016/j.molcel.2009.01.033; PMID: 19328066
- Köhler A, Zimmerman E, Schneider M, Hurt E, Zheng N. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 2010; 141:606 - 17; http://dx.doi.org/10.1016/j.cell.2010.04.026; PMID: 20434206
- Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 2010; 328:1025 - 9; http://dx.doi.org/10.1126/science.1190049; PMID: 20395473
- Geyer PK, Corces VG. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev 1987; 1:996 - 1004; http://dx.doi.org/10.1101/gad.1.9.996; PMID: 3123324
- Comet I, Schuettengruber B, Sexton T, Cavalli G. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc Natl Acad Sci U S A 2011; 108:2294 - 9; http://dx.doi.org/10.1073/pnas.1002059108; PMID: 21262819
- Kyrchanova O, Ivlieva T, Toshchakov S, Parshikov A, Maksimenko O, Georgiev P. Selective interactions of boundaries with upstream region of Abd-B promoter in Drosophila bithorax complex and role of dCTCF in this process. Nucleic Acids Res 2011; 39:3042 - 52; http://dx.doi.org/10.1093/nar/gkq1248; PMID: 21149269
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 2011; 42:330 - 41; http://dx.doi.org/10.1016/j.molcel.2011.03.025; PMID: 21549310
- Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins 2003; 52:80 - 7; http://dx.doi.org/10.1002/prot.10389; PMID: 12784371
- Pierce B, Weng Z. ZRANK: reranking protein docking predictions with an optimized energy function. Proteins 2007; 67:1078 - 86; http://dx.doi.org/10.1002/prot.21373; PMID: 17373710
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Current protocols in bioinformatics / editoral board, Andreas D Baxevanis [et al] 2006; Chapter 5:Unit 5 6.
- Karess RE, Rubin GM. Analysis of P transposable element functions in Drosophila. Cell 1984; 38:135 - 46; http://dx.doi.org/10.1016/0092-8674(84)90534-8; PMID: 6088058
