Abstract
MicroRNAs (miRNAs) are short non-coding RNA molecules that regulate post-transcriptional gene expression. They influence a wide range of physiological functions, including neuronal processes, and are regulated by various mechanisms, such as DNA methylation. This epigenetic mark is recognized by transcriptional regulators such as the methyl CpG binding protein Mecp2. Rett syndrome is a complex neurological disorder that has been associated with mutations in the gene coding for Mecp2. Thus, we examined the possible miRNA misregulation caused by Mecp2 absence in a mouse model of Rett syndrome. Using miRNA expression microarrays, we observed that the brain of Rett syndrome mice undergoes a disruption of the expression profiles of miRNAs. Among the significantly altered miRNAs (26%, 65 of 245), overall downregulation of these transcripts was the most common feature (71%), whilst the remaining 30% were upregulated. Further validation by quantitative RT-PCR demonstrated that the most commonly disrupted miRNAs were miR-146a, miR-146b, miR-130, miR-122a, miR-342 and miR-409 (downregulated), and miR-29b, miR329, miR-199b, miR-382, miR-296, miR-221 and miR-92 (upregulated). Most importantly, transfection of miR-146a in a neuroblastoma cell line caused the downregulation of IL-1 receptor-associated kinase 1 (Irak1) levels, suggesting that the identified defect of miR-146a in Rett syndrome mice brains might be responsible for the observed upregulation of Irak1 in this model of the human disease. Overall, we provide another level of molecular deregulation occurring in Rett syndrome that might be useful for understanding the disease and for designing targeted therapies.
Introduction
MicroRNAs (miRNAs) are short (approximately 22-nucleotide long) endogenous non-protein-coding RNA molecules responsible for post-transcriptional regulation of gene expression.Citation1 The regulation mediated by miRNAs has begun to be elucidated. Their target recognition mechanism, for instance, has already been identified; they interact with partially complementary target sites in messenger RNAs (mRNAs),Citation2,Citation3 which allows each miRNA to control many different mRNAs through incomplete base pairing recognition.Citation4 Hence, multiple pathways are affected by their regulation. Our knowledge about the mechanisms by which these molecules exert their regulatory function is growing quickly. First, miRNAs were described as binding the 3′-untranslated region (UTR) of their target mRNAs,Citation5 although some examples of 5′-UTR binding have recently been found.Citation6–Citation8 Furthermore, it was widely accepted that they inhibited transcription or directly degraded their targetsCitation9 until the recent discovery that they enhance translation.Citation8,Citation10 Consequently, miRNA functions and their repercussions need to be explored in detail for a correct understanding of the processes in which they are involved. Expression patterns of miRNAs may be tissue-specific, related to development or ubiquitious. Increasing evidence shows the importance of miRNAs in regulating many aspects of cell developmentCitation11,Citation12 and the great variety of normal physiological functions that they influence have begun to be revealed by studies in human cells and model organisms. Therefore, deregulation of miRNAs is involved in a wide range of human diseases, including cancer.Citation13–Citation15
miRNAs have many roles, but many are essential for the correct function of the nervous system,Citation16 and several of them are highly and specifically expressed in brain.Citation4,Citation16,Citation17 In mammals, conditional ablation of essential components of the miRNA machinery, such as the miRNA-generating enzyme Dicer, appears to be critical for brain function. For instance, a study by Kim et al.Citation18 showed the importance of miRNAs for terminal differentiation and maintenance of multiple neuron types. They also showed that the reduction of the miRNA level caused by Dicer deletion in midbrain dopaminergic neurons in vivo reduced locomotion in mice. In another report, Dicer deficiency and the resulting miRNA depletion in Purkinje cells led to a slow degeneration of these post-mitotic cells. Although Dicer ablation had no immediate impact on Purkinje cell function, the long-term absence of Dicer resulted in a neurodegenerative process leading to ataxia, motor impairment and severe cerebellar dysfunction.Citation19 Nevertheless, a severe phenotype has been described as a result of inactivation of Dicer in excitatory forebrain neurons in vivo. The consequences included microcephaly, reduced dendritic branch elaboration and large increases in dendritic spine length but no changes in spine density or cortical lamination.Citation20 All the observations found in these different models are reminiscent of the phenotype of different human patients with neurological diseases. These studies highlight the importance of components of the miRNA processing machinery that interfere with the miRNA pathway in the central nervous system and demonstrate that mis-regulation of this pathway results in gross abnormalities that may contribute to a variety of neurological disorders.
Considering the importance of miRNAs in controlling all stages of cell physiology, it is not surprising that their own expression needs careful fine-tuning. Epigenetic regulation contributes to this process. DNA methylation of specific sites in CpG islands is an important epigenetic mechanism used to regulate expression of coding genesCitation21 and miRNAs.Citation22 The crosstalk between DNA methylation and chromatin remodeling, which leads to expression regulation, is established by different nuclear factors, among which are the family of proteins that contain a methyl-CpG-binding domain, commonly known as MBD proteins.Citation23–Citation25 The methyl CpG binding protein 2 (Mecp2) was the first member of this family to be characterized and is able to recognize a single methylated CpG dinucleotide.Citation26–Citation28 In human cancer cells, the binding of MBD proteins to the hypermethylated CpG islands of tumor suppressor genes is associated with transcriptional silencing.Citation29–Citation31 The recent discovery that DNA hypermethylation of CpG islands within the transcription start site of miRNAs causes its transcriptional silencing in cancer cellsCitation32–Citation36 provides a link between classical epigenetic marks, transcriptional repressors and miRNA expression. Most importantly, Lujambio et al.Citation33 have shown that the hypermethylated CpG island of miRNA-124a is occupied by Mecp2.
The relationship between Mecp2 mutations and Rett syndrome (RTT, Online Mendelian Inheritance in Man Database 312750) was discovered in 1999.Citation37 This X-linked neurological disorder causes a severe phenotype in one out of every 10,000–15,000 live births, making it the second most frequent cause of mental retardation in females. Girls with Rett syndrome have apparently normal development for their first 6–18 months of life, after which they gradually lose their faculty of speech, while purposeful hand use is replaced by stereotypical movements consisting of midline hand-wringing, clapping and hand-mouthing. Although head size is normal at birth, its rate of growth decelerates, leading to acquired microcephaly by 2–3 years of age. RTT patients lose their language, cognitive, social and motor abilities, acquiring an initial autistic behavior that later develops to become severe mental retardation. The motor dysfunction includes ataxia, gait apraxia, tremors and spasticity. Other common characteristics include seizures and respiratory abnormalities such as hyperventilation and apnea.Citation38,Citation39 Loss of ambulation and development of scoliosis accompany survival to adulthood. Neuropathological analysis of brains usually reveals a normal structure except for the number and density of synapses and a reduction in the number of dendritic spines as well as the volume of individual neurons.Citation38,Citation39 Given the relevance of miRNAs in neural development, their regulation by DNA methylation and the importance of Mecp2 in brain function, we wondered if Mecp2 may regulate miRNAs involved in nervous development and, thus, explain different features of RTT.
Results and Discussion
Identification of candidate miRNAs misregulated in RTT.
We studied a well-established mouse model of RTT that mimics the human disease.Citation40 We performed miRNA microarray expression analyses using six pairs of wild-type (WT) and Mecp2-null (KO) littermates from the described RTT mouse model. RNA was extracted from total brain and hybridized on a platform containing 365 specific oligonucleotide probes generated from 245 miRNAs.Citation41 We analyzed the resulting data using two approaches. First, we compared all WT mice as a group with the group of all KO mice (unpaired comparison). This assumes that the similarity between the members of each class is greater than that between sibling pairs (see Methods for details). Second, we also made a comparison considering that the sibling pairs might be more similar to each other than to other couples (paired comparison). In this approach we compared the microarray results from each KO animal with its corresponding WT littermate and then analyzed the data from all pairs (see Methods for details). Combining both approaches, we found that 65 of 245 (26%) miRNAs had a different expression pattern in WT and Mecp2-null brain samples, while similar expression was observed in the remaining 180 miRNAs (74%). Among these significantly altered miRNAs, overall downregulation of transcripts was the most common feature (45 of 65, 71%), while upregulation occurred in the other 30% (18 of 65). shows the list of downregulated and upregulated miRNAs in the described RTT mouse model.
We further validated a representative number of deregulated candidate miRNAs obtained by the expression microarray approach using quantitative RT-PCR in a set of eight pairs of WT and KO mouse brains. The most commonly disrupted microRNAs were miR-146a, miR-146b, miR-130, miR-122a, miR-342 and miR-409 (downregulated) and miR-29b, miR329, miR-199b, miR-382, miR-296, miR-221 and miR-92 (upregulated). shows representative qRT-PCR data.
DNA methylation and chromatin immunoprecipitation data.
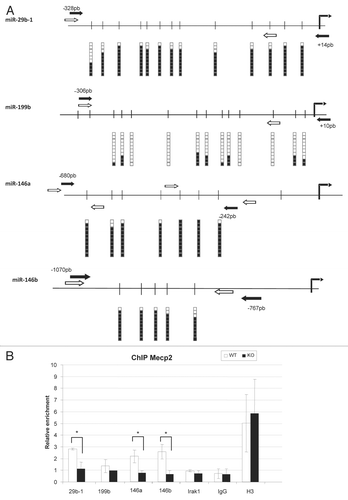
Most of the scientific literature concerning Mecp2, with the intriguing exception of the study by Chahrour et al.,Citation42 suggests that the protein acts as a transcriptional repressorCitation22–Citation24,Citation29,Citation43 that binds with high affinity to methylated DNA.Citation44 Thus, we wondered how miRNA expression, DNA methylation of the miRNA 5′-end loci and Mecp2 occupancy were interconnected in the aforementioned mouse brain context. We focused our study on two upregulated (miR-29b and miR-199b) and two downregulated (miR-146a and miR-146b) RTT-associated miRNAs that were identified and validated in our approach. DNA methylation status of the miRNA 5′-ends was determined by bisulfite genomic sequencing of multiple clones and Mecp2 occupancy was established by quantitative chromatin immunoprecipitation (qChIP). DNA methylation differences in the four studied miRNA 5′-ends were not detected in the WT vs. KO samples (data not shown), highlighting the role of Mecp2 as a “reader” of the methylcytosine signal rather than as a “writer,” the latter function being fulfilled instead by DNA methyltransferases. Most importantly, we found that all three miRNA hypermethylated 5′-end loci (miR-29b, miR-146a and miR-146b) were occupied by the Mecp2 protein, while the protein was absent from the unmethylated 5′-end of miR-199b (). Thus, these results reinforce the notion that Mecp2 has a great “appetite” for methylated cytosine residues. A different matter is the association with the expression of these particular miRNAs. Although the hypermethylated and Mecp2 bound 5′-end of miR-29b fits the long-proposed transcriptional repressor role,Citation22–Citation24,Citation29,Citation43 the findings in miR-146a and miR-146b link to those reported by Chahrour et al.Citation42 In the case of miR-199, which has an unmethylated and Mecp2-unbound 5′-end, the observed upregulation in RTT could be due to indirect mechanisms. Thus, Mecp2 in the context of brain cells might be a versatile protein with a high affinity for methylated DNA that has various transcriptional effects on the underlying genomic sequence. Most importantly, for the identified Rett deregulated miRNAs, such as miR-29 and miR-146b, important roles in the biology neural and glial cell and their associated diseases have been described.Citation45–Citation47
Downregulated miR-146a and miR-146b target the IL-1 receptor-associated kinase 1 (Irak1), an upregulated gene in RTT.
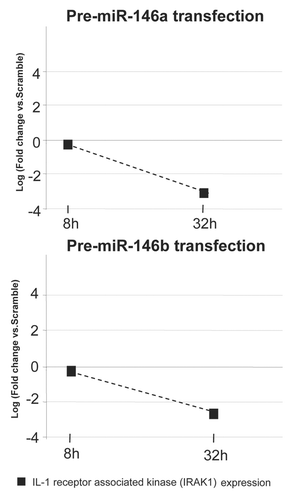
Two independent sources of information provide further clues about the role of two of the identified RTT downregulated miRNAs, miR-146a and miR-146b, in the biology of the disease: first, the observation that miR-146a and miR-146b base paired with sequences in the 3′-UTR of the IRAK1 and post-transcriptionally repressed the gene in generated reporter constructs transfected into 293 cells,Citation48 and second, our previous observation that the IRAK1 gene is upregulated in this RTT mouse model in the absence of Mecp2 directly binding to its promoter in normal brain cells.Citation49 To assess functionally the possible regulatory role of these miRNAs on IRAK1 in a neuron-like context, we transiently transfected miR-146a and miR-146b in neuroblastoma cells. We observed that, upon transfection, IRAK1 expression became downregulated, as measured by conventional and quantitative RT-PCR (). Thus, it is likely that in normal brain one of the functions of miR-146a and miR-146b is to repress IRAK1 expression, whilst in Rett syndrome the downregulation of miR-146a and miR-146b causes the overexpression of their target gene IRAK1, an inflammatory state of the brain tissueCitation50 and contributes to the phenotype of the disease.
Materials and Methods
Animal model.
Four-week-old B6.129P2(C)-Mecp2tm.11Bird/J (stock number: 003890) heterozygous females (Mecp2+/−) were acquired from the Jackson Laboratory (Bar Harbor, ME, USA). In brief, the mutant strain was generated by replacing exons 3 and 4 of Mecp2 in embryonic stem cells with the same exons flanked by loxP sites. Homozygous Mecp2lox/lox females were mated with mice with ubiquitous Cre expression to bring about gene disruption.Citation40 The offspring from the crosses of Mecp2+/− females with C57BL/6J males were genotyped by PCR, as described elsewhere.Citation51
Mouse housing and tissue collection.
Mice were kept under specific pathogen-free conditions in accordance with the recommendations of the Federation of European Laboratory Animal Science Associations. Lighting conditions (lights on from 08:00 h to 20:00 h) and temperature (22°C) were kept constant. Animals were allowed ad libitum access to food and water and were inspected every day. Tissue samples were obtained from hemizygous Mecp2-null males (Mecp2−/y, KO) and their wild-type (WT) littermates after establishing RTT-like symptoms in the defective animals (at about 6–11 weeks of age). Mice were euthanized in accordance with the Guidelines for Humane Endpoints for Animals Used in Biomedical Research. Brains were frozen on dry ice immediately after removal and stored at −80°C until use. WT and KO mouse tissues were extracted and handled under identical conditions.
RNA isolation and hybridization.
Total RNA was extracted by TRIzol treatment (Invitrogen Corp., Carlsbad, CA, USA) following the manufacturer's instructions. miRNA microarray profiling was performed as previously described.Citation41 Briefly, 5 µg of total RNA were biotin-labeled and hybridized on a custom miRNA microarray platform. These arrays contain 365 probes generated from 245 miRNAs from human and mouse and 15 tRNAs (eight human and seven mouse). Every specific 40-mer oligonucleotide probe was spotted in triplicate.Citation41 Biotin-containing transcripts were directly detected by Streptavidin-Alexa Fluor 647 conjugation. Processed slides were scanned on an Axon 4000B microarray scanner and images were quantified using GENEPIX PRO 6.0 (Axon Instruments). Twelve microarrays were performed, from six WT and KO sibling pairs each.
miRNA microarray analyses.
Two different computational analyses of the microarrays were performed. In the first one, average values of the replicate spots of each miRNA were background subtracted, normalized using the quantiles method calculated with Bioconductor package (www.bioconductor.org) and further analyzed selecting the miRNAs measured as present in at least as many samples as the smallest class in the data set (50%); a miRNAs was excluded under the following conditions: less than 20% of expression data values have at least a 1.5-fold change in either directions from the microRNA's median value and missed values were replaced with 4.5 (basic expression value log2). MiRNA signatures were determined by the Class Prediction [BRB-ArrayTools developed by Dr. Richard Simon and Amy Peng Lam (Version: 3.7.0, National Cancer Institute)]. Six methods of prediction were used: compound covariate predictor, diagonal linear discriminant analysis, k-nearest neighbor (using k = 1 and 3), nearest centroid and support vector machines. The output contains the result of the permutation test on the crossvalidated misclassification rate and a list of genes that comprise the predictor, with parametric p values for each gene and the CV-support percent (percent of times when the gene was used in the predictor for a leave-one-out cross-validation procedure). In the second analysis of microarrays, statistical comparisons were performed using Class Comparison t-test [Type of univariate test used: two-sample t-test (with random variance model)]. In this way, the miRNAs able to best separate the groups were identified. Paired class comparison t-test was performed to test the statistically significant comparisons between couples of WT and KO paired arrays. The main difference between these two approaches is that the first one evaluates the data comparing the group of WT mice vs. the group of KO mice without considering that each mouse has its related sibling. And the second analysis takes into account this closer relationship between the couples and compares miRNAs that appear to be different in each couple with those from the other couples.
Quantification of miRNA expression by quantitative realtime polymerase chain reaction (qPCR).
TaqMan MicroRNA assays (Applied Biosystems, Foster City, CA, USA) were used to quantify mature miRNAs as described previously.Citation52 Each specific reverse transcriptase (RT) reaction (Applied Biosystems) contained 15 ng of purified, DNase-treated (turbo DNA-free, Ambion) total RNA. The product was diluted and 2 µl were added for each real-time PCR (1:15 final dilution). Reactions (10 µl) were incubated in an Applied Biosystems 7900HT Fast Real-Time PCR system in 384-well plates. Three reactions were performed to obtain each measurement. Total RNA from 15–25 WT/KO pairs was extracted and studied, and the whole PCR assay was performed at least twice for each miRNA and sample. We used the 2(−ΔΔCt) methodCitation53 to determine relative quantitative expression of individual miRNAs, using sno-142 for normalization. The results were expressed as the relative change (KO/WT). Genes were considered to be upregulated or downregulated if the ratio was greater than 1.5 or less than 0.67, respectively.
Gene expression study by quantitative real-time reverse transcriptase PCR (qRT-PCR).
Total RNA (1 µg) was reverse-transcribed using SuperScript™ II reverse transcriptase (Invitrogen Corp.). A negative control reaction was included to confirm the absence of contamination. Quantitative real-time PCRs were performed as previously described (Urdinguio et al. 2008). Briefly, each reaction mixture contained 25 ng of cDNA, 500 nM of each primer and 10 µl of 2x SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). All measurements were performed in triplicate, including non-template controls. Separate calibration curves were determined for each analysis. PCR reactions were run and analyzed using the 7,900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA). qRT-PCRs were carried out using brain samples from three WT/KO pairs and cell line samples to quantify the expression of the mouse gene Irak1 (forward: 5′-GAC CAA GTA TTT GAA AGA CCT G-3′, reverse: 5′-GTA GTG CCT CCC TGG GTA CA-3′). Expression values were normalized with respect to mouse Gapdh expression (forward: 5′-GGA GCC AAA CGG GTC ATC ATC TC-3′; reverse: 5′-AGA AGA CTG TGG ATG GCC CCT C-3′). Primers were acquired from Sigma-Aldrich.
Chromatin immunoprecipitation assays and PCR quantification (qChIP).
All procedures were performed on ice, with buffers containing protease inhibitors (Complete EDTA-free, Roche Diagnostics, Indianapolis, IN, USA). Brain tissues from WT or KO mice were homogenized in cold phosphate-buffered saline (PBS) with 1% formaldehyde and incubated with agitation for 30 min in a cold room. Chromatin crosslinking was stopped by addition of glycine to a volume of 125 mM and cells were washed in PBS. Subsequent procedures were performed as described elsewhere.Citation54 Chromatin was sheared with a Bioruptor (Diagenode) to an average length of 300–600 bp. The following antibodies were used: rabbit polyclonal anti-Mecp2 (Abcam), anti-total histone H3 (Abcam), negative control IgG antiserum (Upstate Biotechnologies). Quantitative real-time PCR was performed for each specific region using SYBR® Green in an ABI 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Three independent experiments from different WT/KO pairs were tested for each sequence. Quantitative real-time PCR data were analyzed as described by Chahrour et al.Citation42
DNA methylation study.
miRNA sequences were analyzed using miRBase (www.mirbase.org/) and the University of California at Santa Cruz Human Genome Browser (http://genome.cse.ucsc.edu). Considering that 90% of human miRNA promoters are predicted to be located 500 bp upstream of the mature miRNA,Citation55 we decided to study 1,000 bp upstream using the CpG Island Searcher ProgramCitation56 to determine which miRNAs were embedded in a CpG island. DNA methylation status was analyzed by PCR analysis of bisulfite-modified genomic DNA, which induces chemical conversion of unmethylated, but not methylated, cytosine to uracil.
Bisulfite sequencing was performed as previously described.Citation48 In brief, genomic DNA from WT and KO mouse brains was bisulfite-modified after extraction.Citation57 Primers for PCR amplification of the corresponding CpG islands from bisulfite-modified DNA were designed using the Methyl Primer Express® program (www.appliedbiosystems.com/methylprimerexpress). PCR amplicons were purified and subcloned. After clone selection and growth, extracted and purified samples were sequenced using a 3130xl Genetic Analyzer (Applied Biosystems). Ten independent clones were studied from three WT/KO pairs.
Transfection of miRNA precursor molecules in a neuroblastoma cell line.
Mouse neuroblastoma Neuro-2a cell line was obtained from the American Type Culture Collection and grown in 10% FBS in DMEM medium at 37°C in a humidified atmosphere of 5% CO2. miR-146a and miR-146b precursor molecules and negative control miRNAs were purchased from Ambion (Oligo1- PM10722, Oligo2- AM10722, Oligo3- PM10105, Oligo4- AM10105). Oligofectamine (Invitrogen) was used at 100 nmol/l RNA duplexes for the transient transfection of pre-miRNAs. Cells were collected 8, 16, 24, 32, 40, 48, 56, 64 and 72 h after transfection and the expression of mature miRNAs and the Irak1 gene was assessed by qPCR and qRT-PCR, respectively, as described above.
Figures and Tables
Figure 1 miRNAs upregulated and downregulated in the brain of a mouse model for Rett syndrome. Validation by quantitative RT-PCR provided the most commonly disrupted miRNAs. Example of the expression of the miR-29b in pairs wild-type/Mecp2 null brain showing upregulation of the transcript in the disease-associated tissue.
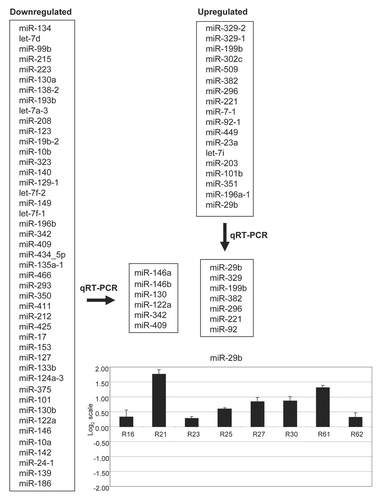
Figure 2 DNA methylation and Mecp2 occupancy analysis for the 5′-end CpG islands of miR-29b, miR-199b, miR-146a and miR-146b. (A) Bisulfite genomic sequencing of multiple clones. Black squares, methylated cytosines; white squares, unmethylated cystosines. Vertical black arrow, transcription start sites of the miRNA. Black arrows, bisulfite genomic sequencing primers; white arrows, chromatin immunoprecipitation primers. (B) Quantitative chromatin immunoprecipitation (ChIP) assays. Mecp2 occupancy is shown in the wild-type brain for the miR-29b-1, miR-146a and miR-146b CpG islands. IgG and H3 are used as negative and positive controls for the ChIP assay.
Acknowledgements
We are grateful to the Catalan and Valencian Rett Syndrome Associations for their support. R.G.U. is funded by the Comunidad de Madrid FPI Programme. The research leading to these results has received funding from the European Community's Sixth Framework Programme (FP7/2007-2013) under grant agreement n° PITN-GA-2009-238242—DISCHROM project, from the Instituto de Salud Carlos III—Ministerio de Sanidad y Consumo Proyecto FIS (Fondo Investigación Sanitaria, Spain) through the E-RARE EuroRETT network, from the Fondation Lejeune (France) and from Grant SAF2007-6-8.
References
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009; 10:126 - 139
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120:15 - 20
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007; 27:91 - 105
- Lim LP, Lau N, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005; 433:769 - 773
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 2003; 115:787 - 798
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA 2007; 104:9667 - 9672
- Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 2008; 4:77 - 85
- Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 2008; 30:460 - 471
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight?. Nat Rev Genet 2008; 9:102 - 114
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can upregulate translation. Science 2007; 318:1931 - 1934
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 2008; 9:219 - 230
- Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet 2008; 9:789 - 796
- Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res 2006; 66:7390 - 7394
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell 2009; 136:26 - 36
- Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet 2008; 24:489 - 497
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci 2006; 7:911 - 920
- Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes and therapies. Lancet Neurol 2009; 11:1056 - 1072
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science 2007; 317:1220 - 1224
- Schaefer A, O'Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, et al. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med 2007; 204:1553 - 1558
- Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci 2008; 28:4322 - 4330
- Esteller M. Epigenetics in cancer. N Engl J Med 2008; 358:1148 - 1159
- Davalos V, Esteller M. MicroRNAs and cancer epigenetics: a macrorevolution. Curr Opin Oncol 2010; 22:35 - 45
- Bienvenu T, Chelly J. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat Rev Genet 2006; 7:415 - 426
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 2006; 31:89 - 97
- Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: reading the wrong words. Br J Cancer 2008; 98:1881 - 1885
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol 1998; 18:6538 - 6547
- Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet 2003; 19:269 - 277
- Dhasarathy A, Wade PA. The MBD protein family-reading an epigenetic mark?. Mutat Res 2008; 647:39 - 43
- Ballestar E, Paz MF, Valle L, Wei S, Fraga MF, Espada J, et al. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J 2003; 22:6335 - 6345
- Lopez-Serra L, Ballestar E, Fraga MF, Alaminos M, Setien F, Esteller M. A profile of methyl-CpG binding domain protein occupancy of hypermethylated promoter CpG islands of tumor suppressor genes in human cancer. Cancer Res 2006; 66:8342 - 8346
- Lopez-Serra L, Ballestar E, Ropero S, Setien F, Billard LM, Fraga MF, et al. Unmasking of epigenetically silenced candidate tumor suppressor genes by removal of methyl-CpG-binding domain proteins. Oncogene 2008; 27:3556 - 3566
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the protooncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 2006; 9:435 - 443
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 2007; 67:1424 - 1429
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA 2008; 105:13556 - 13561
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res 2008; 68:4123 - 4132
- Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res 2009; 69:9038 - 9046
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 1999; 23:185 - 188
- Zoghbi HY. Mecp2 dysfunction in humans and mice. J Child Neurol 2005; 20:736 - 740
- Neul JL, Zoghbi HY. Rett syndrome: a prototypical neurodevelopmental disorder. Neuroscientist 2004; 10:118 - 128
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet 2001; 27:322 - 326
- Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA 2004; 101:9740 - 9744
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. Mecp2, a key contributor to neurological disease, activates and represses transcription. Science 2008; 320:1224 - 1229
- Ballestar E, Ropero S, Alaminos M, Armstrong J, Setien F, Agrelo R, et al. The impact of MECP2 mutations in the expression patterns of Rett syndrome patients. Hum Genet 2005; 116:91 - 104
- Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res 2003; 31:1765 - 1774
- Jiao J, Herl LD, Farese RV, Gao FB. microRNA-29b regulates the expression level of human progranulin, a secreted glycoprotein implicated in frontotemporal dementia. PLoS One 2010; 5:10551
- Xia H, Qi Y, Ng SS, Chen X, Li D, Chen S, et al. microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res 2009; 1269:158 - 165
- Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci USA 2008; 5:6415 - 6420
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NFkappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 2006; 103:12481 - 12486
- Urdinguio RG, Lopez-Serra L, Lopez-Nieva P, Alaminos M, Diaz-Uriarte R, Fernandez AF, et al. Mecp2-null mice provide new neuronal targets for Rett syndrome. PLoS ONE 2008; 3:3669
- Su J, Richter K, Zhang C, Gu Q, Li L. Differential regulation of interleukin-1 receptor associated kinase 1 (IRAK1) splice variants. Mol Immunol 2007; 44:900 - 905
- Miralves J, Magdeleine E, Joly E. Design of an improved set of oligonucleotide primers for genotyping Mecp2tm1.1Bird KO mice by PCR. Mol Neurodegener 2007; 2:16
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 2005; 11:1737 - 1744
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402 - 408
- Deng V, Matagne V, Banine F, Frerking M, Ohliger P, Budden S, et al. FXYD1 is an Mecp2 target gene overexpressed in the brains of Rett syndrome patients and Mecp2-null mice. Hum Mol Genet 2007; 16:640 - 650
- Zhou X, Ruan J, Wang G, Zhang W. Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol 2007; 3:37
- Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol 2003; 3:235 - 240
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996; 93:9821 - 9826