Abstract
Melanoma is an aggressive malignancy that is resistant to current therapy, and the most lethal of all human skin cancers. It is characterized by several genetic alterations that lead to changes in gene expression and tumorigenesis by triggering alterations in the normal transcriptional circuitry. Transformation and tumor progression are thought to be promoted by a complex interplay between the accumulation of genetic alterations and epigenetic changes. In this review, we discuss recent studies that have implicated SWI/SNF chromatin remodeling enzymes as epigenetic regulators of a transcriptional circuit that operates within the context the genetic alterations that frequently occur in melanoma.
Introduction
Despite significant progress made in reducing mortality from many forms of cancer, the incidence and mortality due to melanoma is rapidly rising. Melanoma most often develops from the transformation of cutaneous melanocytes and is an aggressive malignancy that is resistant to current therapy, making it the most lethal of all human skin cancers.Citation1 A better understanding of the molecular mechanisms that regulate melanomagenesis and progression is needed in order to develop effective therapeutic options. Several genetic alterations have been associated with melanoma development and progression, including activating mutations in the gene encoding the BRAF component of the ERK-mitogen activated protein kinase pathway, inactivating mutations in the gene that encodes the p16INK4A cyclin dependent kinase inhibitor, and amplification of the gene encoding Microphthalmia Associated Transcription Factor (MITF).Citation2–Citation5 However, none of these genetic alterations can single-handedly transform melanocytes. Transformation and tumor progression are thought to be promoted by a complex interplay between the accumulation of genetic alterations and epigenetic changes that involve DNA methylation or changes in chromatin structure.Citation6 Recent studies have implicated SWI/SNF chromatin remodeling enzymes as epigenetic regulators of a transcriptional circuit that operates within the context of these genetic alterations.Citation7–Citation10
SWI/SNF Chromatin Remodeling Enzymes
SWI/SNF chromatin remodeling enzymes are evolutionarily conserved, multisubunit complexes of 1–2 MDa that utilize the energy derived from ATP hydrolysis to alter nucleosome structure and either activate or repress transcription.Citation11–Citation13 Each of these complexes contains a central catalytic domain which is BRG1 or BRM and 8–12 accessory proteins (BAFs) ( and B). In vitro, chromatin remodeling is achieved by a core complex containing BRG1 or BRM, BAF47, BAF 170 and BAF 155. In vivo, additional BAFs are required for interactions with transcriptional activators and repressors.Citation14–Citation19
Diverse SWI/SNF complexes with a distinct central ATPase and BAF composition may be present in different cell types and developmental stages ().Citation20 The two ATPases, BRG1 and BRM, are 70% identical at the amino acid level and have similar chromatin remodeling activity in vitro and overlapping as well as distinct functions in vivo (). However, BRG1 but not BRM is essential for mouse development.Citation21,Citation22 Furthermore, BRG1 and BRM can have antagonistic effects on the regulation of some genes.Citation23 The association of BRG1/BRM associated factors also influences the transcriptional outcome.Citation24 Thus, SWI/SNF subunit composition, determined by the identity of the particular ATPase and the association of a varied assortment of BAFs, is an important determinant that specifies positive or negative effects on particular promoters. A recent study suggested that in addition to each of these preassembled SWI/SNF complexes, heterogeneous sub-complexes can be recruited to different promoters.Citation25 The diversity in SWI/SNF subunit composition introduces the potential for intrinsically determined mechanisms that regulate SWI/SNF function at different sites in the genome. Thus, the combinatorial assembly of diverse SWI/SNF complexes has been proposed to form a code that dictates the transcriptional outcome within a particular chromatin context.Citation26
SWI/SNF enzymes lack DNA binding specificity and are thought to be recruited to genomic loci by interacting with gene specific regulators.Citation27 Interactions between the components of the SWI/SNF complex and a multitude of regulators have been characterized, including interactions with master regulators of differentiation, nuclear hormone receptors, and cancer associated transcriptional regulators, such as AP-1, c-MYC, β-catenin, RB, BRCA-1 and p53.Citation20,Citation28–Citation32 Although SWI/SNF complexes interact with many transcriptional regulators, genome wide analysis indicates that inactivation of SWI/SNF function in normal cells or reconstitution of SWI/SNF subunits in deficient cancer cells results in modulation of the expression of a limited set of genes.Citation33,Citation34 Thus, the requirement for SWI/SNF is likely to be programmatically determined by the cellular context and SWI/SNF subunit composition. Furthermore, signal transduction pathways play a key role in modulating interactions between SWI/SNF complexes and gene specific transcription factors, thereby altering the recruitment of SWI/SNF complexes to target promoters and/or modulating SWI/SNF activity.Citation20
Loss of the expression of several SWI/SNF subunits has been associated with malignant transformation.Citation35 The BAF47 subunit frequently undergoes bialleleic deletion in malignant rhabdoid tumors and mice that are heterozygous for BAF47 develop tumors with rhabdoid features that display loss of heterozygosity.Citation36–Citation39 Thus, BAF47 acts as a classic tumor suppressor. Downregulation of other BAFs as well as downregulation of the ATPase subunits has been demonstrated for a wide variety of human cancers.Citation35 Reconstitution of the missing SWI/SNF subunit in deficient cancer cell lines typically results in cell cycle arrest, apoptosis or differentiation.
Numerous studies strongly suggest that downregulation of SWI/SNF chromatin remodeling components contributes to tumorigenesis.Citation35 However, BRG1 heterozygous mice develop tumors that are different from BAF47 tumors and these tumors do not display loss of heterozygosity.Citation40 Furthermore, in gastric and prostate cancer, overexpression of the SWI/SNF subunits, BRG1 and BAF57, has been associated with tumor progression.Citation18,Citation41,Citation42 A recent study indicated that tumorigenesis caused by loss of BAF47 is dependent on continued BRG1 activity.Citation43 In combination, these observations suggest that in addition to downregulation of SWI/SNF activity, alteration of SWI/SNF specificity by de-regulation of particular subunits may be a key event that drives tumorigenesis. Several recent studies suggest that de-regulation of SWI/SNF function may play a critical role in melanoma.Citation7–Citation10
SWI/SNF Chromatin Remodeling Enzymes in Melanoma
Downregulation of the SWI/SNF component, BAF47 allows bypass of oncogenic BRAF induced apoptosis and senescence.
Approximately 50–70% of melanomas have activating mutations in the BRAF gene, most of which involve a valine to glutamic acid substitution at position 600 (BRAFV600E), resulting in constitutive BRAF-MEK-ERK signaling and tumorigenesis.Citation2,Citation44,Citation45 However, expression of BRAF(V600E) in primary melanocytes induces growth arrest that leads to senescence rather than transformation, suggesting the requirement for additional disruptions that allow bypass of BRAF(V600E) induced senescence.Citation46
The BAF47 component of the SWI/SNF complex was recently identified in a screen for factors whose downregulation allows bypass of BRAF(V600E) induced apoptosis and senescence.Citation7 BAF47 expression was found to be activated by a secreted factor, insulin-like growth factor binding protein 7 (IGFBP7) that is required for BRAF(V600E) induced apoptosis and senescence in melanocytes. However, in melanoma cells that harbor the BRAFV600E oncogene, IGFBP7 expression is lost by promoter hypermethylation and BAF47 expression may be compromised. BAF47 expression was activated by treating melanoma cells with recombinant IGFBP7. Increased expression of BAF47 then allowed recruitment of BRG1 based SWI/SNF complexes to the promoter of pro-apoptotic BNIP3L, activation of BNIP3L expression, and induction of apoptosis.Citation7 This study elegantly demonstrated a mechanism by which tumorigenicity can result from the failure of SWI/SNF recruitment to a specific locus by loss of a single SWI/SNF subunit. The observation that SWI/SNF complexes lacking BAF47 are fully functional at some promoters but not others, defines a specificity role for BAF47 in regulating SWI/SNF activity.Citation47 The recent observation that BAF47 expression is downregulated in a significant number of primary and metastatic melanomas and that downregulation of BAF47 correlates with a poor prognosis and increased chemosensitivity supports the hypothesis that BAF47 has an important role in melanomagenesis.Citation48
BRG1 interacts with p16INK4A.
Inactivation of the cyclin dependent kinase inhibitor, p16INK4A by point mutation, deletion or promoter hypermethylation occurs at high frequency in melanoma.Citation49,Citation50 Furthermore, loss of p16INK4A allows bypass of BRAF(V600E) induced senescence.Citation51 In malignant rhabdoid tumor derived cell lines lacking BAF47, p16INK4A expression is compromised.Citation52 In these cells, ectopic expression of BAF47 restored p16INK4A expression by promoting recruitment of BRG1 based SWI/SNF complexes to the p16INK4A promoter.Citation53 Once on the promoter, SWI/SNF complexes activate p16INK4A expression through a transcriptional mechanism that involves eviction of repressive complexes and promoter demethylation.Citation53,Citation54 Thus, loss of BAF47 in melanoma may also compromise the expression of p16INK4A and other senescence promoting genes by preventing SWI/SNF recruitment to the promoters of these genes. The possibility also exists that in addition to the failure to activate tumor suppressive pathways, BAF47 loss may lead to aberrant recruitment of BRG1 to alternative sites and to the activation of genes that promote tumorigenesis.
A novel mechanism by which SWI/SNF chromatin remodeling activity may be intertwined with p16INK4A function in melanoma is through direct interactions between BRG1 and p16INK4A.Citation8 Interestingly, BRG1 was found not to be required for p16INK4A induced senescence in melanoma cells, thus the interaction with p16INK4A may modulate SWI/SNF chromatin remodeling activity and impact gene expression. SWI/SNF complexes are known to interact with two important transcriptional regulators of p16INK4A: MITF, an activator, and β-catenin, a repressor of p16INK4A expression.Citation30,Citation55–Citation57 An intriguing possibility is that p16INK4A and BRG1 are involved in a feedback loop that regulates the activities of these transcriptional regulators.
SWI/SNF chromatin remodeling enzymes promote MITF mediated transcription.
MITF is a basic helix loop helix leucine zipper transcription factor that is developmentally required for the commitment of neural crest precursors to the melanocyte lineage, for survival, proliferation, and differentiation into functional melanocytes that synthesize melanin.Citation58 Thus, MITF has been designated the master regulator of melanocyte differentiation and can activate expression of genes required for melanin synthesis when introduced into non-melanocytic cells.Citation59
In melanoma, MITF is considered a lineage survival oncogene that is amplified in 10–20% of human melanomas and can cooperate with BRAF(V600E) to transform immortalized human melanocytes.Citation60 MITF promotes melanoma proliferation by directly activating the expression of cyclin dependent kinase 2 (CDK2) and the transcriptional regulator, TBX2, a suppressor of senescence.Citation61–Citation63 MITF also promotes melanoma survival by directly activating expression of the pro-survival genes, BCL2 and MLIAP.Citation64,Citation65 By promoting cell survival pathways, MITF may contribute to melanoma chemoresistance.Citation60 However, MITF can inhibit cell cycle progression by directly activating p21CIP1 and p16INK4A expression.Citation56,Citation66 A rheostat model has been proposed to reconcile these contrasting MITF activities such that a low level of MITF activity is required to prevent apoptosis, an intermediate level to promote proliferation, and a higher level to promote terminal differentiation and cell cycle arrest.Citation67 MITF activity is likely to be determined by expression levels, post-translational modifications, and interactions with co-regulatory proteins.
We previously determined that in a tissue culture model of differentiation, SWI/SNF chromatin remodeling enzymes promote MITF mediated activation of melanocyte specific gene expression.Citation55 We found that MITF promoted the recruitment of SWI/SNF components to melanocyte specific promoters on which SWI/SNF function was required to remodel chromatin structure. The observed requirement for SWI/SNF in the co-activation of MITF target genes that are involved in melanin synthesis raised the hypothesis that SWI/SNF enzymes are also MITF transcriptional partners in melanoma cells and thereby regulate melanoma proliferation and survival. However, because SWI/SNF components are frequently mutated or downregulated in many human cancers, it was not clear whether SWI/SNF function was compromised in melanoma.
In a survey of established melanoma cell lines, we determined that BRG1 or BRM expression was deficient in a subset (apporoximately 10%) of cells compared to normal melanocytes and HeLa cells but that other SWI/SNF components were highly expressed.Citation9 Importantly, we found that the expression of one SWI/SNF ATPase was retained in all melanoma cell lines. An independent survey of additional melanoma cell lines also found that most melanoma cell lines expressed high levels of all SWI/SNF components.Citation10 This suggested that downregulation but not absolute loss of SWI/SNF function may occur in a subset of melanomas. Interestingly, in melanoma tumors, downregulation of BRG1 expression was most striking in primary melanomas compared to metastatic melanomas, suggesting the possibility of stage specific modulation of BRG1 expression.Citation8
Restoration of BRG1 in other BRG1 deficient cancer cells was previously shown to induce cell cycle arrest through an RB dependent mechanism.Citation31,Citation68 In contrast to the severe proliferative effects that occur upon restoration of BRG1 in other deficient cancer cells, we found that restoration of BRG1 in BRG1 deficient melanoma cells had a mild effect on proliferation.Citation9 The most striking phenotypic change that occurred upon expression of BRG1 in melanoma cells was an increase in pigmentation. Investigation into the molecular mechanisms by which BRG1 promoted pigmentation revealed that BRG1 interacted with endogenous MITF and promoted increased expression of a subset of MITF target genes.Citation9
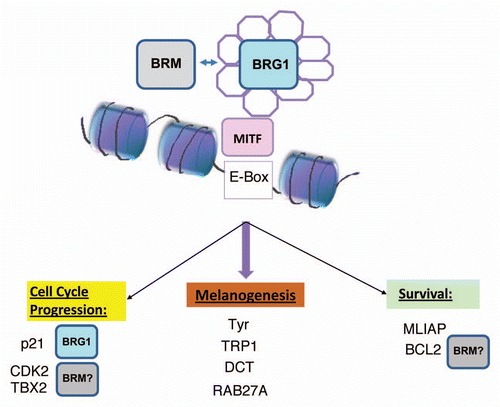
Interestingly, the genes affected by BRG1 were those that regulate melanin synthesis and melanosome function (tyrosinase, TRP1, DCT and RAB27) and cell cycle inhibition (p21CIP1) as well as the inhibitor of apoptosis, MLIAP.Citation9 However, the expression of pro-proliferative MITF target genes, including TBX2 and CDK2, was not affected by BRG1. BRG1 was recruited to the promoters of MITF target genes in a manner that was partially dependent on MITF and promoted altered histone methylation and increased association of phosphorylated RNA polymerase II on MITF target promoters, indicating activated transcription. Interestingly, BRG1 mediated activation of MITF target genes was correlated with increased chemoresistance.Citation9
The alternative SWI/SNF ATPase, BRM also interacted with MITF, however ectopic expression of BRM did not significantly alter MITF target gene expression, suggesting that BRM does not compensate for BRG1 deficiency in these cells.Citation9 Downregulation of BRG1/BRM in BRG1 deficient melanoma cells resulted in decreased expression of MITF target genes except p21CIP1 and compromised melanoma tumorigenicity. Thus, at least two different SWI/SNF complexes that contain BRG1 and BRM may differentially regulate several MITF target genes important for tumorigenicity (). However, BRM could not compensate for BRG1 in promoting maximal activation of melanocyte specific genes and p21CIP1 expression. The inability of BRM to fully compensate for BRG1 loss in the co-activation of MITF target genes may have important implications for MITF-mediated transcription.
The differential regulation of a subset of MITF target genes by BRG1 and BRM was puzzling since both BRG1 and BRM were found to interact with MITF.Citation9 Of particular interest was that BRM did not compensate for BRG1 deficiency in enhancing expression of the melanogenic proteins, tyrosinase, tyrosinase related protein 1 (TRP1), and dopachrome tautomerase (DCT), nor could it induce pigmentation. Interestingly, the requirement for MITF in the recruitment of either BRG1 or BRM to the promoters of the melanogenic genes was partial, raising the possibility that additional factors may promote SWI/SNF recruitment to MITF target promoters. Indeed, although MITF can activate expression of melanocyte specific genes when introduced in non-melanocytic cells, the levels of expression are well below levels in melanocytes, suggesting that MITF is not sufficient for maximal activation of differentiation related genesCitation55 (unpublished data).
Although BRG1 and BRM are highly similar, there are divergent regions between the two ATPases that mediate interactions with a distinct set of transcriptional regulators ().Citation70 Thus, it is conceivable that other factors differentially interact with BRG1 and BRM and enhance MITF mediated activation of these genes by either cooperatively or independently recruiting distinct SWI/SNF complexes with either BRG1 or BRM.
Factors implicated in the transcriptional regulation of the genes encoding the melanogenic enzymes include β-catenin, SOX9, SOX10, the paired box PAX3, SP1 and USF1.Citation29,Citation30,Citation71–Citation74 The amino-terminal region of BRG1 encompasses sequences that are not highly conserved in BRM, and that were previously determined to interact with β-catenin.Citation30 Furthermore, BRG1 selectively interacts with zinc finger transcription factors such as SP1.Citation70 Although there have not been any reports of SWI/SNF interactions with SOX10 or PAX3, BRG1 has been demonstrated to interact with other SRY-Box (SOX) proteins and paired box transcription factors (PAX).Citation75,Citation76 Many of these transcriptional regulators also activate expression of MITF and may interact with BRG1 to promote MITF expression.Citation10 Thus, BRG1 may mediate interactions with a set of distinct regulators that modulate the output resulting from MITF transcriptional activity.
An additional mode by which the two SWI/SNF ATPases may differentially regulate gene expression is through associations with a different assortment of BAFs. PBAF complexes are BRG1 containing complexes that are distinguished from BAF complexes by the presence of BAF180 and BAF200 and the absence of BAF250 Citation77 (). Interestingly, we found that p21CIP1 expression was activated by exogenous expression of BRG1 but not by exogenous expression of BRM.Citation9 Furthermore, only BRG1 was detected at the p21CIP1 promoter. BAF180 has been shown to activate p21CIP1 expression when expressed in BAF180 deficient breast cancer cell lines, presumably by interacting with one or more of the transcription factors that regulate p21CIP1 expression.Citation78 If the function of PBAF complexes is selectively restored by BRG1 in BRG1 deficient melanoma cells, it is plausible that BAF180 would provide an additional interface for interactions with transcriptional regulators that may selectively recruit or increase the stability of SWI/SNF complexes containing BRG1 at the p21CIP1 promoter. Interestingly, BRG1 is preferentially recruited by the tumor suppressor, p53 to the p21CIP1 promoter and to a subset of other p53 dependent promoters.Citation32
SWI/SNF complexes have a longstanding relationship with p53, being co-activators required for p53 mediated tumor suppression.Citation79,Citation80 However recent studies suggest that SWI/SNF complexes can also curtail p53 activity.Citation81,Citation82 SWI/SNF inactivation causes prolonged p53 activation following DNA damage and it was recently demonstrated that in the absence of DNA damage, BRG1 exists in a complex with CBP that functions to destabilize p53.Citation81 Interestingly, we found that downregulation of BRG1/BRM in BRG1 deficient melanoma cell lines resulted in a small increase in p21CIP1 expression that was not significant.Citation9 However, downregulation of BRG1/BRM in a p53 wild type melanoma cell line (B16) that expresses high levels of both BRG1 and BRM, resulted in a significant increase in p21CIP1 expression and expression of other p53 target genesCitation9 (unpublished observations). Thus, a critical level of BRG1 and/or SWI/SNF complex activity may be required to curtail p53 activity. This aspect of SWI/SNF function may be essential for the proliferation and survival of a cancer type, such as melanoma, that is not frequently characterized by p53 mutations.
We found that downregulation of BRG1/BRM in BRG1 deficient melanoma cells significantly inhibited the expression of most MITF target genes, including the pro-proliferative and survival genes, CDK2, TBX2 and BCL2, compromising tumorigenicity.Citation9 However, it is not clear whether BRM preferentially regulates pro-proliferative MITF target genes. In normal tissues, BRM is primarily expressed in differentiated cells whereas BRG1 is primarily expressed in proliferating cells.Citation83 Multiple studies show that overexpression of BRM has a strong anti-proliferative effect.Citation84,Citation85 However, the ability of BRM to inhibit proliferation may be modulated by oncogenes that perturb signaling pathways as well as by acetylation of carboxy terminal sitesCitation86 (). Thus, depending on the context, low levels of BRM may be required to maintain a critical level of SWI/SNF activity in order to prevent cessation of proliferation and its anti-proliferative effect suppressed by post-translational modifications. Other SWI/SNF components have been demonstrated to be post-translationally modified in response to aberrant signaling pathways in cancer.Citation87 Post-translational modifications in response to aberrant signaling pathways may alter important aspects of SWI/SNF function in melanoma.
Conclusions
The melanoma transcriptional circuitry is defined by multiple components which revolve around MITF, the master regulator of melanocyte differentiation and a designated lineage survival oncogene in melanoma. MITF is essential for the commitment, survival and specialization of the melanocyte lineage. In melanoma, MITF functions as a rheostat by integrating the appropriate signals to elicit a variable biological output in terms of tumor proliferation and metastasis.Citation67 Other components of the circuit ensure efficient operation and functionality.
We and others have determined that SWI/SNF enzymes regulate MITF activity as well as other components of the transcriptional circuitry in melanoma.Citation7–Citation10 Based on these studies, we propose that SWI/SNF chromatin remodeling enzymes be designated as on/off switches in this circuitry. The switch is one of the most basic yet important electrical devices. Although many switches appear to be the same, there are several types of switches that can function very differently and that have varying capacities (e.g., single pole, double pole, 3-way, 4-way). Likewise, heterogeneous SWI/SNF complexes can switch the transcriptional circuitry on and off by different mechanisms, providing distinct levels and modes of regulation. Just as each of the different types of switches must be correctly wired into the circuitry to function properly, each of the different SWI/SNF complexes must be correctly connected to the other transcriptional components. Perturbations in SWI/SNF function either through mis-expression of individual SWI/SNF components, through alterations in transcription factor interactions, or through aberrant activation of signaling pathways would be expected to distort the functioning of the normal circuitry. A better understanding of how SWI/SNF complexes are connected to the melanoma transcriptional circuitry may provide the necessary information to effectively disconnect the necessary components.
Abbreviations
| MITF | = | microphthalmia-associated transcription factor |
| BAF | = | BRG1/BRM associated factor |
| PBAF | = | polybromo associated factor |
| IGFBP7 | = | insulin-like growth factor binding protein 7 |
| MLIAP | = | melanoma inhibitor of apoptosis |
| CBP | = | CREB-binding protein |
Figures and Tables
Figure 1 (A) List of Mammalian SWI/SNF components including gene names and other commonly used designations. (B) Diverse SWI/SNF complexes containing either BRG1 or BRM and associated factors exist in mammalian cells. BRG1 and BRM are mutually exclusive. (C) Schematic comparison of BRG1 and BRM structure. The two ATPases are 70% identical, containing conserved as well as unique sequences. Stars represent lysine residues in BRM that can be post-translationally modified by acetylation.
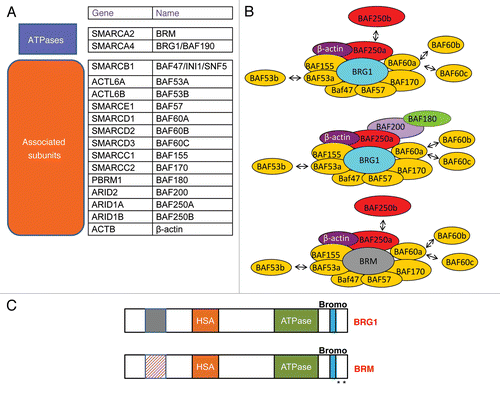
Figure 2 Potential contribution of distinct SWI/SNF complexes containing either BRG1 or BRM in the regulation of different classes of MITF target genes.Citation9
Acknowledgements
I.L.D. was supported by the National Institute of Environmental Health Sciences; Grant number: 5K22ES12981, Ohio Cancer Research Associates, American Cancer Society, Ohio Division.
References
- Jhappan C, Noonan FP, Merlino G. Ultraviolet radiation and cutaneous malignant melanoma. Oncogene 2003; 22:3099 - 3112
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature 2002; 417:949 - 954
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet 2009; 41:544 - 552
- Goldstein AM, Chan M, Harland M, Gillanders EM, Hayward NK, Avril MF, et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res 2006; 66:9818 - 9828
- Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer 2006; 6:593 - 602
- Rothhammer T, Bosserhoff AK. Epigenetic events in malignant melanoma. Pigment Cell Res 2007; 20:92 - 111
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF Induces Senescence and Apoptosis through Pathways Mediated by the Secreted Protein IGFBP7. Cell 2008; 132:363 - 374
- Becker TM, Haferkamp S, Dijkstra MK, Scurr LL, Frausto M, Diefenbach E, et al. The chromatin remodelling factor BRG1 is a novel binding partner of the tumor suppressor p16INK4a. Mol Cancer 2009; 8:4
- Keenen B, Qi H, Saladi SV, Yeung M, de la Serna IL. Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia-associated transcription factor target genes in melanoma. Oncogene 29:81 - 92
- Vachtenheim J, Ondrusova L, Borovansky J. SWI/SNF chromatin remodeling complex is critical for the expression of microphthalmia-associated transcription factor in melanoma cells. Biochem Biophys Res Commun 392:454 - 459
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev 1999; 13:2339 - 2352
- Varga-Weisz P. ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene 2001; 20:3076 - 3085
- Urnov FD, Wolffe AP. Chromatin remodeling and transcriptional activation: the cast (in order of appearance). Oncogene 2001; 20:2991 - 3006
- Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell 1999; 3:247 - 253
- Belandia B, Orford RL, Hurst HC, Parker MG. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J 2002; 21:4094 - 4103
- Hsiao PW, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol 2003; 23:6210 - 6220
- Ito T, Yamauchi M, Nishina M, Yamamichi N, Mizutani T, Ui M, et al. Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J Biol Chem 2001; 276:2852 - 2857
- Link KA, Burd CJ, Williams E, Marshall T, Rosson G, Henry E, et al. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol Cell Biol 2005; 25:2200 - 2215
- Park J, Wood MA, Cole MD. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol Cell Biol 2002; 22:1307 - 1316
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet 2006; 7:461 - 473
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell 2000; 6:1287 - 1295
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J 1998; 17:6979 - 6991
- Flowers S, Nagl NG Jr, Beck GR Jr, Moran E. Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. J Biol Chem 2009;
- Nagl NG Jr, Wang X, Patsialou A, Van Scoy M,, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell cycle control. EMBO J 2007; 26:752 - 763
- Ryme J, Asp P, Bohm S, Cavellan E, Farrants AK. Variations in the composition of mammalian SWI/SNF chromatin remodelling complexes. J Cell Biochem 2009; 108:565 - 576
- Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell 2009; 136:200 - 206
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell 2007; 128:707 - 719
- Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet 1999; 22:102 - 105
- Galibert MD, Carreira S, Goding CR. The Usf-1 transcription factor is a novel target for the stress-responsive p38 kinase and mediates UV-induced Tyrosinase expression. EMBO J 2001; 20:5022 - 5031
- Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J 2001; 20:4935 - 4943
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, et al. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 1994; 79:119 - 130
- Xu Y, Zhang J, Chen X. The activity of p53 is differentially regulated by Brm- and Brg1-containing SWI/SNF chromatin remodeling complexes. J Biol Chem 2007; 282:37429 - 37435
- Liu R, Liu H, Chen X, Kirby M, Brown PO, Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 2001; 106:309 - 318
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, et al. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol 2005; 25:3997 - 4009
- Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene 2009; 28:1653 - 1668
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 1998; 394:203 - 206
- Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, Webster W, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol 2001; 21:3598 - 3603
- Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci USA 2000; 97:13796 - 13800
- Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep 2000; 1:500 - 506
- Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, et al. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene 2008; 27:460 - 468
- Sun A, Tawfik O, Gayed B, Thrasher JB, Hoestje S, Li C, et al. Aberrant expression of SWI/SNF catalytic subunits BRG1/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate 2007; 67:203 - 213
- Sentani K, Oue N, Kondo H, Kuraoka K, Motoshita J, Ito R, et al. Increased expression but not genetic alteration of BRG1, a component of the SWI/SNF complex, is associated with the advanced stage of human gastric carcinomas. Pathobiology 2001; 69:315 - 320
- Wang X, Sansam CG, Thom CS, Metzger D, Evans JA, Nguyen PT, et al. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res 2009; 69:8094 - 8101
- Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Res 2003; 63:5198 - 5202
- Sharma A, Trivedi NR, Zimmerman MA, Tuveson DA, Smith CD, Robertson GP. Mutant V599EB-Raf regulates growth and vascular development of malignant melanoma tumors. Cancer Res 2005; 65:2412 - 2421
- Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet 2003; 33:19 - 20
- Doan DN, Veal TM, Yan Z, Wang W, Jones SN, Imbalzano AN. Loss of the INI1 tumor suppressor does not impair the expression of multiple BRG1-dependent genes or the assembly of SWI/SNF enzymes. Oncogene 2004; 23:3462 - 3473
- Lin H, Wong RP, Martinka M, Li G. Loss of SNF5 expression correlates with poor patient survival in melanoma. Clin Cancer Res 2009; 15:6404 - 6411
- Bartkova J, Lukas J, Guldberg P, Alsner J, Kirkin AF, Zeuthen J, et al. The p16-cyclin D/Cdk4-pRb pathway as a functional unit frequently altered in melanoma pathogenesis. Cancer Res 1996; 56:5475 - 5483
- Walker GJ, Flores JF, Glendening JM, Lin AH, Markl ID, Fountain JW. Virtually 100% of melanoma cell lines harbor alterations at the DNA level within CDKN2A, CDKN2B, or one of their downstream targets. Genes Chromosomes Cancer 1998; 22:157 - 163
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 2005; 436:720 - 724
- Chai J, Charboneau AL, Betz BL, Weissman BE. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res 2005; 65:10192 - 10198
- Betz BL, Strobeck MW, Reisman DN, Knudsen ES, Weissman BE. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene 2002; 21:5193 - 5203
- Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol 2008; 28:3457 - 3464
- de la Serna IL, Ohkawa Y, Higashi C, Dutta C, Osias J, Kommajosyula N, et al. The microphthalmia-associated transcription factor (MITF) requires SWI/SNF enzymes to activate melanocyte specific genes. J Biol Chem 2006;
- Loercher AE, Tank EM, Delston RB, Harbour JW. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J Cell Biol 2005; 168:35 - 40
- Delmas V, Beermann F, Martinozzi S, Carreira S, Ackermann J, Kumasaka M, et al. Beta-catenin induces immortalization of melanocytes by suppressing p16INK4a expression and cooperates with N-Ras in melanoma development. Genes Dev 2007; 21:2923 - 2935
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet 2004; 38:365 - 411
- Tachibana M, Takeda K, Nobukuni Y, Urabe K, Long JE, Meyers KA, et al. Ectopic expression of MITF, a gene for Waardenburg syndrome type 2, converts fibroblasts to cells with melanocyte characteristics. Nat Genet 1996; 14:50 - 54
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005; 436:117 - 122
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, et al. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 2004; 6:565 - 576
- Carreira S, Dexter TJ, Yavuzer U, Easty DJ, Goding CR. Brachyury-related transcription factor Tbx2 and repression of the melanocyte-specific TRP-1 promoter. Mol Cell Biol 1998; 18:5099 - 5108
- Vance KW, Carreira S, Brosch G, Goding CR. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res 2005; 65:2260 - 2268
- McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 2002; 109:707 - 718
- Dynek JN, Chan SM, Liu J, Zha J, Fairbrother WJ, Vucic D. Microphthalmia-associated transcription factor is a critical transcriptional regulator of melanoma inhibitor of apoptosis in melanomas. Cancer Res 2008; 68:3124 - 3132
- Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, et al. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature 2005; 433:764 - 769
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev 2006; 20:3426 - 3439
- Wong AK, Shanahan F, Chen Y, Lian L, Ha P, Hendricks K, et al. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res 2000; 60:6171 - 6177
- Keenen B, de la Serna IL. Chromatin remodeling in embryonic stem cells: regulating the balance between pluripotency and differentiation. J Cell Physiol 2009; 219:1 - 7
- Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell 2003; 11:377 - 389
- Passeron T, Valencia JC, Bertolotto C, Hoashi T, Le Pape E, Takahashi K, et al. SOX9 is a key player in ultraviolet B-induced melanocyte differentiation and pigmentation. Proc Natl Acad Sci USA 2007; 104:13984 - 13989
- Jiao Z, Mollaaghababa R, Pavan WJ, Antonellis A, Green ED, Hornyak TJ. Direct interaction of Sox10 with the promoter of murine Dopachrome Tautomerase (Dct) and synergistic activation of Dct expression with Mitf. Pigment Cell Res 2004; 17:352 - 362
- Lang D, Lu MM, Huang L, Engleka KA, Zhang M, Chu EY, et al. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 2005; 433:884 - 887
- Bentley NJ, Eisen T, Goding CR. Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol Cell Biol 1994; 14:7996 - 8006
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA 2009; 106:5187 - 5191
- Yang Y, Stopka T, Golestaneh N, Wang Y, Wu K, Li A, et al. Regulation of alphaA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J 2006; 25:2107 - 2118
- Xue Y, Canman JC, Lee CS, Nie Z, Yang D, Moreno GT, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci USA 2000; 97:13015 - 13020
- Xia W, Nagase S, Montia AG, Kalachikov SM, Keniry M, Su T, et al. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res 2008; 68:1667 - 1674
- Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, et al. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 2000; 102:257 - 265
- Isakoff MS, Sansam CG, Tamayo P, Subramanian A, Evans JA, Fillmore CM, et al. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc Natl Acad Sci USA 2005; 102:17745 - 17750
- Park JH, Park EJ, Hur SK, Kim S, Kwon J. Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair (Amst) 2009; 8:29 - 39
- Naidu SR, Love IM, Imbalzano AN, Grossman SR, Androphy EJ. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene 2009;
- Reisman DN, Sciarrotta J, Bouldin TW, Weissman BE, Funkhouser WK. The expression of the SWI/SNF ATPase subunits BRG1 and BRM in normal human tissues. Appl Immunohistochem Mol Morphol 2005; 13:66 - 74
- Muchardt C, Bourachot B, Reyes JC, Yaniv M. ras transformation is associated with decreased expression of the brm/SNF2alpha ATPase from the mammalian SWI-SNF complex. EMBO J 1998; 17:223 - 231
- Glaros S, Cirrincione GM, Muchardt C, Kleer CG, Michael CW, Reisman D. The reversible epigenetic silencing of BRM: implications for clinical targeted therapy. Oncogene 2007; 26:7058 - 7066
- Bourachot B, Yaniv M, Muchardt C. Growth inhibition by the mammalian SWI-SNF subunit Brm is regulated by acetylation. EMBO J 2003; 22:6505 - 6515
- Foster KS, McCrary WJ, Ross JS, Wright CF. Members of the hSWI/SNF chromatin remodeling complex associate with and are phosphorylated by protein kinase B/Akt. Oncogene 2006; 25:4605 - 4612