Abstract
Regulation of Set1-COMPASS-mediated H3K4 methylation and Dot1-mediated H3K79 methylation by H2BK123 ubiquitination (H2Bub1) is an evolutionarily conserved trans-histone crosstalk mechanism. How H2Bub1 impacts chromatin structure and affects Set1-COMPASS/Dot1 functions has not been fully defined. Ubiquitin was proposed to bind proteins to physically bridge H2Bub1 with Set1-COMPASS/Dot1. Alternatively, the bulky ubiquitin was thought to be a “wedge” that loosens the nucleosome for factor access. Contrary to the latter possibility, recent discoveries provide evidence for nucleosome stabilization by H2Bub1 via preventing the constant H2A-H2B eviction. Recent data has also uncovered a “docking-site” on H2B for Set1-COMPASS. Collectively, these findings invoke a model, where ubiquitin acts as a “glue” to bind the nucleosome together for supporting Set1-COMPASS/Dot1 functions. This review provides an overview of these novel findings. Additionally, how H2Bub1 and its deubiquitination might alter the chromatin dynamics during transcription is discussed. Possible models for nucleosome stabilization by ubiquitin are also provided.
Introduction
Eukaryotic DNA is compressed within the nucleus by virtue of its tight packaging into chromatin, which is made up of repeating units called nucleosomes. Each nucleosome is comprised of ∼147 bp of DNA wrapped around a histone octamer, containing a tetramer core of H3–H4 dimers flanked by two H2A–H2B dimers.Citation1 Post-translational histone modification is one of the mechanisms employed by cellular processes to remodel chromatin and gain access to the underlying DNA template.Citation2,Citation3 Histones undergo a wide variety of reversible covalent modifications, including arginine (R) methylation, serine/threonine phosphorylation and several types of modifications on their lysine (K) residues (acetylation, methylation, ubiquitination, sumoylation and ADP-ribosylation).Citation2,Citation4,Citation5 Histone modifications act as binding sites for specialized recognition modules on effector proteins, which either directly or indirectly alter the nucleosome or chromatin structure.Citation6,Citation7 Alternatively, some modifications can directly affect the chromatin structure by charge neutralization (e.g., H4K16 acetylation).Citation8 In a complex regulatory network, termed as a “crosstalk”, modification on one histone can either support or prevent the establishment of another modification within the same histone (in cis) or at a distant site on a different histone (in trans).Citation9–Citation11 In budding yeast, H3K4 methylation is affected in cis by H3R2 methylationCitation9 and in a trans-histone crosstalk, both H3K4 and -K79 methylation are regulated by H2BK123 ubiquitination (H2Bub1)Citation12,Citation13 (). In recent years, this trans-histone pathway has garnered much attention, since both H3K4 and -K79 methylation have been implicated to play a role in many nuclear processes, such as transcription (activation/repression), replication, recombination and repair.Citation14,Citation15 Considerable information from different model systems has been gathered regarding the catalytic enzymes and factors that regulate the addition and removal of these modifications.Citation12,Citation13,Citation16 However, the fundamental mechanism by which H2Bub1 regulates H3K4 and -K79 methylation has remained elusive. Although H2B ubiquitination was discovered some 30 years ago,Citation17 whether conjugation of ubiquitin onto H2B has any effect on nucleosome structure has remained a longstanding question in chromatin biology. Here, we will focus on recent findings that revealed a novel role for H2Bub1 and its deubiquitination in the dynamic regulation of nucleosome stability and chromatin structure, and the implications of this regulation on the trans-histone H3 methylation.
The Trans-Histone Crosstalk Between H2Bub1 and H3K4/-K79 Methylation
In budding yeast, the ubiquitin-conjugating enzyme Rad6 and the E3-ligase Bre1 covalently attach ubiquitin onto K123 in H2B.Citation12,Citation13,Citation16 H3K4 and -K79 methylation can exist in mono- (me1), di- (me2) or tri-methylated (me3) state and are catalyzed by methyltransferases Set1 and Dot1, respectively. While Dot1 is a stand-alone enzyme, Set1 associates with regulatory subunits to form a multi-protein complex (COMPASS)Citation4,Citation13 (). Yeast mutants lacking H2Bub1 (rad6Δ, bre1Δ or ubiquitination-site mutant, H2B-K123R) revealed the presence of a trans-histone crosstalk pathway, since H3K4me2, H3K4me3 and H3K79me3 are abolished in the absence of H2Bub1.Citation12,Citation13,Citation16 Additionally, H3K4me1 and H3K79me2 are also severely reduced. In contrast, increase in H2Bub1 levels, due to the loss of H2Bub1-specific deubiquitinases (Ubp8 and Ubp10), causes an increase in H3K4 and -K79 methylation.Citation18–Citation20
Understanding the Language of the Trans-Histone Crosstalk
Models proposed to explain the mechanism of H2Bub1-mediated H3K4 and -K79 methylation include a possibility, where the conjugated ubiquitin acts as a “bridge” to directly recruit the methyltransferases.Citation21 Since Set1 and Dot1 associate with chromatin even in the absence of H2Bub1,Citation22,Citation23 their recruitment is not the primary function of H2Bub1. Alternatively, H2Bub1 might act as a “bridge” to recruit a regulatory factor to mediate the crosstalk. Swd2, a Set1-COMPASS component, has been implicated as a key link between H2Bub1 and H3K4 methylation by two independent studies. In the first study, H2Bub1 is required for the association of Swd2 with Set1-COMPASS, and Swd2 was proposed to stimulate Set1 activity via a conformational change resulting from its direct or indirect interaction with H2Bub1.Citation24 Additionally, Swd2 was also shown to interact with Dot1 and affect H3K79 methylation. In striking contrast, a second study suggested that the Rad6/Bre1-mediated H2Bub1 leads to Swd2 ubiquitination and the subsequent recruitment of Spp1 (a COMPASS subunit required for H3K4me3) stimulates the catalytic activity of Set1.Citation25 However, Spp1 interacts with chromatin independent of H2Bub1.Citation26 Therefore, given the conflicting findings of these two studies, precisely how Swd2 participates in the crosstalk remains to be ascertained. The 19S regulatory complex of the proteasome has also been implicated to mediate the trans-histone crosstalk, but the underlying mechanism is not understood.Citation27 Given that addition of ubiquitin is a relatively bulky modification and since H2Bub1 is closely associated with transcriptionally active regions, H2Bub1 was also postulated to act as a “wedge” to non-specifically open-up the chromatin and allow access to the histone modifying enzymes or their regulatory factors.Citation21,Citation28
Holding the Fort: H2Bub1 and Nucleosome Stability
We tested the “Wedge” model by substituting ubiquitin with a bulkier SUMO moiety at the H2B C-terminal region using yeast cells expressing a chimeric H2B, which was engineered with consensus sumoylation sites in place of the site of ubiquitination. Although the occupancy and distribution of the induced H2B sumoylation on chromatin were similar to H2Bub1, it was unable to support H3K4 and -K79 methylation.Citation29 Recently, McGinty et al. (2009)Citation30 also showed that, unlike semisynthetic ubiquitinated H2B, sumoylated H2B reconstituted into nucleosome in vitro does not stimulate human Dot1(hDot1L)-mediated H3K79 methylation. Therefore, these in vivo and in vitro studies establish that ubiquitin does not mediate the trans-histone crosstalk simply by virtue of its bulkiness.
It has been proposed that H2Bub1 exerts its regulatory functions by affecting chromatin structure,Citation31 but the nature of this structural change was never defined. Availability of discrete yeast mutants, which either completely lack H2Bub1 (rad6Δ, bre1Δ or H2B-K123R) or contain very high levels of H2Bub1 (ubp8Δ and/or ubp10Δ), prompted us to reinvestigate the effect of H2Bub1 on overall chromatin structure using micrococcal nuclease (MNase). Surprisingly, high levels of H2Bub1 rendered the chromatin resistant to nuclease digestion and the absence of H2Bub1 caused increased nuclease sensitivity.Citation29 This finding suggested that, contrary to its supposed role in opening up the chromatin, presence of H2Bub1 makes the chromatin compact and in fact, it is the absence of H2Bub1 that leads to an “open” or loose chromatin. Correlating well with this possibility, genes suppressed by RNF20 (the human homolog of yeast Bre1) are enriched in H2Bub1 and are present in a “closed” or compact chromatin configuration.Citation32 In addition, in vitro reconstituted nucleosome containing H2Bub1 showed significantly slower rate of DNase I digestion compared to those containing only H2B.Citation33 The decreased sensitivity to MNase and DNase I digestion in the presence of H2Bub1 might allude to a structural change within the nucleosome, a likely alteration in the histone-DNA or histone-histone interactions that restricts nuclease accessibility. Using salt-dependent nucleosome disruption assays, we confirmed that H2Bub1 stabilizes the nucleosome by affecting the electrostatic interactions, since histones were easily extractable in low salt solutions in the absence of H2Bub1 and high salt solutions were needed to solubilize H2Bub1-containing chromatin.Citation29 Our observation that histone amounts were reduced on native chromatin in the absence H2Bub1, but not in ChIP assays (where the chromatin is subjected to formaldehyde cross-linking, which can trap any weak histone-histone and histone-DNA interactions), further supports the need for the presence of H2Bub1 in stabilizing intra-nucleosomal interactions. Indeed, histone amounts were increased on native chromatin in the presence of high levels of H2Bub1, due to the generally enhanced nuclesome stability.Citation29 Interestingly, the ChIP assays also revealed that H2Bub1 stabilizes the nucleosome primarily by affecting H2A–H2B, since only H2B but not H3 levels are increased over active and repressed genes upon increase in H2Bub1 levels.Citation29
Biochemical and structural studies of the yeast nucleosome have revealed that it is intrinsically less stable than those from other eukaryotesCitation34,Citation35 (). In addition, H2A-H2B is constantly evicted from and reassembled into chromatin in yeast,Citation36,Citation37 even in the absence of DNA replication and transcription.Citation38 Therefore, based on these studies and our findings, we propose that the conjugation of ubiquitin onto H2B prevents H2A–H2B eviction and stabilizes the nucleosome, and removal of the conjugated ubiquitin by deubiquitinases causes nucleosome destabilization (). Therefore, H2Bub1 and its deubiquitination might function as an on/off switch similar to histone acetylation and deacetylation. Interestingly, the deubiquitinase Ubp8 is part of the SAGA coactivator complex, which also contains the histone acetyltransferase Gcn5.Citation18,Citation19 In addition, Rad6 and histone deacetylase Rpd3 were identified as negative regulators of transcription in a bypass suppression screen; since the deletion of RAD6 or RPD3 could rescue the lethality of BUR1 deletion [Bur1, a cyclin-dependent kinase (CDK) required for efficient transcription elongation by RNA polymerase II (RNA Pol II)].Citation39 Thus, a scenario can be envisaged, wherein histone deubiquitination and acetylation might function in tandem to counter the nucleosome stabilized by histone ubiquitination and deacetylation during transcription.
Modulation of Chromatin Dynamics by H2Bub1 and its Deubiquitination During Transcription
Maintaining the dynamic equilibrium between ubiquitination and deubiquitination is also critical for cell growth. Mutations that abolish H2Bub1 (bre1Δ, H2B-K123R) or lead to high levels of H2Bub1 (ubp8Δubp10Δ) adversely affect cell growth.Citation29,Citation40 Microarray-based studies have shown that both the loss and excessive levels of H2Bub1 can reduce the transcript levels of several genes.Citation20,Citation41,Citation42 Additionally, these changes in H2Bub1 levels can also result in de-repression and/or poor activation of inducible genes.Citation20,Citation41 Therefore, H2Bub1 plays both positive and negative roles in the regulation of gene transcription. This leads to the question, how does the nucleosome stabilization or destabilization mediated by H2Bub1 or its deubiquitination exert an effect on gene expression?
H2Bub1 and Transcriptional Repression
H2Bub1 plays a role in the repression of inducible genes both in yeast and in mammals. In yeast, absence of H2Bub1 (rad6Δ or H2B-K123R) causes de-repressed transcription of ARG1 under non-inducing condition with a concomitant increase in TBP binding to the promoter.Citation43 In contrast, ARG1 transcript levels are reduced by elevated levels of H2Bub1 in ubp8Δubp10Δ.Citation20 Also, the basal expression of several repressed/inducible genes (PHO5, GAL1 and ADH2) is reduced due to increased H2Bub1 levels (ubp8Δ and/or ubp10Δ).Citation20,Citation42 Correlating well with these transcript analyses, our ChIP assays revealed the presence of H2Bub1 on the ARG1 promoter even under non-inducing condition and the H2B levels on ARG1 TATA region are enhanced when H2Bub1 levels are increased (ubp8Δ and/or ubp10Δ).Citation29 Similar results were obtained for the well-characterized, positioned nucleosomes that encompass TATA boxes and/or upstream activating sequences present in the promoters of PHO5, GAL10 and ADH2. H2A-H2B is constantly evicted from and reassembled into the chromatin of repressed genes even in the absence of DNA replication.Citation38 This eviction might provide a window-of-opportunity for TBP or activators to gain access to their binding sites and thereby, allowing basal transcription. Therefore, it can be envisaged that Rad6/Bre1-mediated H2Bub1 might repress the basal transcription by stabilizing the nucleosome positioned over the promoter region to prevent TBP or activator binding (). Given that SAGA complex is involved in the regulation of these inducible genes,Citation18,Citation44 Ubp8-mediated deubiquitination might then destabilize the nucleosomes to allow TBP binding or to promote RNA Pol II progression following activation ().
Human Bre1 (hBre1/RNF20) is involved in the regulation of epidermal growth factor (EGF) inducible genes and in the constitutive suppression of a subset of genes that includes proto-oncogenes (c-myc and c-fos).Citation32 The RNF20-suppressed genes contain high levels of H2Bub1, H3K4me3, H3K9/14ac (“marks” of transcription) and RNA Pol II occupancy, and are significantly enriched in compact chromatin (). These observations have led Shema et al. (2008)Citation32 to propose that even though the RNF20-suppressed genes are in a transcriptional “on” state, the unfavorable chromatin structure, presumably mediated by H2Bub1, likely hinders transcription leading to polymerase pausing. The human homolog of yeast Ubp8 (USP22 in human SAGA) is also involved in the regulation of inducible genes, as seen from its role in androgen receptor- and estrogen receptor-mediated transactivation.Citation45 Collectively, based on these observations, Espinosa (2008)Citation46 proposed a model, wherein USP22 might deubiquitinate H2Bub1 in RNF20-suppressed genes including the EGF-inducible genes, and promote RNA Pol II progression. Based on our findings, it is tempting to further speculate that H2Bub1 stabilizes the nucleosomes in RNF20-suppressed genes to maintain the “closed”/compact chromatin, which results in a paused RNA Pol II. Subsequently, in the presence of the ligand, deubiquitination of H2Bub1 by USP22 might destabilize the nucleosome allowing the promoter escape or progression of RNA Pol II into the elongation phase ().
H2Bub1 and Transcriptional Elongation
Both Rad6/Bre1 and Ubp8 are recruited to the coding regions of genes.Citation47,Citation48 Moreover, their mutations confer slow growth in the presence of 6-azauracil (indicative of transcriptional elongation defects)Citation47,Citation48 and reduce the expression of several subsets of genes.Citation41,Citation42 Therefore, maintenance of the equilibrium between the multiple rounds of ubiquitination and deubiquitination of H2B is critical for the efficient progression of transcription. Increased H2Bub1 levels in ubp8Δ causes an increase in H2B levels across the transcribed regions, but not in the promoters of constitutively expressed genes,Citation29 suggesting that nucleosome stabilization and destabilization also occurs during transcription elongation. H2Bub1 also affects transcription elongation and chromatin architecture by regulating the nucleosome assembly/disassembly functions of the histone chaperone FACT (comprised of Spt16 and Pob3).Citation49,Citation50 A stable nucleosome would pose a formidable barrier to RNA Pol II progression. This raises the question, how does H2Bub1 stabilize nucleosomes and yet, facilitates transcription elongation?
Nucleosomes immediately adjacent to the transcribing RNA Pol II might be inherently highly unstable, as they undergo constant histone eviction and deposition, and are subjected to the torsional stress imposed by the ongoing transcription. The following model can be proposed for the function of H2Bub1 during elongation (): Rad6/Bre1-mediated H2Bub1 might initially stabilize the nucleosomes to enhance Spt16 binding to chromatin in front of the polymerase. Deubiquitination by Ubp8 might then occur to destabilize the nucleosomes and enhance the disassembly activity of Spt16 that allows RNA Pol II progression. Since a single H2A–H2B dimer is displaced from the nucleosome by FACT in vitro,Citation51 it is tempting to speculate that Ubp8-mediated deubiquitination and the disassembly of the two H2A–H2B dimers occur in a step-wise fashion. Subsequently, during the Spt6 and Spt16-mediated nucleosome reassembly in the wake of RNA Pol II progression,Citation37,Citation52,Citation53 H2Bub1 stabilizes the newly deposited nucleosomes. This step-wise, cyclical event of nucleosome destabilization-stabilization and nucleosome disassembly-assembly is supported by the observation that chromatin-bound Spt16 levels are reduced in the absence of H2Bub1 and H2Bub1 levels are reduced in an Spt16 mutant with defective nucleosome reassembly function.Citation37 Loss of H2Bub1 and/or the reduced histone deposition by the defective Spt16 also result in cryptic transcription initiation from within the coding regions.Citation37,Citation54 Collectively, these findings suggest an important role for H2Bub1-mediated nucleosome stabilization in maintaining the chromatin structure for the fidelity of transcriptional initiation and the proper progression of RNA Pol II ().
Yeast nucleosome is intrinsically less stable than those from other eukaryotesCitation34,Citation35 and 85% of the yeast genome is transcribed, unlike only 30% of the genome transcribed in humans.Citation55 The high level of transcription and intrinsic nucleosome instability might explain the >10% H2B existing as H2Bub1 in yeast as opposed to the 1–2% H2Bub1 in mammals.Citation56 To explain the mechanism of RNF20-mediated gene repression, it was proposed that the “negative effects” of H2Bub1 acts as a “checkpoint” during the early stages of elongation causing RNA Pol II to pause for the coordination of elongation with RNA processing.Citation46 Based on our findings, it can be further proposed that this “negative effect”, which includes a compact chromatin formation, is likely due to the nucleosome stabilization by H2Bub1. RNA Pol II pausing is not prevalent in yeast.Citation57 However, deubiquitination of H2Bub1 by Ubp8 is required for the chromatin association of Ctk1 (a CDK required during the later stages of elongation) (), which demonstrates another regulatory role for H2Bub1 during transcription.Citation48 Therefore, modulating and coordinating the FACT-mediated chromatin dynamics and regulating the association of factor(s) with chromatin are likely to be the “checkpoint” functions for H2Bub1-mediated nucleosome stability during eukaryotic transcription.
SETting the Stage and Connecting the DOTs: H2Bub1-Mediated Nucleosome Stability and the trans-Histone H3 Methylation
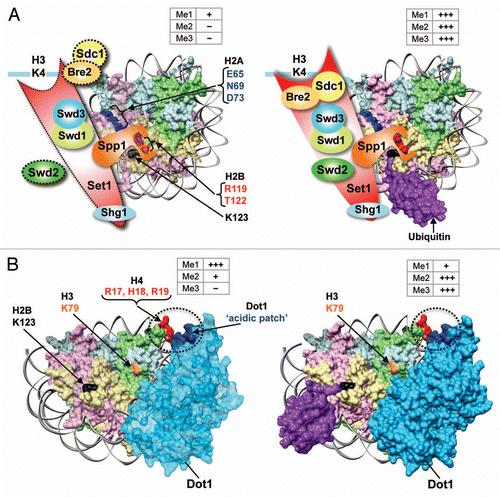
Modifications and mutations in the H3 N-terminal residues adjacent to K4 reduce Set1-COMPASS-mediated methylation by affecting the binding of regulatory subunits or by directly affecting the substrate recognition by the SET domain.Citation9 Alanine-mutagenesis of histones showed that mutations in three H2A residues (E65, N69 and D73) and one H2B residue (R119) affect H3K4 methylation without affecting the steady state H2Bub1 levels.Citation58 Additionally, we found mutations in two H2B C-terminal helix residues (H2B-R119A and/or H2B-T122D) increase global H2Bub1 levels, but reduce H3K4 methylation without affecting H3K79 methylation.Citation59 These H2A and H2B residues are all present facing away from the nucleosome core and are easily accessible for interaction (). Therefore, it can be envisaged that, following H2B ubiquitination, these H2A and H2B residues along with the residues in H3 N-terminal tail might constitute a composite interaction surface (“docking-site”) for the association of Set1-COMPASS with chromatin. Indeed, we find H2B R119 and T122 are the “docking-site” for Set1-COMPASS on chromatin as they physically interact with Spp1.Citation59 Moreover, H2B-R119A and/or H2B-T122D also affect both global and chromatin-bound levels of Sdc1, revealing a role for H2B C-terminal helix in controlling the complex integrity.Citation59 Therefore, it is conceivable that by preventing H2A–H2B eviction and enhancing nucleosome stability, H2Bub1 supports the retention of the composite “docking-site” for Set1-COMPASS on chromatin ( and ). Future investigations are needed to address whether the H2A residues affect the binding of Set1-COMPASS subunits. Unlike H3K4 methylation, Dot1-mediated H3K79 methylation is not reduced in H2B-R119A and/or H2B-T122D.Citation59 How is this uncoupling achieved? First, these residues reside on a helical phase that faces away from H3K79 (). Second, they do not reduce Dot1 binding (Chandrasekharan and Sun, unpublished observation), but instead serve as a distinct “docking-site” for only Set1-COMPASS. Collectively, based on our findings, it can be proposed that H2Bub1-mediated nucleosome stabilization retains the binding site for Set1-COMPASS in the H2B C-terminal helix and supports the stability, integrity and processivity of Set1-COMPASS by affecting the chromatin association of its subunits, at least in part, by directly affecting the binding of Spp1 ().
Given the proximity between H3K79 and the H2B C-terminal, several possible scenarios can be envisioned as to how H2Bub1-mediated nucleosome stability can support H3K79 methylation by Dot1. One possibility is that, similar to Set1-COMPASS, H2Bub1 might affect the chromatin binding of Dot1. However, this seems unlikely, since the chromatin-binding and H3K79 methylation of Dot1 is controlled in a novel trans-histone pathway by the basic patch in the H4 N-terminal tailCitation60 and Dot1 binds chromatin normally even in the absence of H2Bub1.Citation23 Alternatively, H2Bub1 might control the catalytic functions of Dot1 via an allosteric mechanism. This possibility stems from the observation that H2Bub1 reconstituted into chemically defined nucleosome can support the direct stimulation of human hDot1L-mediated H3K79 methylation in vitro.Citation61 It is conceivable that this allosteric change might be induced by a direct interaction between ubiquitin and hDot1L. However, free ubiquitin provided in trans cannot stimulate hDot1L-mediated H3K79 methylation.Citation61 Therefore, an attractive alternative possibility is that the nucleosome stabilized by H2Bub1 might directly induce an allosteric change in Dot1 to support its ability to catalyze H3K79me2 and -K79me3 (). Further studies will uncover the precise mechanism by which H2Bub1 stimulates Dot1 functions.
Mechanism of Nucleosome Stabilization by Conjugation of Ubiquitin onto H2B
At present, it is not known how the conjugation of ubiquitin at H2BK123 stabilizes nucleosomes. However, a recent study assessing the ability of hDot1L to function on nucleosomes reconstituted in vitro using chemically synthesized H2Bub1 provides several valuable insights into the mechanism.Citation62 First and foremost, the anomalous gel migration of the nucleosomes reconstituted in vitro with H2B monoubiquitinated at different lysine residues in its C-terminal helix provides a compelling evidence that the conjugated ubiquitin does affect mononucleosome structure and/or surface charge in a position-dependent manner. Second, the ubiquitin fold per se is not important, but a set of charged residues on ubiquitin is essential for hDot1L function. Moreover, these residues are not the canonical residues involved in the interactions between ubiquitin and ubiquitin-binding proteins. Third, proximity of the conjugated ubiquitin to H2B is also critical, as hDot1L-mediated H3K79 methylation is reduced upon altering the length of glycine linker between ubiquitin and H2B. Collectively, these observations are consistent with the two scenarios that were previously proposed:Citation29 First, H2Bub1 might stabilize the nucleosome by the interactions of the surface charged residues of ubiquitin with the DNA and/or the histones. Second, conjugation of ubiquitin might indirectly alter the histone-DNA and/or histone-histone interactions, in particular, the interactions of the “unstable” H2A–H2B dimer with the DNA and/or the H3–H4 tetramer core. Further studies are needed to test these possibilities.
Concluding Remarks
Recent findings of a role for H2Bub1 in modulating nucleosome stability and chromatin dynamics provide valuable insight to the function of this modification during transcription.Citation29,Citation37 Furthermore, the identification of a distinct “docking-site” for Set1-COMPASS on the H2B C-terminal helix sheds light on the mechanism by which H2Bub1-mediated nucleosome stabilization controls H3K4 and -K79 methylation.Citation59 Collectively, these studies provide a novel working model for the regulation of the trans-histone crosstalk between H2Bub1 and H3K4/-K79 methylation. Eventually, a solved structure of the nucleosome containing H2Bub1 might determine any changes in the intra-nucleosomal interactions that occur upon conjugation of ubiquitin to H2B. Recent advances in synthetic protein chemistry have enabled the production of large amounts of ubiquitinated H2B containing nucleosomes and these approaches hold promise in solving the elusive crystal structure.Citation30,Citation62 Nevertheless, future investigations using the in vitro H2Bub1-reconstituted nucleosome combined with the in vivo studies using histone mutants in yeast should uncover the mechanistic details of H2Bub1-mediated nucleosome stabilization and its function in regulating H3 methylation and gene expression.
Abbreviations
| COMPASS | = | complex associated with Set1 |
| ChIP | = | chromatin immunoprecipitation |
| TBP | = | TATA binding protein |
| SAGA | = | Spt-Ada-Gcn5-acetyltransferase |
| FACT | = | facilitates chromatin transcription |
| SET | = | suppressor of variegation-enhanser of zeste-trithorax |
Figures and Tables
Figure 1 The trans-histone crosstalk between histone H2B ubiquitination and H3K4/-K79 methylation in budding yeast. A simulated structure for an ubiquitinated nucleosome is shown by placing the C-terminal residue of ubiquitin (PDB: 1UBQ) close to lysine 123 (K123) of H2B present in the yeast nucleosome (PDB 1ID3; H3, pale blue; H4, pale green; H2A, pale pink; H2B, pale yellow). Rad6 (the ubiquitin-conjugating enzyme) and Bre1 (the E3 ligase) conjugate ubiquitin onto K123 in the H2B C-terminal helix. The conjugated ubiquitin is removed by two deubiquitinases, Ubp8 and Ubp10. Histone methyltransferases Set1 and Dot1 catalyze mono- (me1), di- (me2) and trimethylation (me3) of H3 K4 and K79 residues, respectively. Set1 is present in a multi-protein complex (COMPASS) along with seven subunits, which regulate the complex stability, integrity and processive methylation.
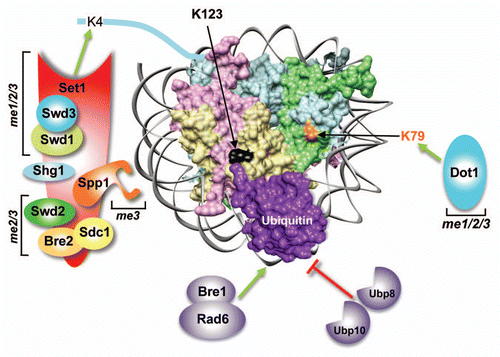
Figure 2 Model for the dynamic changes in yeast nucleosome stability mediated by H2B ubiquitination and deubiquitination. The innate weak intra-nucleosomal interactions and the constant H2A–H2B eviction lead to an unstable yeast nucleosome. Conjugation of ubiquitin stabilizes the nucleosome by negating these destabilizing events. On the contrary, removal of ubiquitin by deubiquitination promotes nucleosome instability. Stable nucleosome might allow or restrict factor binding to chromatin. By preventing H2A–H2B eviction, H2Bub1 retains the binding surfaces for Set1-COMPASS in H2A and H2B on chromatin. The composite “docking-site” for Set1-COMPASS on the nucleosome is predicted to be made up of the three residues in H2A (E65, N69 and D73) and the two experimentally confirmed residues in the H2B C-terminal helix (R119 and T122). Modified from Chandrasekharan et al.Citation59
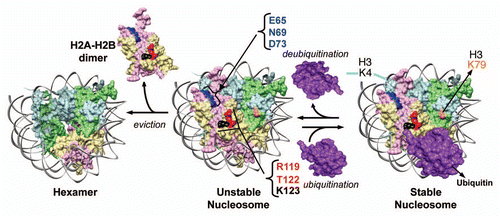
Figure 3 Model for the regulation of inducible gene expression by H2B ubiquitination and deubiquitination. (A) In budding yeast, conjugation of ubiquitin prevents H2A–H2B eviction and stabilizes the nucleosome positioned over the promoters of inducible genes in repressed state. The stable nucleosome might restrict the binding of activators (Act) and/or the TATA binding protein (TBP) to their cognate sites and prevents basal transcription. Following induction, DNA-bound activators recruit the SAGA complex containing the deubiquitinase Ubp8. Removal of the conjugated ubiquitin by Ubp8 causes nucleosome instability and promotes nucleosome disassembly by histone chaperones and/or other remodeling machineries. In turn, this allows the binding of TBP and recruitment of RNA Pol II for activated transcription. UAS, upstream activating sequence; TATA, the TATA box; INR, initiator sequence. (B) In mammals, inducible genes repressed by the E3 ligase RNF20 contain high levels of H2Bub1 and a poised RNA Pol II, and are enriched in the “marks” of active transcription [H3K9/14 acetylation (Ac, yellow circle) and H3K4me3 (cyan flag)]. H2Bub1 occupancy is high immediately downstream of the transcription start site and RNF20-repressed genes are in “closed”/compact chromatin configuration. Therefore, H2Bub1 mediated by RNF20 and possible E2-conjugating enzymes (UbcH6 or human homolog of Rad6, HR6A/B) might stabilize the nucleosome (PDB: 1KX5) and block transcription. USP22, the deubiquitinase in human SAGA (hSAGA), is required for the activation of many inducible genes. Therefore, following induction by activating stimuli, activators recruit hSAGA to the promoters of RNF-20 repressed genes to remove H2Bub1 and destabilize the nucleosome. This is followed by nucleosome disassembly, which results in “open”/loose chromatin formation. Removal of the stable “impeding” nucleosome allows the progression of paused RNA Pol II into the elongation phase. Adapted from Espinosa (2008).Citation46
![Figure 3 Model for the regulation of inducible gene expression by H2B ubiquitination and deubiquitination. (A) In budding yeast, conjugation of ubiquitin prevents H2A–H2B eviction and stabilizes the nucleosome positioned over the promoters of inducible genes in repressed state. The stable nucleosome might restrict the binding of activators (Act) and/or the TATA binding protein (TBP) to their cognate sites and prevents basal transcription. Following induction, DNA-bound activators recruit the SAGA complex containing the deubiquitinase Ubp8. Removal of the conjugated ubiquitin by Ubp8 causes nucleosome instability and promotes nucleosome disassembly by histone chaperones and/or other remodeling machineries. In turn, this allows the binding of TBP and recruitment of RNA Pol II for activated transcription. UAS, upstream activating sequence; TATA, the TATA box; INR, initiator sequence. (B) In mammals, inducible genes repressed by the E3 ligase RNF20 contain high levels of H2Bub1 and a poised RNA Pol II, and are enriched in the “marks” of active transcription [H3K9/14 acetylation (Ac, yellow circle) and H3K4me3 (cyan flag)]. H2Bub1 occupancy is high immediately downstream of the transcription start site and RNF20-repressed genes are in “closed”/compact chromatin configuration. Therefore, H2Bub1 mediated by RNF20 and possible E2-conjugating enzymes (UbcH6 or human homolog of Rad6, HR6A/B) might stabilize the nucleosome (PDB: 1KX5) and block transcription. USP22, the deubiquitinase in human SAGA (hSAGA), is required for the activation of many inducible genes. Therefore, following induction by activating stimuli, activators recruit hSAGA to the promoters of RNF-20 repressed genes to remove H2Bub1 and destabilize the nucleosome. This is followed by nucleosome disassembly, which results in “open”/loose chromatin formation. Removal of the stable “impeding” nucleosome allows the progression of paused RNA Pol II into the elongation phase. Adapted from Espinosa (2008).Citation46](/cms/asset/35f07e0e-71f0-4ae7-a988-012468d935c4/kepi_a_10912314_f0003.gif)
Figure 4 Model for the regulation of chromatin dynamics by H2B ubiquitination and deubiquitination during transcription elongation. (A) Rad6 and Bre1 associate with the Paf1 complex and travel with the elongating form of RNA Pol II, which is characterized by the phosphorylation (red oval) of serine residues in the heptad repeats that constitute the C-terminal domain (CTD, thick wavy line). (i and ii) Rad6/Bre1-mediated H2Bub1 stabilizes the nucleosome in front of the polymerase to counteract any torsional stress and might acts as a “checkpoint” to coordinate transcription-coupled events, such as, allowing the binding of Spt16/FACT and restricting Ctk1 binding to chromatin. (B) In (i), Ubp8, a component of the SAGA sub-complex that travels with RNA Pol II, removes the conjugated ubiquitin to destabilize the nucleosome. (ii) This facilitates nucleosome disassembly by Spt16/FACT. (iii) Deubiquitination by Ubp8 allows association of Ctk1 with chromatin for the phosphorylation of serine 2 in RNA Pol II CTD. (C) In (i), nucleosomes are reassembled behind the elongating RNA Pol II likely by the step-wise initial addition of H3-H4 by Spt6 followed by the addition of H2A–H2B by Spt16/FACT. (ii) Rad6/Bre1-mediated H2Bub1 stabilizes the nucleosome to facilitate nucleosome reassembly and to prevent any backtracking by RNA Pol II. (iii) Stable nucleosome also prevents any promiscuous transcription that might occur by the binding of TBP to cryptic TATA-like sequences within the coding region.
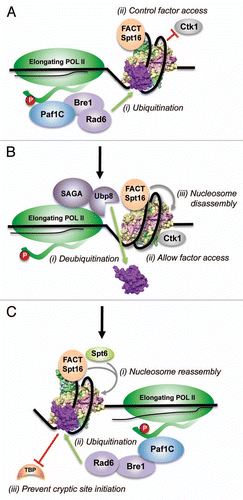
Figure 5 Model for the trans-histone crosstalk mechanism between H2Bub1 and H3K4/-K79 methylation. (A) Set1-COMPASS binds to the nucleosome via the interaction of Spp1 with the H2B C-terminal and likely by other hitherto unknown interactions with H2A. In the absence of H2Bub1, the constant H2A–H2B eviction and intrinsic nucleosome instability might disrupt or disassemble the composite “docking-site” for Set1-COMPASS on chromatin, leading to dissociation and/or weakened inter-subunit interactions within the complex and a catalytically-unfavorable conformation for Set1 (dotted lines). Together, these defects adversely affect the processive H3K4 methylation by Set1-COMPASS. In the presence of H2Bub1, H2A–H2B eviction is prevented and the stable nucleosome retains the binding surface for Set1-COMPASS on chromatin. This leads to increased chromatin association, inter-subunit interactions, and overall stability of Set1-COMPASS resulting in high levels of processive methylation. Modified from Chandrasekharan et al.Citation59 (B) Dot1 binds chromatin via the interaction of an acidic patch in its C-terminal region (EDVDE, amino acids 557–561) with a basic patch in H4 (RHR, amino acids 17–19). In the absence of H2Bub1, chromatin-bound Dot1 cannot catalyze H3K79me3 due to an inhibitory conformation (light blue). In the presence of H2Bub1, the stable nucleosome promotes an allosteric change in Dot1 that allows the production of high levels of H3K79me2 and -K79me3. +, low levels of methylation; +++, high levels of methylation; −, absence of methylation. Weak H2A–H2B dimer subjected to eviction, transparent and light colored; stable H2A–H2B dimer, bright colored.
Acknowledgements
We apologize for not citing other relevant articles due to space constraints. We thank Brian Strahl for his valuable suggestions. We thank The Vanderbilt-Ingram Cancer Center, The Robert J. and Helen C. Kleberg Foundation, NCI SPORE in Breast Cancer (5P0CA098131) and National Institutes of Health (RO1CA109355) for the financial support.
References
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 1999; 98:285 - 294
- Kouzarides T. Chromatin modifications and their function. Cell 2007; 128:693 - 705
- van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 2009; 19:207 - 217
- Berger SL. The complex language of chromatin regulation during transcription. Nature 2007; 447:407 - 412
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet 2009; 43:559 - 599
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol 2007; 8:983 - 994
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 2007; 14:1025 - 1040
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006; 311:844 - 847
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, et al. Arginine methylation at histone H3R2 controls deposition of H3K4 tri-methylation. Nature 2007; 449:928 - 932
- Latham JA, Dent SYR. Cross-regulation of histone modifications. Nat Struct Mol Biol 2007; 14:1017 - 1024
- Fingerman IM, Du HN, Briggs SD. Controlling histone methylation via trans-histone pathways. Epigenetics 2008; 35:237 - 242
- Osley MA. H2B ubiquitylation: The end is in sight. Biochim Biophys Acta 2004; 1677:74 - 79
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 2006; 75:243 - 269
- Wood A, Schneider J, Shilatifard A. Cross-talking histones: implications for the regulation of gene expression and DNA repair. Biochem Cell Biol 2005; 83:460 - 467
- Pinskaya M, Morillon A. Histone H3 lysine 4 di-methylation: a novel mark for transcriptional fidelity?. Epigenetics 2009; 4:302 - 306
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell 2008; 29:653 - 663
- West MH, Bonner WM. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res 1980; 8:4671 - 4680
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 2003; 17:2648 - 2663
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR 3rd, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem 2004; 279:1867 - 1871
- Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol 2005; 25:6123 - 139
- Henry KW, Berger SL. Trans-tail histone modifications: wedge and bridge?. Nat Struct Biol 2002; 9:565 - 566
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 2003; 11:709 - 719
- Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol Cell 2005; 19:271 - 277
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 2007; 131:1084 - 1096
- Vitaliano-Prunier A, Menant A, Hobeika M, Géli V, Gwizdek C, Dargemont C. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol 2008; 10:1365 - 1371
- Takahashi YH, Lee JS, Swanson SK, Saraf A, Florens L, Washburn MP, et al. Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol Cell Biol 2009; 29:3478 - 3486
- Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol Cell 2004; 13:435 - 442
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 2002; 18:104 - 108
- Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci USA 2009; 106:16686 - 16691
- McGinty RK, Köhn M, Chatterjee C, Chiang KP, Pratt MR, Muir TW. Structure-activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem Biol 2009; 4:958 - 968
- Jason LJ, Moore SC, Lewis JD, Lindsey G, Ausió J. Histone ubiquitination: a tagging tail unfolds?. Bioessays 2002; 24:166 - 174
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 2008; 22:2664 - 2676
- Davies N, Lindsey GG. Histone H2B (and H2A) ubiquitination allows normal histone octamer and core particle reconstitution. Biochim Biophys Acta 1994; 1218:187 - 193
- Piñeiro M, Puerta C, Palacián E. Yeast nucleosomal particles: structural and transcriptional properties. Biochemistry 1991; 30:5805 - 5810
- White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J 2001; 20:5207 - 5218
- Formosa T, Eriksson P, Wittmeyer J, Ginn J, Yu Y, Stillman DJ. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J 2001; 20:3506 - 3517
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 2008; 31:57 - 66
- Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell 2007; 25:345 - 355
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 2005; 123:593 - 605
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 2003; 11:267 - 274
- Zhang X, Kolaczkowska A, Devaux F, Panwar SL, Hallstrom TC, Jacq C, et al. Transcriptional regulation by Lge1p requires a function independent of its role in histone H2B ubiquitination. J Biol Chem 2005; 280:2759 - 2770
- Mutiu AI, Hoke SM, Genereaux J, Liang G, Brandl CJ. The role of histone ubiquitylation and deubiquitylation in gene expression as determined by the analysis of an HTB1(K123R) Saccharomyces cerevisiae strain. Mol Genet Genomics 2007; 277:491 - 506
- Turner SD, Ricci AR, Petropoulos H, Genereaux J, Skerjanc IS, Brandl CJ. The E2 ubiquitin conjugase Rad6 is required for the ArgR/Mcm1 repression of ARG1 transcription. Mol Cell Biol 2002; 22:4011 - 4019
- Huisinga KL, Pugh BF. A TATA binding protein regulatory network that governs transcription complex assembly. Genome Biol 2007; 8:46
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 2008; 29:92 - 101
- Espinosa JM. Histone H2B ubiquitination: the cancer connection. Genes Dev 2008; 22:2743 - 2749
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, et al. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol 2005; 25:637 - 651
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, et al. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell 2007; 27:275 - 288
- Laribee RN, Fuchs SM, Strahl BD. H2B ubiquitylation in transcriptional control: a FACT-finding mission. Genes Dev 2007; 21:737 - 743
- Hartzog GA, Quan TK. Just the FACTs: histone H2B ubiquitylation and nucleosome dynamics. Mol Cell 2008; 31:2 - 4
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science 2003; 301:1090 - 1093
- Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science 2003; 301:1096 - 1099
- Adkins MW, Tyler JK. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell 2006; 21:405 - 416
- Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol 2003; 23:8323 - 8333
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 2007; 17:877 - 885
- Kao CF, Osley MA. In vivo assays to study histone ubiquitylation. Methods 2003; 31:59 - 66
- Wade JT, Struhl K. The transition from transcriptional initiation to elongation. Curr Opin Genet Dev 2008; 18:130 - 136
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol 2008; 15:881 - 888
- Chandrasekharan MB, Huang F, Chen YC, Sun ZW. Histone H2B C-terminal helix mediates trans-histone H3K4 methylation independent of H2B ubiquitination. Mol Cell Biol 2010; 30:3216 - 3223
- Fingerman IM, Li HC, Briggs SD. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev 2007; 21:2018 - 2029
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 2008; 453:812 - 816
- Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol 2010; 6:267 - 269