Abstract
Environmental exposures in-utero may alter the epigenome, thus impacting chromosomal stability and gene expression. We hypothesized that in utero exposures to maternal smoking and perfluoroalkyl compounds (PFCs) are associated with global DNA hypomethylation in umbilical cord serum. Our objective was to determine if global DNA methylation could be used as a biomarker of in utero exposures to maternal smoking and PFCs. Using an ELISA-based method, global DNA methylation was quantified in umbilical cord serum from 30 newborns with high (>10 ng/ml, mean 123.8 ng/ml), low (range 1-10 ng/ml, mean 1.6 ng/ml) and very low (<1 ng/ml, mean 0.06 ng/ml) cord serum cotinine levels. Y chromosome analysis was performed to rule out maternal DNA cross-contamination. Cord serum global DNA methylation showed an inverse dose response to serum cotinine levels (p<0.001). Global DNA methylation levels in cord blood were the lowest among newborns with smoking mothers (mean=15.04%; 95% CI, 8.4, 21.7) when compared to babies of mothers who were second-hand smokers (21.1%; 95% CI, 16.6, 25.5) and non-smokers (mean=29.2%; 95% CI, 20.1, 38.1). Global DNA methylation was inversely correlated with serum PFOA (r= -0.72, p <0.01) but not PFOS levels. Serum Y chromosome analyses did not detect maternal DNA cross-contamination. This study supports the use of global DNA methylation status as a biomarker of in utero exposure to cigarette smoke and PFCs.
Introduction
Epigenomics provides a window through which we can understand the impact of the environment, nutrition and lifestyle choices on chronic disease susceptibility and risk.Citation1 Life-long effects of in utero exposures may be mediated through alterations in the best-understood epigenomic mark, DNA methylation. Global DNA methylation, one of the earliest molecular changes observed in the transition from a normal to a diseased cell, may be a good epigenomic bio-dosimeter of prenatal exposures to environmental toxicants that lead to impaired health throughout the life-course.Citation2 Global DNA hypomethylation largely affects intergenic and intronic regions, particularly repeat sequences and transposable elements and is believed to result in chromosomal instability and increased mutation events.
Environmental exposures have been shown to be associated with epigenetic changes. Arsenic, one of the heavy metals found in cigarette smoke, has been shown to be associated to global DNA hypomethylation in vitroCitation3 and in animal modelsCitation4 and to be associated with in utero DNA hypomethylation.Citation5 Cadmium, a metal associated with smoking and second hand smoke exposure has been associated with DNA hypomethylation along with induction of cell proliferation.Citation6 Cadmium accumulates in the placenta, reduces birthweight and may also play a role in uterine growth restriction.Citation7 Polycyclic aromatic hydrocarbons (PAH), also components of cigarette smoke, have been associated with altered global DNA methylation in peripheral blood leucocytes of male nonsmoking coke-oven workers.Citation8
A handful of studies have examined the association between tobacco smoke exposure and global DNA methylation. Active smoking is known to be associated with global DNA hypomethylation in the aero-digestive mucosa.Citation9 In addition, two studies have reported inconsistent results documenting associations between prenatal tobacco smoke exposure and global DNA methylation. Terry et al.Citation10 reported that prenatal exposure resulted in higher levels of DNA methylation in mononuclear cells from whole blood of adult women. Breton et al.Citation11 reported decreased global DNA methylation in buccal cells from kindergarten and first grade children who were exposed to maternal smoking in utero. However, these studies evaluated DNA methylation in different tissues and both studies are limited by having used surrogate measurements of global DNA methylation, having determined in utero smoking exposure retrospectively by questionnaires and not having controlled for exposure to second hand smoke over time.Citation12 At this time there are no published studies that examine the association between global DNA methylation at birth and in utero exposures to smoking or to persistent environmental contaminants.
To determine if global DNA methylation in newborns is associated with exposure to prenatal maternal smoking we measured global DNA methylation in cord blood serum of full term babies with adequate birthweight, and high, low and very low cotinine levels in cord serum. As a secondary analysis we also examined the association between levels of DNA methylation and persistent perfluoroalkyl compounds (PFCs) in cord serum: perfluorooctane sulfonate (PFOS) and perfluorooctanoate (PFOA).
Results
Among the 30 babies included in this study, 15 were male and 15 were female. As shown in , those included in this study were similar to all eligible babies except that there was by design oversampling of babies whose mothers were active and passive smokers, a tendency for the mothers in this sub study to be somewhat less educated and a trend toward these mothers having been less likely to have received prenatal care in the first four months of pregnancy. As expected, nonsmokers in the sample had somewhat lower levels of serum cotinine than in all eligible babies. Maternal age ranged from 16–37 years and paternal age ranged from 15–49 years. Seven mothers were <18 years of age and seven were 30 or older. One mother was Asian, two were Caucasian and the other 27 were African American. Of the 11 smokers that were selected, two had reported that they did not smoke but were classified as smokers on the basis of having very high serum cotinine levels (117 and 118 ng/ml). Means, medians and ranges of DNA methylation index results (%5 mC), PFOS and PFOA levels, gestational age and growth parameters also are presented in . The subjects selected for global methylation analysis were similar in all ways to the rest of the eligible infants with the exception of gestational age, which was on average 3.5 days longer.
shows the relationships between DNA methylation index results and selected maternal and infant characteristics. Maternal age did not show a linear relationship to DNA methylation. However, both younger (<20 y) and older (30+ y) mothers had babies with reduced global DNA methylation (NS). Likewise there was a non-significant trend toward increased DNA methylation with increased gestational age within this group of term births (p = 0.19). There was no evidence that DNA methylation was associated with pre-pregnancy BMI or sex of the baby. Measures of infant growth—birthweight, length, head circumference and ponderal index—also were not associated with global DNA methylation.
also shows results of multivariate linear regression models for global DNA methylation in relation to exposures to cotinine levels, smoking status and levels of PFOS and PFOA. Cotinine, the natural log of cotinine, active smoking and passive smoking were significantly associated with global DNA hypomethylation. The associations for PFOA and the natural log of PFOA were of borderline statistical significance. PFOS was not associated with changes in global DNA methylation. Multiple regression models were done first including maternal age (in categories) and then gestational age as covariates. Inclusion of either maternal age or gestational age somewhat attenuated the effect estimates across the board. However, in both sets of multivariate models the associations between cotinine and smoking status and DNA methylation were statistically significant.
presents the DNA methylation data by smoking status. The global methylation index was significantly (p < 0.01) lower for 11 newborns exposed to maternal smoking (mean = 15.04%; 95% CI, 8.4, 21.7) than among eight newborns with nonsmoking mothers (mean = 29.2%; 95% CI, 20.1, 38.1). The serum DNA global methylation index also was significantly (p = 0.01) lower for 11 newborns whose mothers were exposed to secondhand smoke during pregnancy (mean = 21.1%; 95% CI, 16.6, 25.5) when compared to newborns not exposed to maternal smoking.
presents the results of linear regression modeling of DNA methylation index results across the three groups, in relation to log-transformed cotinine, PFOA and PFOS levels. There was a statistically significant inverse relationship between the natural log of cotinine measured in cord blood serum and the global DNA methylation index () and an inverse relationship for PFOA (p = 0.06, ) but not PFOS (). Serum PFOS and PFOA levels were not related to maternal smoking status or to serum cotinine levels (data not shown).
To ensure that we were measuring global DNA methylation changes in fetal DNA and not in maternal DNA, we quantified the Y chromosome content in umbilical cord serum DNA for a subset of eight male infants. One sample was not quantifiable leaving seven for statistical analysis. The ratios of Y Human DNA to total Human DNA (mean = 6.7) would reflect the amount of male infants' DNA relative to the amount of maternal DNA that might be present in cord blood of the same patient. Any ratio above one is indicative that the sample is from a male infant without the presence of female DNA. Results range from 1.6–15.5, so that there is no measurable level of maternal DNA in cord blood serum, implying that the observed global methylation changes are of fetal and not maternal origin ().
Discussion
Our results suggest that circulating fetal DNA already can exhibit aberrant methylation patterns in response to environmental exposures. To our knowledge this is the first study to report global DNA hypomethylation in umbilical cord serum significantly associated, in a dose-response fashion, to in utero exposure to tobacco smoke. These results are consistent with Breton et al.Citation11 who reported association between tobacco smoke and DNA global hypomethylation, albeit in a different life stage and a different systemic compartment. Additionally, our secondary analyses raise the possibility that DNA global hypomethylation in cord serum is associated with pre-natal exposures to perfluoroalkyl compounds, especially PFOA.
Cord blood irrigates all the cells of the fetus collecting cell free DNA that can be isolated from cord serum. Therefore, DNA extracted from cord serum provides us a genomic footprint of fetal cellular turnover. However, since there have been sporadic reports of maternal DNA in cord bloodCitation22 we tested the precedence of the DNA we extracted from cord blood. Serum Y chromosome analyses did not detect maternal DNA cross contamination in cord blood. Therefore, we are measuring global methylation levels in fetal DNA isolated from cord serum.
Global DNA methylation levels have been found to be inversely associated with blood plasma levels for several persistent organic pollutants (POP) in Greenlandic InuitCitation23 and in tissues from animals exposed to ionizing radiation.Citation24 Low-dose exposure to POPs has recently been associated with global DNA hypomethylation in a healthy adult population.Citation25 POPs such as organochlorine (OC) pesticides, which are xenobiotics that accumulate in adipose tissue, may be chronically exerting their deleterious effect on the epigenomic machinery. These recent findings have profound implications for future generations also, since chemical-induced epigenetic changes are heritable through generations.Citation26 In addition, in utero exposures to toxic compounds may lead to deleterious effects on the epigenomic molecular machinery and subsequent adverse reproductive outcomes.
It has been documented that in utero exposure to dioxins, PCBs, OC pesticides or bisphenol-A may cause adverse pregnancy outcomes such as low birth weight, preterm delivery, or intrauterine growth retardation.Citation27,Citation28 For example, there is already evidence that in utero exposure to bisphenol-A leads to DNA hypomethylation of Hoxa10, with a subsequent alteration of the developmental programming of uterine estrogen response in an animal model.Citation29 Global DNA hypomethylation is commonly found in most types of cancer. Higher degrees of genome-wide hypomethylation have been found to be strongly associated with late grades and with larger tumor sizes, suggesting that genome-wide de-methylation occurs simultaneously with tumor progression.Citation30 Emerging evidence indicates that such alterations also occur in other chronic diseases and may be associated to environmental exposures in utero.Citation31
Dietary-induced epigenetic changes are also heritable through generations.Citation32 Methyl groups from S-adenosyl-methionine (SAM) are needed for DNA methylation. Diets low in sources of methyl groups can lead to global DNA hypomethylation by impairing synthesis of SAM.Citation33 Folic acid depleted dietary intake has been shown to lead to global DNA hypomethylation in animal studies. The loss of global DNA methylation can be reversed if folate is added to the diet on or before the ninth week. If folate is introduced afterwards, DNA methylation loss and progression to cancer can be stopped, but not reversed.Citation34 Interestingly, preconceptional exposure to famine during the Dutch Hunger Winter of 1944–5 was recently shown to be associated with hypomethylation of the insulin-like growth factor 2 (IGF2) and (INSIGF) gene six decades later.Citation35,Citation36
Epigenomic alterations associated to in utero exposures may thus have life-course effects. The epigenome can be visualized as an interface between the dynamic environment and the inherited static genome, configured during development to shape the diversity of gene expression programs for different cell types by a highly organized process.Citation37 Cell differentiation during embryonic-fetal development involves different epigenetic processes which, if altered, may affect either somatic or germ cells.Citation38 Epigenetic alterations can occur in somatic cells at different stages of life, from fecundation to adulthood, and when germ cells are affected, such changes can even be passed on to future generations.Citation39 Physical, biological and chemical factors, as well as social factors, such as maternal care, modify the epigenome. Thus, exposures to environmental agents such as tobacco smoke in utero may influence susceptibility to disease throughout the life-course, via epigenetic alterations, which can be used as biomarkers of exposure and risk. Epigenomic biomarkers may lead to advancements in the fields of developmental diseases biology and molecular epidemiology, where the search for molecular biomarkers of exposure-related effects that can be used for screening, regulatory and risk prediction purposes has proven elusive. The main challenge has been to identify molecular biomarkers that are part of the downstream effects triggered after the fetus is exposed to the toxic environmental contaminant.
Global DNA methylation status is, probably, not a specific biomarker of in utero exposure to cigarette smoke and PFCs only, but to many other environmental exposures, such as air and water pollution, and dietary intake as well. In addition global DNA hypomethylation is known to be associated with a series of chronic diseases, including diabetes, bipolar disorders, obesity, schizophrenia and cancer.Citation40,Citation41 Thus establishing a link between exposure and disease is not as important as detecting the loss of methylation to ascertain risk and determine therapeutic and behavior modification alternatives. In many cases, epigenetic modifications are reversible, thus providing an opportunity to reverse the chronic disease process and understand the impact of lifestyle choices on chronic disease susceptibility and risk.Citation42 In the case of PFOS and PFOA, no studies on DNA methylation have been reported; however, exposure to peroxisome proliferators have been found to cause global hypomethylation in the rat liver.Citation43
This study has several limitations. The sample size limited the possibility of performing multivariable regression analyses with several predictors simultaneously, which may have elucidated associations of global DNA hypomethylation levels in cord serum with maternal and paternal variables. The detection limit for the Imprint® Methylated DNA Quantification Kit is 5 ng of methylated DNA and input DNA used may be as low as 10–200 ng. The sensitivity of the ELISA assay, limited by antibody affinity and substrate availability may have led to a slight under quantification of global DNA methylation levels. However this sensitivity issue should not affect the relative difference global DNA methylation across cotinine or perfluorinated alkyl levels in serum. Finally, lack of information on other environmental factors known to affect DNA methylation status, such as folate, arsenic and cadmium levels may be confounding the associations reported in this study. Both environmental and dietary factors have been shown to modulate global DNA methylation levels, yet their combined effect has not been elucidated.Citation44 These questions should be pursued in follow-up studies.
Altogether the findings of this proof-of-principle study suggest that differential global DNA methylation levels may be used as a bio-dosimeter of fetal exposure and of exposure associated biological effects on the fetus in cord serum. Furthermore, if the association between global DNA hypomethylation and cotinine levels in cord serum is validated in a larger cohort, smoking cessation efforts among pregnant women should be strengthened. Finally, global biomarkers of epigenomic alterations may be useful biomarkers for chronic diseases related to persistent environmental exposures. The results of this study suggest that a detailed examination of these possibilities is warranted.
Materials and Methods
Subject selection.
Thirty subjects were sampled for this proofof- principle study from 113 subjects who met the criteria of having been born term (≥37 weeks gestation), of normal weight (≥2,500 grams) and having cotinine levels and adequate amounts of serum. The sample stratified on cord serum cotinine levels: (1) High cotinine-Serum cotinine levels indicative of recent maternal active smoking (>10 ng/ml) regardless of smoking history; (2) Low cotinine-Serum cotinine levels considered to be clearly within a passive smoking range (1–5 ng/ml) among mothers reporting no smoking during pregnancy; (3) Very low cotinine-Serum cotinine levels consistent with nonsmokers (<1 ng/ml) among mothers reporting no smoking during pregnancy. Only term and normal weight newborns were selected to avoid potential interactions between epigenomic levels and factors leading to pre-term birth and or low-birthweight, which may confound the association between global DNA methylation and cotinine or PFC levels in serum. Sampling was not done at random but rather the nonsmokers were selected from among those that had both, no history of smoking and the lowest cotinine values to increase statistical power. Sampling was blinded to levels of polyfluorinated chemicals PFOS and PFOA.
Subjects were selected from the Baltimore THREE (Tracking Health Related to Environmental Exposures) Study, a cross-sectional study of newborn deliveries at the Johns Hopkins Hospital in Baltimore, MD, USA designed to determine cord blood levels of a number of environmental chemicals, along with their relationship to thyroid hormone status and birth outcomes. Data from this study have been reported in several peer-reviewed publications.Citation13–Citation18 The study received approval from the Johns Hopkins Medicine Institutional Review Board and was determined to be HIPAA exempt.
Umbilical cord blood samples were collected immediately after birth using the Witter cord cradle (Witter et al. 2001), and stored at 4°C for <3 h before further processing. Of these, 41 samples had insufficient cord blood volume for laboratory analyses, leaving a total of 300 samples available for chemical laboratory analyses. Once collected, umbilical cord and cord blood samples were placed in the Labor and Delivery Suite refrigerators. Within a few hours of birth, samples were collected and transported by study personnel to a laboratory at the Johns Hopkins Bloomberg School of Public Health for processing. Umbilical cord samples were flash frozen. Blood specimens were centrifuged at 1,000 g for 15 min and serum was aliquoted into 2 mL polypropylene cryovials. Tissue and blood samples were then stored at −80°C.
Frozen samples were transferred on dry ice to laboratories at the Centers for Disease Control and Prevention in Atlanta for cotinine analyses while other aliquots were archived. A total of 286 samples had sufficient volume for measurement of cotinine was conducted using positive-ion atmospheric pressure chemical ionization tandem mass spectrometry as previously described.Citation19 Likewise the CDC laboratory analyzed serum samples for PFOS and PFOA, using negative-ion TurboIonSpray ionization, a variant of electrospray ionization, tandem mass spectrometry.Citation20
DNA isolation from cord blood serum.
Of 286 samples analyzed by the CDC, 113 met the inclusion criteria (above), of which 30 one ml cord serum samples were digested with 50 ug/mL proteinase K (Boehringer Mannheim, Germany) in the presence of 1% sodium dodecyl sulfate (SDS) at 48°C for 3 days, followed by phenol/chloroform extraction and ethanol precipitation and finally dissolved in 30 µL of LoTE (2.5 mmol/L EDTA and 10 mmol/L Tris-HCL) as previously described.Citation21
Global DNA methylation assays.
The global DNA methylation levels in umbilical cord serum samples were obtained with an ELISA based commercial kit (MDQ1, Imprint® Methylated DNA Quantification Kit, Sigma Aldrich). The MDQ1 kit is a high-throughput, molecular biology kit, which uses a 96-well plate format to provide accurate differential global DNA methylation absorbance readings with as little as 50 ng of genomic DNA. Two microliters (µl) of DNA at a concentration of 100 ng/µl were diluted with 28 µl of lysis and binding buffers and incubated at 60µC. The samples were incubated with capture and detection antibodies and absorbance was read at 450 nanometers. Quantification of global DNA methylation was obtained from calculating the amount of methylated cytosines in the sample (5 mC) relative to global cytidine (5 mC + dC) in a positive control that had been previously methylated. All samples were analyzed in duplicate.
Y chromosome quantification.
The Quantifiler Y Human Male DNA Quantification Kit (Applied Biosystems) and Quantifiler Human DNA Quantification Kit (Applied Biosystems) were used to quantify the amount of total amplifiable human DNA and Y Human Male that was present in eight cord blood DNA samples obtained from male infants. The quantification assay combines three 5' nuclease assays, namely: target-specific human DNA assay and target specific Y human male DNA assay, plus an internal PCR control (IPC) for both chemistries. The total human target in this assay is a 62-base Human Telomerase reverse transcriptase gene (hTERT) located at 5p15.33, which is detected by a TaqMan MGB probe labeled with FAM dye. The male target, the 64-base sex-determining region Y (SRY) located at Yp11.3, is detected by a TaqMan® MGB probe labeled with FAM dye. The IPC, 130-base PCR control, a synthetic sequence not found in nature, is detected by a TaqMan® MGB probe labeled with VIC dye. The targets are measure by Forster-type energy transfer (FRET) technique. The ratio of human male DNA to human DNA in each sample is calculated by dividing the amount of Y human male DNA by the amount of human DNA present in each sample. This test is enriched to show a high male signal among a large amount of female DNA. Therefore, the ratio of human male DNA to human DNA that rules out the presence of female DNA can be equal to one or higher.
Statistical analysis.
Correlation analyses, Welch t-tests and Wilcoxon rank tests were used to evaluate the differences in the global DNA methylation index across levels of exposure to cigarette smoke and PFCs in cord serum DNA; and to evaluate the differences in the global DNA methylation index across levels of exposure to cigarette smoke in cord tissue DNA. Bivariate (unadjusted) linear regression models on DNA methylation index were carried out to evaluate relationships to cotinine, PFOA and PFOS and the natural logs of cotinine, PFOA and PFOS as well as smoking status. Other potential independent variables were explored including maternal age (linear and categorical), maternal prepregnancy body mass index (BMI) (linear), sex, race and gestational age. On the basis of this exploration, separate multivariate models were constructed. Models were constructed using Stata version 10.
Abbreviations
| BMI | = | body mass index |
| PFCs | = | perfluorinated alkyl compounds |
| PFOA | = | perfluorooctanoate |
| PFOS | = | perfluorooctane sulfonate |
| THREE | = | tracking health related to environmental exposures |
Figures and Tables
Figure 1 Global DNA methylation index in cord blood serum DNA of 11 newborns exposed in utero to tobacco smoke constituents from maternal smoking, 11 newborns whose mothers were passive smokers and eight newborns whose mothers were nonsmokers. (Box plots).
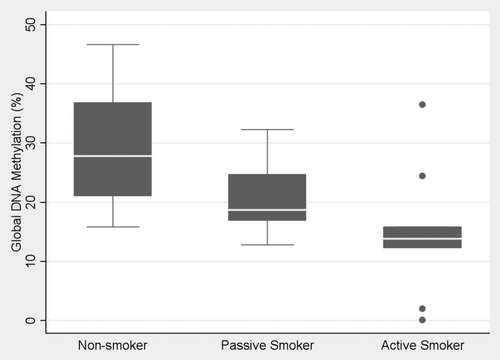
Figure 2 (A) Linear relationship between the natural log of cotinine in cord blood serum and the global DNA methylation index in 30 newborns with high (11), low (11) and very low (8) cotinine serum levels. (B) Linear relationship between the natural log of PFOA in cord blood serum and the global DNA methylation index in 30 newborns. (C) Linear relationship between the natural log of PFOS in cord blood serum and the global DNA methylation index in 30 newborns.
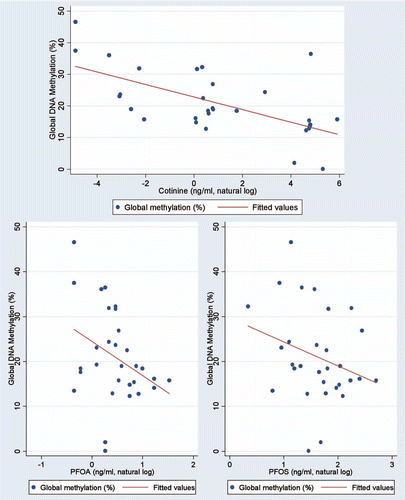
Table 1 Characteristics of infants selected for epigenetics study compared with all eligible infants, Baltimore THREE study
Table 2 Results of linear models (pearson's correlations and slope beta values) for cotinine, PFOA and PFOS concentrations (log-transformed) and other selected study variables on %5-mC (n = 30), Baltimore THREE study
Table 3 Quantification of serum Y chromosome human DNA (ng/ul) and serum total human DNA (ng/ul) obtained from umbilical cord blood of seven male infants, Baltimore THREE study
Acknowledgements
This research was supported in part by the NCI 5U01CA13-8. It also received support from the Maryland Cigarette Restitution Program Research Grant given to the Johns Hopkins Medical Institutions and by funding from the NIOSH Education and Research Center for Occupational Safety and Health at the Johns Hopkins Bloomberg School of Public Health (#T42CCT310419). R.U.H. was supported in part by grant 1R01ES015445 of the National Institute of Environmental Health Sciences (NIEHS). The authors wish to thank Jose Rodriguez-Orengo for assisting with genetic analyses, John Bernert and the CDC Tobacco Exposure Biomarkers laboratory for performing the cotinine analyses and Antonia Calafat and the CDC Organic Analytical Toxicology laboratory for performing analyses of PFCs, as well as reviewing the draft manuscript. We also thank Larry Needham for helpful comments and administrative support of this study. Many thanks to Julie Herbstman and Jochen Heidler for data and umbilical cord sample collection and Ruth Quinn for study coordination.
References
- Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis 2007; 22:91 - 103
- Guerrero-Preston R. Global epigenetic screening technologies: a novel tool to address cancer health disparities in high-risk population groups. P R Health Sci J 2008; 27:350 - 356
- Jensen TJ, Novak P, Wnek SM, Gandolfi AJ, Futscher BW. Arsenicals produce stable progressive changes in DNA methylation patterns that are linked to malignant transformation of immortalized urothelial cells. Toxicol Appl Pharmacol 2009; 241:221 - 229
- Chen H, Li S, Liu J, Diwan BA, Barrett JC, Waalkes MP. Chronic inorganic arsenic exposure induces hepatic global and individual gene hypomethylation: implications for arsenic hepatocarcinogenesis. Carcinogenesis 2004; 25:1779 - 1786
- Vahter M. Effects of arsenic on maternal and fetal health. Annu Rev Nutr 2009; 29:381 - 399
- Huang D, Zhang Y, Qi Y, Chen C, Ji W. Global DNA hypomethylation, rather than reactive oxygen species (ROS), a potential facilitator of cadmium-stimulated K562 cell proliferation. Toxicol Lett 2008; 179:43 - 47
- Ronco AM, Urrutia M, Montenegro M, Llanos MN. Cadmium exposure during pregnancy reduces birth weight and increases maternal and foetal glucocorticoids. Toxicol Lett 2009; 188:186 - 191
- Pavanello S, Bollati V, Pesatori AC, Kapka L, Bolognesi C, Bertazzi PA, et al. Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int J Cancer 2009; 125:1692 - 1697
- Smith IM, Mydlarz WK, Mithani SK, Califano JA. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer 2007; 121:1724 - 1728
- Terry MB, Ferris JS, Pilsner R, Flom JD, Tehranifar P, Santella RM, et al. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol Biomarkers Prev 2008; 17:2306 - 2310
- Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med 2009; 180:462 - 467
- Perera F, Tang WY, Herbstman J, Tang D, Levin L, Miller R, et al. Relation of DNA methylation of 5'-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS One 2009; 4:4488
- Apelberg BJ, Goldman LR, Calafat AM, Herbstman JB, Kuklenyik Z, Heidler J, et al. Determinants of fetal exposure to polyfluoroalkyl compounds in Baltimore, Maryland. Environmental Science &; Technology 2007; 41:3891 - 3897
- Apelberg BJ, Witter FR, Herbstman JB, Calafat AM, Halden RU, Needham LL, et al. Cord serum concentrations of perfluorooctane sulfonate (PFOS) and perfluorooctanoate (PFOA) in relation to weight and size at birth. Environ Health Perspect 2007; 115:1670 - 1676
- Herbstman JB, Sjodin A, Apelberg BJ, Witter FR, Patterson DG, Halden RU, et al. Determinants of prenatal exposure to polychlorinated biphenyls (PCBs) and polybrominated diphenyl ethers (PBDEs) in an urban population. Environ Health Perspect 2007; 115:1794 - 1800
- Colquhoun DR, Goldman LR, Cole RN, Gucek M, Mansharamani M, Witter FR, et al. Global screening of human cord blood proteomes for biomarkers of toxic exposure and effect. Environ Health Perspect 2009; 117:832 - 838
- Herbstman J, Apelberg BJ, Witter FR, Panny S, Goldman LR. Maternal, infant and delivery factors associated with neonatal thyroid hormone status. Thyroid 2008; 18:67 - 76
- Herbstman JB, Sjodin A, Apelberg BJ, Witter FR, Halden RU, Patterson DG, et al. Birth delivery mode modifies the associations between prenatal polychlorinated biphenyl (PCB) and polybrominated diphenyl ether (PBDE) and neonatal thyroid hormone levels. Environ Health Perspect 2008; 116:1376 - 1382
- Bernert JT Jr, Turner WE, Pirkle JL, Sosnoff CS, Akins JR, Waldrep MK, et al. Development and validation of sensitive method for determination of serum cotinine in smokers and nonsmokers by liquid chromatography/atmospheric pressure ionization tandem mass spectrometry. Clin Chem 1997; 43:2281 - 2291
- Kuklenyik Z, Needham LL, Calafat AM. Measurement of 18 perfluorinated organic acids and amides in human serum using on-line solid-phase extraction. Anal Chem 2005; 77:6085 - 6091
- Hoque MO, Lee CC, Cairns P, Schoenberg M, Sidransky D. Genome-wide genetic characterization of bladder cancer: a comparison of high-density single-nucleotide polymorphism arrays and PCR-based microsatellite analysis. Cancer Res 2003; 63:2216 - 2222
- Bauer M, Orescovic I, Schoell WM, Bianchi DW, Pertl B. Detection of maternal DNA in umbilical cord plasma by fluorescent PCR amplification of short tandem repeat sequences. Ann N Y Acad Sci 2001; 945:161 - 163
- Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect 2008; 116:1547 - 1552
- Pogribny I, Raiche J, Slovack M, Kovalchuk O. Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochem Biophys Res Commun 2004; 320:1253 - 1261
- Kim KY, Kim DS, Lee SK, Lee IK, Kang JH, Chang YS, et al. Association of low-dose exposure to persistent organic pollutants with global DNA hypomethylation in healthy koreans. Environ Health Perspect 118:370 - 374
- Anway MD, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors. Endocrinology 2006; 147:43 - 49
- Wigle DT, Arbuckle TE, Turner MC, Berube A, Yang Q, Liu S, et al. Epidemiologic evidence of relationships between reproductive and child health outcomes and environmental chemical contaminants. J Toxicol Environ Health B Crit Rev 2008; 11:373 - 517
- Windham G, Fenster L. Environmental contaminants and pregnancy outcomes. Fertil Steril 2008; 89:111 - 116
- Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J 2010; 24:2273 - 2280
- Esteller M. Epigenetics in cancer. N Engl J Med 2008; 358:1148 - 1159
- Reamon-Buettner SM, Borlak J. A new paradigm in toxicology and teratology: altering gene activity in the absence of DNA sequence variation. Reprod Toxicol 2007; 24:20 - 30
- Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect 2006; 114:567 - 572
- Davis CD, Uthus EO. DNA methylation, cancer susceptibility and nutrient interactions. Exp Biol Med (Maywood) 2004; 229:988 - 995
- Pogribny IP, Ross SA, Wise C, Pogribna M, Jones EA, Tryndyak VP, et al. Irreversible global DNA hypomethylation as a key step in hepatocarcinogenesis induced by dietary methyl deficiency. Mutat Res 2006; 593:80 - 87
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA 2008; 105:17046 - 17049
- Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet 2009; 18:4046 - 4053
- Dolinoy DC, Jirtle RL. Environmental epigenomics in human health and disease. Environ Mol Mutagen 2008; 49:4 - 8
- Szyf M. The early life environment and the epigenome. Biochim Biophys Acta 2009; 1790:878 - 885
- Martinez-Frias ML. Can our understanding of epigenetics assist with primary prevention of congenital defects?. J Med Genet 2009;
- Pogribny IP, Beland FA. DNA hypomethylation in the origin and pathogenesis of human diseases. Cell Mol Life Sci 2009; 66:2249 - 2261
- Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta 2007; 1775:138 - 162
- Herranz M, Esteller M. DNA methylation and histone modifications in patients with cancer: potential prognostic and therapeutic targets. Methods Mol Biol 2007; 361:25 - 62
- Pogribny IP, Tryndyak VP, Boureiko A, Melnyk S, Bagnyukova TV, Montgomery B, et al. Mechanisms of peroxisome proliferator-induced DNA hypomethylation in rat liver. Mutat Res 2008; 644:17 - 23
- Lee DH, Jacobs DR Jr, Porta M. Hypothesis: a unifying mechanism for nutrition and chemicals as lifelong modulators of DNA hypomethylation. Environ Health Perspect 2009; 117:1799 - 1802