Abstract
The ability of environmental factors to shape health and disease involves epigenetic mechanisms that mediate gene-environment interactions. Metastable epiallele genes are variably expressed in genetically identical individuals due to epigenetic modifications established during early development. DNA methylation within metastable epialleles is stochastic due to probabilistic reprogramming of epigenetic marks during embryogenesis. Maternal nutrition and environment have been shown to affect metastable epiallele methylation patterns and subsequent adult phenotype. Little is known, however, about the role of histone modifications in influencing metastable epiallele expression and phenotypic variation. Utilizing chromatin immunoprecipitation followed by qPCR, we observe variable histone patterns in the 5’ long terminal repeat (LTR) of the murine viable yellow agouti (Avy) metastable epiallele. This region contains 6 CpG sites, which are variably methylated in isogenic Avy/a offspring. Yellow mice, which are hypomethylated at the Avy LTR and exhibit constitutive ectopic expression of agouti (a), also display enrichment of H3 and H4 di-acetylation (p=0.08 and 0.09, respectively). Pseudoagouti mice, in which Avy hypermethylation is thought to silence ectopic expression, exhibit enrichment of H4K20 tri-methylation (p=0.01). No differences are observed for H3K4 tri-methylation (p=0.7), a modification often enriched in the promoter of active genes. These results show for the first time the presence of variable histone modifications at a metastable epiallele, indicating that DNA methylation acts in concert with histone modifications to affect inter-individual variation of metastable epiallele expression. Therefore, the potential for environmental factors to influence histone modifications, in addition to DNA methylation, should be addressed in environmental epigenomic studies.
Introduction
The phrase “metastable epiallele” was first coined by Rakyan and colleagues in 2002 to describe genes that are variably expressed in genetically identical individuals as a result of epigenetic modifications established during early development.Citation1 The term “epiallele” refers to “an allele that can stably exist in more than one epigenetic state, resulting in different phenotypes,” while “metastable” highlights the “labile nature of the epigenetic state of these particular alleles.”Citation1 Metastable epialleles are most often associated with retroelements and transgenesis, resulting in ectopic or aberrant transcription of nearby genes. Furthermore, the extent of DNA methylation at metastable epialleles is stochastic due to probabilistic reprogramming of epigenetic marks during embryogenesis, resulting in variable phenotypes across genetically identical individuals.Citation2 Important to the emerging field of environmental epigenomics, these stochastic methylation patterns may be influenced by maternal nutrition and environmental exposures during early development.Citation3–Citation7 Little is known, however, about the role of chromatin structure and histone modifications in influencing metastable epiallele expression and subsequent phenotypic variation.
Three of the identified murine metastable epialleles (Avy, AxinFu, CabpIAP) are associated with contraoriented intracisternal A particle (IAP) proviral insertions.Citation8–Citation10 Approximately 1,000 copies of IAP retrotransposons are present in the mouse genomeCitation11 and about 40% of the human genome is comprised of transposable elements, of which approximately 9% are retrotransposons.Citation12 The murine viable yellow agouti (Avy) allele is the most extensively studied metastable epiallele.Citation9 The Avy mouse model, in which coat color variation is correlated to epigenetic marks established early in development, has been used to investigate the impacts of nutritional and environmental influences on the fetal epigenome. The wildtype murine Agouti gene encodes for a paracrine signaling molecule that produces either black eumelanin (a) or yellow phaeomelanin (A). Both A and a transcription are initiated from a hair-cycle specific promoter in exon 2. Transient A expression in hair follicles during a specific stage of hair growth results in a sub-apical yellow band on each black hair shaft, causing the brown (agouti) coat color of wild-type mice.Citation9 The Avy metastable epiallele resulted from the insertion of an IAP murine retrotransposon upstream of the transcription start site of the Agouti gene ().Citation6,Citation9 A cryptic promoter in the proximal end of the Avy IAP promotes constitutive ectopic Agouti transcription, leading to yellow fur, obesity and tumorigenesis.Citation13,Citation14 CpG methylation in the Avy IAP correlates inversely with ectopic Agouti expression. The degree of methylation at six CpG sites within the 5′ long terminal repeat (LTR) of the Avy IAP varies dramatically among individual isogenic Avy/a mice, causing a wide variation in coat color ranging from yellow (unmethylated) to pseudoagouti (methylated) ().Citation13 Yet, the types and patterns of histone modifications in this or any other metastable epiallele, remains undefined.
Chromatin consists of a nucleoprotein complex that packages linear genomic DNA via an array of nucleosomes. Each nucleosome consists of ∼147 base pairs of DNA coiled around an octamer of core histone proteins (H2A, H2B, H3 and H4). In addition to their structural role, histones play an important regulatory role in local gene activities, including transcription and DNA repair as well as more global functions such as DNA replication. At least eight types of histone modifications exist, of which histone acetylation, methylation, phosphorylation and ubiquitination are the most extensively studied.Citation15 These modifications occur at over 60 different residues, rendering a full characterization of the histone codeCitation16 an enormous task. Histone acetylation is usually associated with transcriptional activation because acetylation of lysine residues neutralizes its basic charge, promoting chromatin relaxation and recruiting bromo-domain proteins that facilitate transcription.Citation17 On the other hand, histone methylation results in various transcriptional consequences; H3K4, H3K36 and H3K79 methylation are most often associated with transcriptional activation, while H3K9, H3K27 and H4K20 methylation are linked to transcriptional repression. Modified histones interact with DNA methylation to recruit multi-subunit chromatin-protein complexes, such as the repressive polycomb group proteins or the activating SWI-SNF proteins.
To investigate the histone profile of the Avy metastable epiallele, we utilize in vivo chromatin immunoprecipitation (ChIP) of Avy/a liver tissue followed by quantitative real-time PCR (qPCR) to characterize four key histone modifications in the 5′ IAP of the Avy metastable epiallele. Within the same series of mice, we also correlate and associate the identified variable histone marks with site-specific Avy CpG methylation profiles and Agouti gene expression. We chose to investigate H3 and H4 lysine (K) di-acetylation, which are both marks associated with transcriptional activation,Citation17 and are thought to be modified by environmental and nutritional agents, including nickel and soy.Citation18,Citation19 We also investigate two histone methylation modifications. H3K4 tri-methylation has been associated with transcriptional activation via methyl binding and chromo-domain protein recruitment (reviewed in ref. Citation20). Conversely, H4K20 tri-methylation has been associated with transcriptional repression.Citation21 It is reported herein that H3 and H4 lysine di-acetylation is enriched within the IAP LTR upstream of the Agouti gene in yellow Avy/a offspring and that H4K20 trimethylation in the same region is enriched within pseudoagouti Avy/a offspring. These findings indicate that in addition to stochastic methylation patterns, variable histone modifications also play a role in the inter-individual epigenetic variation of metastable epiallele expression in Avy mice.
Results
Site-specific Avy histone marks, Avy CpG DNA methylation and quantitative Agouti gene expression were assessed in individual (non-pooled) Avy/a animals of the yellow and pseudoagouti coat color classes. Liver tissues for these analyses were collected from eight yellow and seven pseudoagouti animals and stored in aliquots. DNA methylation and expression studies were conducted on all 15 animals, though ChIP histone experiments were performed on only six to 12 animals due to tissue quantity limitations or technical difficulty.
Variable histone modifications in the Avy metastable epiallele.
We adapted the ChIP protocol for use in intact liver tissue from d22 yellow and pseudoagouti Avy/a mice using four histone modification antibodies of interest plus negative and positive antibody controls. Primers were designed to investigate antibody enrichment of histone modifications in the 5′ LTR of the Avy IAP. This region contains 6 CpG sites, which have been shown to be variably methylated in isogenic Avy/a offspring of different coat color phenotypes.Citation4 Specifically, primers were chosen to amplify sites 4 to 9, a region postulated from previous DNA methylation studies to contain the highest probability of modified histone binding (indicated by a * in ).Citation4,Citation5 As a control, a second set of primers were designed to evaluate histone structure and variability in the pseudoexon 1A (PS1A) genomic region located 300 bp downstream of the Avy IAP (indicated by a ** in ).
Following ChIP and qPCR, we observed Avy IAP enrichment of H3 and H4 di-acetylation in yellow offspring compared to genetically identical pseudoagouti offspring reared in similar maternal and postnatal environments ( and B). A marginally statistically significant increase in H3 di-acetylation immunoprecipitated DNA was seen in yellow animals compared to pseudoagouti animals following normalization with Input DNA (p = 0.09; n = 6 per group) (). Mean H3 di-acetylation binding activity in yellow mice was 23.4 ± 12.0% compared to 1.3 ± 0.75% in pseudoagouti mice. Similarly, for the H4 di-acetylation mark, a marginally statistically significant increase was seen in yellow animals compared to pseudoagouti animals following normalization with Input DNA (p = 0.08; n = 3 per group) (). Mean H4 di-acetylation binding activity in yellow mice was 70.1 ± 19.9% compared to 4.6 ± 4.5% in pseudoagouti mice. No differences in H3 and H4 acetylation marks were observed in the PS1A genomic region located 300 bp downstream of the Avy IAP (p = 0.4 and p = 0.4; n = 6 and n = 3 per group, respectively), indicating that the observed effects in histone variability are specific to the 5′ LTR of the Avy IAP ( and D). Mock immunoprecipitation with negative control mouse IgG displayed a percent of input binding activity of 1.72% and it did not differ between yellow and pseudoagouti animals.
Interestingly, we did not observe a statistically significant difference in Avy IAP or genomic PS1A H3K4 tri-methylation enrichment in yellow compared to pseudoagouti animals (p = 0.7 and p = 0.9; n = 6 and n = 5 per group, respectively) ( and C). For the Avy IAP, average binding activity within yellow mice was 13.6 ± 3.5% compared to 16.3 ± 6.1% in pseudoagouti animals (). We did, however, observe enrichment of H4K20 tri-methylation in pseudoagouti offspring compared to genetically identical yellow offspring (). A statistically significant increase in immunoprecipitated DNA levels was demonstrated in pseudoagouti animals compared to yellow animals (p = 0.01, n = 6 per group). Average binding activity for yellow mice was 3.7 ± 1.5% compared to 14.3 ± 3.6% in pseudoagouti mice (). No difference in H4K20 tri-methylation was observed in the PS1A genomic region downstream of the Avy IAP (p = 0.8, n = 5 per group), indicating that the observed effects in histone variability are specific to the 5′ LTR of the Avy IAP (). Mock immunoprecipitation with negative control mouse IgG displayed a percent of input binding activity of 1.38% and did not differ between yellow and pseudoagouti animals.
DNA methylation and agouti (a) expression.
The 5′ Avy cryptic promoter contains 6 CpG sites, which are variably methylated in isogenic Avy/a offspring and can be modified according to maternal nutritional statusCitation4 and environmental exposureCitation5 (). Quantitative liver DNA methylation levels at Avy CpG sites 4–9 are correlated with coat color class-specific variable histone marks. Yellow animals, which exhibit increased enrichment of H3 and H4 histone di-acetylation at the 5′ LTR of the Avy IAP, display mean CpG site 4–9 methylation levels of 8.8% (n = 8) (). Alternatively, pseudoagouti animals, exhibiting enhanced H4K20 tri-methylation modifications at the 5′ LTR of the Avy IAP, have an average CpG site 4–9 methylation level of 87.0% (n = 7; p = 0.0001 compared to yellow animals) (). Analysis of individual CpG sites, revealed significantly (p < 0.0001) lower methylation in the yellow animals at all 6 sites (). Similar Avy DNA methylation patterns across yellow and pseudoagouti animals were observed for kidney and brain tissue (data not shown).
Consistent with CpG methylation levels and displaying the expected direction, quantitative Agouti (a) gene expression levels are associated with coat color class-specific variable histone marks and quantitative DNA methylation levels. Average a expression levels in liver tissue from yellow animals (n = 8) was greater than 600-fold higher compared to pseudoagouti animals (n = 7) (). Similar correlations between methylation levels and expression are observed for Avy/a kidney and brain tissues (data not shown).
Discussion
These results show for the first time the presence of variable histone modifications at the Avy metastable epiallele in genetically identical Avy/a mice. Yellow mice that are hypomethylated at the DNA level in the Avy IAP LTR and exhibit constitutive ectopic agouti expression, also display enrichment of activating histone acetylation modifications (H3 and H4 di-acetylation). Pseudoagouti mice, in which DNA hypermethylation at the Avy IAP is thought to silence ectopic agouti expression, display enrichment of the repressive histone H4K20 tri-methylation modification. These variable histone marks were not observed in the PS1A genomic region 300 bp downstream of the Avy IAP, indicating that variable histone marks are specific to the 5′ LTR of the Avy IAP and may be contributing to the metastable nature of this gene. Furthermore, the presence of variable histone marks and their associated function were correlated with DNA methylation and Agouti gene expression. For example, yellow animals, exhibiting histone marks associated with open chromatin configuration, had much higher levels of Agouti gene expression and much lower levels of DNA methylation. The opposite effects (lower Agouti gene expression and higher DNA methylation) were observed for pseudoagouti animals with histone marks associated with repressive chromatin states.
Together, these data indicate that DNA methylation acts in concert with histone modifications to affect variable expression of the Avy metastable epiallele. Interestingly, enrichment differences were not observed between yellow and pseudoagouti offspring following chromatin immunoprecipitation with the activating chromatin modification, H3K4 tri-methylation. Recently, genome-wide epigenomic mapping studies have revealed that promoter regions associated with high CpG content (CpG islands) are more likely to contain H3K4 methylation, whereas promoters with lower CpG content lack this mark.Citation22,Citation23 Since the LTR of the Avy IAP is not a CpG island, it is therefore reasonable to conclude that H3K4 tri-methylation may not contribute to variable expressivity of the Avy metastable epiallele. Results from this study indicate that a more comprehensive analysis of histone modifications within the Avy IAP LTR is warranted to elucidate the full histone codeCitation16 associated with this metastable epiallele. For example, within pericentric heterochromatin regions, it is known that the repressive H3K9 tri-methylation mark is associated with, if not required for, H4K20 tri-methylation.Citation24,Citation25 Given the enrichment of H4K20 tri-methylation in the Avy IAP of pseudoagouti mice, it is important to assess whether H3K9 methylation is also present within metastable epialleles. Finally, since in vivo ChIP analysis of intact tissue can be technically challenging and enrichment differences for H3 and H4 acetylation only reached marginal significance in our study, it will be important to ensure adequate tissue collection and biological replicates in future analyses of metastable epiallele histone profiles.
The characterization of histone modifications at metastable epialleles is an important step in identifying mechanisms of action promoting environmentally-induced alterations within the epigenome. Compared to the number of studies investigating changes in DNA methylation following environmental or nutritional exposures, the number of studies devoted to assessing environmentally induced changes in histone modifications is relatively few. Histone modifications play an important role in delayed plant flowering in response to prolonged cold weather.Citation26 Costa and colleagues have reported decreased histone acetylation, increased histone methylation and subsequent decreased gene expression following nickel exposure.Citation18,Citation27 Furthermore, chromium exposure has been linked to epigenetically controlled gene expression alterations via interactions with histone acetyltransferase and histone deacetylase enzymes.Citation28 Finally, a study of human identical twins observed epigenetic differences in H3 and H4 histone modifications in 35% of twin pairs.Citation29
Previously, the Avy model has been used as an epigenetic biosensor for determining whether maternal environmental and nutritional factors affect the fetal epigenome.Citation5,Citation6,Citation30,Citation31 For example, recently, weCitation4 demonstrated that maternal dietary supplementation of mice with the phytoestrogen genistein shifts the coat color of offspring toward pseudoagouti by increasing methylation of the Avy retrotransposon. As phytoestrogens are not methyl donors, we hypothesized genistein might influence chromatin structure and subsequent DNA methylation and gene expression via altered histone modifications.Citation4 Emerging data now suggest epigenetic modifications are inherited not only during mitosis, but also can be transmitted transgenerationally.Citation7,Citation13,Citation32,Citation33 However, when Whitelaw et al.Citation34 investigated DNA methylation as the inherited epigenetic mark within Avy gametes, zygotes and blastocysts, DNA methylation was entirely absent from the blastocyst, indicating that CpG methylation is not the inherited mark. Thus, though it is becoming clearer that environmental effects on the epigenome may be inherited in the mammalian germ-line, the exact mechanisms mediating this inheritance are unknown. Histone modifications and non-coding RNAs are two potential mechanisms mediating transgenerational epigenetic inheritance, although their exact roles remain to be elucidated.
Materials and Methods
Viable yellow agouti (Avy) mice.
Liver, kidney and brain tissue from yellow (n = 8) and pseudoagouti (n = 7) Avy/a mice from different litters were collected on post-natal-day 22 (d22) mice and flash frozen in three aliquots with liquid nitrogen prior to storage at −80°C. One liver aliquot each was used for histone, methytion and expression analysis. DNA methylation and expression levels were analyzed in liver, kidney and brain tissue, while histone marks were analyzed in d22 liver tissue only. Mice were obtained from a colony that has been maintained with sibling mating and forced heterozygosity for the Avy allele for over 220 generations, resulting in a genetically invariant background.Citation6 Animals used in this study were maintained in accordance with the Guidelines for the Care and Use of Laboratory Animals and the study protocol was approved by the University of Michigan and Duke University Committees on Use and Care of Animals.
Chromatin isolation and immunoprecipitation (ChIP) assay.
We adapted the ChIP-IT Express protocol provided by Active Motif (Carlsbad, CA) for use on frozen tissue. Briefly, 50 mg of liver tissue (approximately 10 × 106 cells) was crushed into small pieces using a pestle and mortar submerged in liquid nitrogen. Ground liver tissue was incubated at room temperature for 10 min in 10 ml of fixation solution (1% formaldehyde in 1X PBS) to cross-link DNA and associated proteins. Cross-linked samples were washed with glycine-stop-fix solution (10X glycine buffer, 10X PBS and dH2O) to halt fixation and followed by washing with cell-scraping solution (10X PBS, dH2O and 100 mM PMSF). Samples were incubated on ice for 30 min in ice-cold lysis buffer (supplemented with 7.5 µl protease cocktail inhibitor (PIC) and 7.5 µl 100 mM PMSF) prior to douncing on ice to release nuclei from cells. Cells were suspended in 330 µl shearing buffer prior to sonication. All reagents were supplied by Active Motif (Carlsbad, CA). DNA was sonicated to shear lengths of 200 to 1,000 bp using a Branson sonicator with a 2 mm microtip (5, 10 sec pulses with 50 sec pause on ice) with an amplitude setting of 35%. The sheared DNA was centrifuged at 13,000 rpm at 4°C for 12 min to remove cellular debris prior to storage at −80°C. 25 µl were removed to check shearing efficiency and concentration.
Chromatin immunoprecipitation (ChIP) was then carried out according to manufacturer's protocol (Active Motif, Carlsbad, CA). Prior to immunoprecipitation, 10 µl of sheared DNA was reserved to serve as a no-antibody “Input” control. Antibodies specific for H3 acetylation (di-acetyl Lys9 and Lys14, catalog number 06-599), H4 acetylation (di-acetyl Lys5, Lys8, Lys12, Lys16, catalong number 17-211), H4 methylation (trimethyl Lys20, catalog number 07-463) and H3 methylation (tri-methyl Lys4, catalog number 07-473) were obtained from Upstate Cell Signaling Solutions (Millipore, Lake Placid, NY, USA). These antibodies have been used successfully in ChIP protocols (data supplied by Millipore/Upstate).
Mock immunoprecipitations with mouse IgG (negative control) and positive control immunoprecipitation with RNA pol II antibody were included as controls (ChIP-IT Control Kit Mouse, Active Motif catalog number 53011) and PCRs were performed per manufacturer's suggestion. Positive control PCRs using the EFI-alpha primers (233 bp product) performed on samples incubated in RNA pol II antibody were confirmed to yield more product than samples incubated in IgG antibody. Negative control PCRs (245 bp product) were confirmed to yield equivalent amounts of product in samples incubated with RNA pol II and IgG.
Real-time PCR for histone modifications.
Primers were designed for qPCR to result in product amplicons of 150–300 bp (Tm = 60°C) using the Primer3 design program (http://frodo.wi.mit.edu/). Specifically, primers were designed to generate amplicons that span the IAP/genomic DNA boundary in order to distinguish between the Avy and a alleles. Prior to use in qPCR, primer pairs were tested in regular PCR for correct size and absence of primer dimer formation using standard PCR reagents and protocols. PCR primers for the 5′ LTR Avy IAP amplify a 251 bp region spanning CpG sites 4–9 of the Avy IAP (located at nucleotide positions 244, 265, 306, 319, 322 and 334 of GenBank accession number AF540972) and are listed in . For analysis of the PS1A genomic region located 300 bp downstream of the Avy IAP, primer pairs amplifying a 186 bp amplicon spanning genomic CpG sites (nucleotide positions 684, 716 and 794 of GenBank accession number AF540972) were designed and are present in .
For qPCR, 2 µl of ChIP DNA were used as a template in 20 µl reactions containing 10 µl of 2X SYBR Green Master Mix (Applied Biosystems, Foster City, CA) and 0.5 uM primers using the Applied Biosystems 7900HT Fast Real-Time PCR System according to manufacturer's protocols. All samples (ChIP and Input) were analyzed in triplicate and no template control samples were included. RQ Manager 1.2 (Applied Biosystems, Foster City, CA, USA) software was used to calculate average threshold cycle (C(t)) values for each triplicate sample. Histone binding activity was calculated as percent of pre-immunoprecipitated Input DNA as represented by exp2(C(t)input − C(t)ChiP) × 100, where (C(t)input − C(t)ChiP) = ΔCT. This measurement controls for differences in post-shearing ChIP concentrations and allows for comparison across treatment (coat color) groups.Citation35 Coat color group comparisons of the percent of Input DNA were assessed by two sample hypothesis testing of means (t-test) and graphed using STATA version 9.1 software (College Station, TX, USA).
DNA isolation and methylation analysis.
Total DNA was isolated from the liver, kidney and brain of the same d22 Avy/a animals assessed for ChIP experiments, as previously described.Citation5 Quantitative DNA methylation levels at individual CpG sites (4–9) within the Avy IAP LTRCitation5 was performed via sodium bisulfite modification, as previously described,Citation5 followed by pyrosequencing.Citation36 Approximately 1 ug of DNA was bisulfite converted using the EpiTect Bisulfite Kit (Qiagen Inc., Valencia, CA, USA) with clean-up automation on the QIAcube® purification system. The six CpG sites studied are located at nucleotide positions 244, 265, 306, 319, 322 and 334 of GenBank accession number AF540972. Bisulfite converted PCR primers as well as sequencing primers for pyrosequecning are listed in . The percentage of cells methylated was quantified using PyroMark software, which computes the degree of methylation as %5-methylated cytosines (%5 mC) over the sum of methylated and unmethylated cytosines. Samples were tested in duplicate and the average of the two replicates was used in the statistical analysis. Coat color group comparisons of the percentage of cells methylated at each of the 6 CpG sites within the Avy allele were performed by two-sample hypothesis testing of means (t-test) using STATA version 8.0 software (College Station, TX, USA).
Total RNA purification and cDNA synthesis.
Total RNA was purified from the same series of mice used for the ChIP and methylation studies via an adapted protocol for the RNeasy® Mini Kit (Qiagen, Valencia, CA, USA) in conjunction with the QIAcube® automated nucleic acid purification system (Qiagen, Valencia, CA, USA). Briefly, approximately 20 mg of tissue from liver, kidney or brain was placed into individual round-bottom 2 mL safe lock tubes with 600 µL of Buffer RLT® containing 1% (v/v) β-mercaptoethanol. A 5 mm stainless steel bead was added to each tube and the tissues were simultaneously disrupted and homogenized using two 30 s bursts at 30 Hz in a TissueLyser® II (Qiagen, Valencia, CA, USA). Residual debris was removed by centrifugation and the cleared crude lysate was transferred to a clean round-bottom 2 mL tube. Crude lysates, along with RNeasy® Mini Kit spin columns and reagents, were loaded into the QIAcube® automated nucleic acid purification system. Samples were processed as per manufacturer's protocol for large volume samples with a 50 µL final elution volume. For liver tissue lysates only, the manufacturer's protocol was modified to use 50% ethanol, rather than 70% ethanol, in the initial precipitation and binding step. Total RNA quality and concentration were assessed using a ND1000 spectrophotometer (NanoDrop Technology, Wilmington, DL, USA). For each sample, a volume equivalent to 1 µg of total RNA was used with the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) to generate DNA complementary to the messenger RNA.
Real-time PCR for expression analysis.
Primers for real-time PCR of the mouse Agouti (a) gene () were designed using GenScript Real Time PCR (TaqMan) Primer Design Tool (GenScript, Piscataway, NJ, USA, www.genscript.com/ssl-bin/app/primer). Primers for real-time PCR of mouse housekeeping gene β-actin () were designed using NCBI Primer BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome). Both primer pairs were designed to have a Tm = 59°C ± 1°C, contain ≤3 Gs or Cs within 5 bases of the 3′ end and produce a product between 50–150 bp in length. Primers were also designed, where possible, to either bind across an exon junction or produce a product containing at least one exon junction. Each real-time PCR amplification was performed as a 25 µL reaction using iQ™ SYBR® Green Supermix (Bio-Rad, Hercules, CA), 2 µL of 10-fold diluted cDNA and a 100 nM final concentration of each primer. Reactions were carried out in triplicate on a CFX96™ Real-Time System (Bio-Rad, Hercules, CA, USA) and included no template control reactions for each primer set. PCR conditions were 95°C for 3 min, 40 cycles of 95°C for 10 s, 55°C for 30 s and 95°C for 10 s. This was followed by a melt curve analysis of 65°C–95°C in 0.5°C increments. Following real-time PCR and melt curve analysis, products were run on a 2% agarose gel against a 25 bp DNA ladder to verify appropriate product size.
CFX Manager™ Version 1.0 software (Bio-Rad, Hercules, CA, USA) was used to calculate the average threshold cycle (C(t)) for each set of triplicate reactions. The C(t) for Agouti (a) was normalized against the C(t) for endogenous β-actin for each sample. From these data, CFX Manager™ software was used to calculate the normalized expression level of the Agouti (a) gene in tissues from yellow-coated agouti mice relative to the normalized expression level in tissues from brown-coated pseudoagouti mice. Results are represented as fold change in expression (2−ΔΔC(t)) on a linear scale, with error bars representing the standard error of the mean for the normalized data.
Abbreviations
| Avy | = | viable yellow agouti |
| BPA | = | bisphenol A |
| ChIP | = | chromatin immunoprecipitation |
| H3 | = | histone 3 |
| H4 | = | histone 4 |
| IAP | = | intracisternal A particle |
| LTR | = | long terminal repeat |
| PIC | = | protease inhibitor cocktail |
| PS1A | = | pseudoexon 1A |
Figures and Tables
Figure 1 The Avy metastable epiallele. (A) The viable yellow agouti (Avy) allele contains a contra-oriented IAP insertion within pseudoexon 1A (PS1A) of the Agouti gene. A cryptic promoter (short arrow labeled Avy) drives constitutive ectopic Agouti expression. Transcription of the Agouti gene normally initiates from a developmentally regulated hair-cycle specific promoter in exon 2 (short arrow labeled A, a). (B) Genetically identical Avy/a offspring representing the yellow and pseudoagouti coat color phenotypes are shown. * indicates 5′LTR of the Avy IAP region and **represents non-IAP genomic PS1A region.
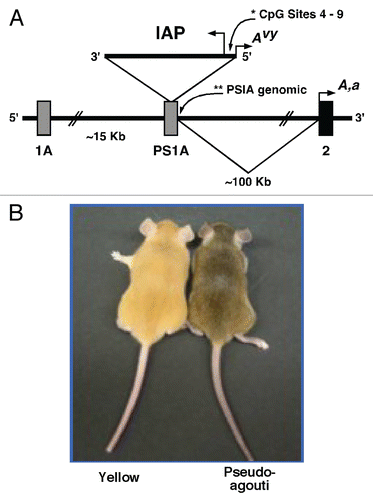
Figure 2 Chromatin precipitation for acetylated histones H3 and H4 in the 5′ LTR of the IAP and in PS1A. Binding activity was calculated as percent of pre-immunoprecipitated input DNA as represented by 2−ΔC(t) X100. (A) DNA precipitated by H3 di-acetylation antibody is enriched in yellow versus pseudoagouti Avy/a mice (p = 0.09; n = 6 per group) within the 5′ LTR of the IAP. (B) DNA precipitated by H4 di-acetylation antibody is enriched in yellow versus pseudoagouti Avy/a mice (p = 0.08; n = 3 per group) within the 5′ LTR of the IAP. (C) DNA precipitated by H3 di-acetylation antibody does not vary by coat color within the PS1A genomic region (p = 0.4; n = 6 per group). (D) DNA precipitated by H4 di-acetylation antibody does not differ by coat color within the genomic PS1A region (p = 0.4; n = 3 per group). * indicates significance at the 0.09 level.
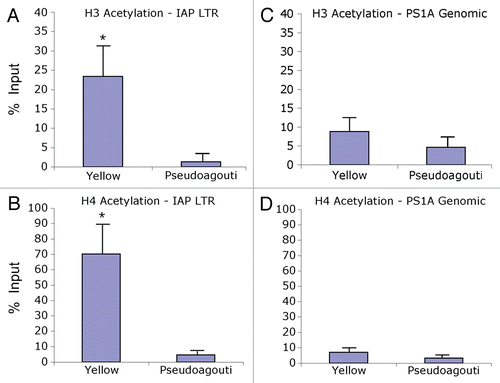
Figure 3 Chromatin precipitation for methylated histones in the 5′ LTR of the IAP and in PS1A. Binding activity was calculated as percent of pre-immunoprecipitated input DNA as represented by 2−ΔC(t) X100. (A) DNA precipitated by H3K4 tri-methylation antibody is not enriched in yellow versus pseudoagouti Avy/a mice (p = 0.7; n = 6 per group) within the 5′ LTR of the IAP. (B) DNA precipitated by H4K20 tri-methylation antibody is enriched in pseudoagouti compared to yellow Avy/a mice (p = 0.01; n = 6 per group) within the 5′ LTR of the IAP. (C) DNA precipitated by H3K4 tri-methylation antibody does not vary by coat color within the PS1A genomic region (p = 0.9; n = 5 per group). (D) DNA precipitated by H4K20 tri-methylation antibody does not differ by coat color within the genomic PS1A region (p = 0.8; n = 5 per group). ** indicates significance at the 0.01 level.
Table 1 DNA methylation and Agouti (a) expression levels by histone and coat color status
Table 2 PCR and pyrosequencing primers
Acknowledgements
This work was supported by NIH grants ES017524, ES13053, ES08823 and ES015165. The authors would like to thank Mary Ellen Koran and Dale Huang for technical assistance.
References
- Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet 2002; 18:348 - 351
- Rakyan VK, Preis J, Morgan HD, Whitelaw E. The marks, mechanisms and memory of epigenetic states in mammals. Biochem J 2001; 356:1 - 10
- Waterland R, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani K. Maternal methyl supplements increase offspring DNA methylation at Axin(fused). Genesis 2006; 44:401 - 406
- Dolinoy DC, Wiedman J, Waterland R, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect 2006; 114:567 - 572
- Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA 2007; 104:13056 - 13061
- Waterland R, Jirtle R. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 2003; 23:5293 - 5300
- Cropley JE, Suter CM, Beckman KB, Martin DIK. Germ-line epigenetic modification of the murine Avy allele by nutritional supplementation. Proc Natl Acad Sci USA 2006; 103:17308 - 17312
- Druker R, Bruxner TJ, Lehrbach NJ, Whitelaw E. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucl Acids Res 2004; 32:5800 - 5808
- Duhl D, Vrieling H, Miller K, Wolff G, Barsh G. Neomorphic agouti mutations in obese yellow mice. Nat Genet 1994; 8:59 - 65
- Vasicek T, Zeng L, Guan X, Zhang T, Costantini F, Tilghman S. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics 1997; 147:777 - 786
- Kuff E, Lueders K. The intracisternal A-particle gene family: structure and functional aspects. Adv Cancer Res 1988; 51:183 - 276
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 2001; 209:860 - 921
- Morgan H, Sutherland H, Martin D, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet 1999; 23:314 - 318
- Miltenberger R, Mynatt R, Wilkinson J, Woychik R. The role of the agouti gene in the yellow obese syndrome. J Nutr 1997; 127:1902 - 1907
- Kouzarides T. Chromatin modifications and their function. Cell 2007; 128:693 - 705
- Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293:1074 - 1080
- Kanno T, Kanno Y, Siegel R, Jang M, Lenardo M, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell 2004; 13:33 - 43
- Yan Y, Kluz T, Zhang P, Chen H, Costa M. Analysis of specific lysine histone H3 and H4 acetylation and methylation status in clones of cells with a gene silenced by nickel exposure. Toxicol Appl Pharmacol 2003; 190:272 - 277
- Hong T, Nakagawa T, Pan W, Kim M, Kraus W, Ikehara T, et al. Isoflavones stimulate estrogen receptor-mediated core histone acetylation. Biochem Biophys Res Commun 2004; 317:259 - 264
- Ruthenburg A, Allis C, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 2007; 25:15 - 30
- Martens J, O'Sullivan R, Braunschweig U, Opravil S, Radolf M, Steinlein P, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 2005; 24:800 - 812
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007; 448:553 - 560
- Karlic R, Chung H, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci USA 2010; 107:2926 - 2931
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 2004; 18:1251 - 1262
- Pannetier M, Julien E, Schotta G, Tardat M, Sardet C, Jenuwein T, et al. PR-SET7 and SUV4-20H regulate H4 lysine-20 methylation at imprinting control regions in the mouse. EMBO Rep 2008; 9:998 - 1005
- Sung S, Amasino RM. Remembering winter: toward a molecular understanding of vernalization. Annu Rev Plant Biol 2005; 56:491 - 508
- Chen H, Ke Q, Kluz T, Yan Y, Costa M. Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol Cell Biol 2006; 26:3728 - 3737
- Wei Y, Tepperman K, Huang M, Sartor M, Puga A. Chromium inhibits transcription from polycyclic aromatic hydrocarbon-inducible promoters by blocking the release of histone deacetylase and preventing the binding of p300 to chromatin. J Biol Chem 2004; 279:4110 - 4119
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 2005; 102:10604 - 10609
- Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J 1998; 12:949 - 957
- Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr 2002; 132:2393 - 2400
- Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KVK, et al. Transgenerational inheritance of epigenetic states at the murine AxinFu allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA 2003; 100:2538 - 2543
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005; 308:1466 - 1469
- Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet 2006; 2:399 - 405
- Chen-Plotkin AS, Sadri-Vakili G, Yohrling GJ, Braveman MW, Benn CL, Glajch KE, et al. Decreased association of the transcription factor Sp1 with genes downregulated in Huntington's disease. Neurobiol Dis 2006; 22:233 - 241
- Colella S, Shen L, Baggerly K. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechnique 2003; 35:146 - 150