Abstract
MyoD is a master regulator of the skeletal muscle gene expression program. ChIP-Seq analysis has recently revealed that MyoD binds to a large number of genomic loci in differentiating myoblasts, yet only activates transcription at a subset of these genes. Here we discuss recent data suggesting that the ability of MyoD to mediate gene expression is regulated through the function of Polycomb and Trithorax Group proteins. Based on studies of the muscle-specific myog gene, we propose a model where the transcriptional activators Mef2d and Six4 mediate recruitment of Trithorax Group proteins Ash2L/MLL2 and UTX to MyoD-bound promoters to overcome the Polycomb-mediated repression of muscle genes. Modulation of the interaction between Ash2L/MLL2 and Mef2d by the p38α MAPK signaling pathway in turns provides fine-tuning of the muscle-specific gene expression program. Thus Mef2d, Six4, and p38α MAPK function coordinately as regulators of a master regulator to mediate expression of MyoD target genes.
Introduction
MyoD is a master regulator of skeletal myogenesisCitation1 due to its ability to initiate the myogenic program in myoblasts,Citation2 fibroblastsCitation3 and a variety of other cell types.Citation4 Upon conditions permissive to myogenesis, MyoD heterodimerizes with the more ubiquitously expressed E-proteinsCitation5 to establish a specific and temporally ordered gene expression programCitation6 giving rise to multinucleated myotubes. This muscle-specific gene expression program is initiated by MyoD and proceeds via a feed-forward mechanismCitation7 to activate genes which possess the consensus E-box sequence VCASCTG (where V is A, C or G while S is C or G) within their promoter/enhancer regions.Citation8–Citation11 Reporter assays performed in vivoCitation12 and in vitroCitation13,Citation14 demonstrate that MyoD directs high level expression of artificial templates containing multimerized E-boxes. This strong transactivation potential is likely empowered through its ability to interact with multiple transcriptional regulatory factors. Indeed, once associated with the promoter region, MyoD recruits the acetyltransferases p300 (leading to acetylation of histones H3 and H4),Citation11,Citation15–Citation18 and pCAF (which acetylates MyoD).Citation14,Citation19 MyoD can also interact with the basal transcriptional machinery,Citation20,Citation21 the ATP-dependent chromatin remodeling factor SWI/SNF,Citation17,Citation22,Citation23 the arginine methyltransferase PRMT5,Citation24 and the transcriptional elongation stimulating factor pTEFb.Citation25,Citation26 Its ability to interact with a number of ubiquitously expressed transcriptional regulators provides insight into the mechanism through which MyoD activates the muscle-specific gene expression program in multiple cell types. However, recent genome-wide analysis has revealed that the binding of MyoD to specific promoters or enhancers is not sufficient to activate gene expression. Indeed, high-throughput ChIP-Seq analysis performed in differentiating myoblasts indicates that MyoD binds to ∼25,000 sites throughout the genome, while only 1,953 genes demonstrate modified expression during myogenesis.Citation11 The limited number of differentially expressed genes observed in response to extensive MyoD occupancy at genomic loci suggests that this master regulator of gene expression must itself be regulated to orchestrate the temporally ordered muscle-specific transcriptional program.
While the mechanism by which the transcriptional activity of MyoD is regulated on a genome-wide scale remains poorly defined, recent studies have implicated Polycomb Group (PcG) and Trithorax Group (TrxG) proteins in regulating MyoD activity to ensure proper temporal and spatial expression of muscle genes.Citation16,Citation17,Citation27,Citation28 For example, YY1 binds to promoters/enhancers of the muscle-specific MHCIIb and CKm genes to mediate the recruitment of the PcG methyltransferase Ezh2, which establishes the repressive histone H3 lysine 27 trimethyl (H3K27me3) mark across these loci.Citation27 The dominant nature of the PcG-mediated H3K27me3 mark may play an important role in ensuring that MyoD is unable to activate transcription in the absence of the antagonizing activity of TrxG proteinsCitation29–Citation31, thereby limiting the number of loci expressed in response to MyoD binding. While genome-wide studies of H3K27me3 at different stages of myogenesis have not yet been performed, it is clear that multiple muscle-specific genes are marked for repression by PcG family members in both growing myoblastsCitation27,Citation28 and fibroblasts.Citation32 Based on these observations, we tested the hypothesis that TrxG proteins antagonize PcG-mediated repression of muscle loci by removing the repressive H3K27me3 mark and replacing it by a transcriptionally permissive histone H3 lysine 4 trimethyl (H3K4me3) mark.
To understand how TrxG proteins antagonize PcG-mediated repression in muscle, the myogenin (myog) gene represents an excellent model since a DNA fragment consisting of the −133 to +18 bp region of the myog promoter is sufficient to confer muscle-specific expression of a lacZ reporter in transgenic mice.Citation33,Citation34 Using the myog promoter as a model system, we found that the homeobox transcription factor Six4 and the MADS-box transcription factor Mef2d collaborate with MyoD to establish transcriptional competency at this gene.Citation16,Citation28 Furthermore, we showed that the role of Six4 is to recruit the histone H3K27 demethylase UTX to mediate a localized demethylation of H3K27me3 within the promoter of the gene.Citation28 Indeed, knockdown of Six4 in differentiating myotubes prevents recruitment of UTX to the myog promoter and inhibits the demethylation of H3K27me3 at this locus.Citation28 Further supporting a role for Six4 in overcoming the repressive effects of PcG proteins is the observation that mutation of the Six4 binding site in the minimal myog promoter leads to loss of lacZ expression in transgenic mouse embryos.Citation35 Taken together, these results provide a mechanism to explain recent findings that Six4 acts synergistically with MyoD to activate the myog promoter during myogenesis.Citation36
While Six4 mediates removal of the H3K27me3 mark, the role of Mef2d is to permit marking of the myog gene with the transcriptionally permissive H3K4me3 modification. Indeed, using a combination of in vitro and in vivo techniques we showed that Mef2d recruits the Ash2L/MLL2 methyltransferase complex to the myog promoter thereby allowing the methylation of H3K4 to permit transcriptional activation.Citation16 Interestingly, we found that the interaction between Ash2L/MLL2 and Mef2d is not constitutive, but instead is the result of a highly regulated cell signaling mechanism that involves direct phosphorylation of Mef2d by the p38α MAP kinase (MAPK).Citation16 Further supporting a major role for p38α-mediated phosphorylation of Mef2d in the activation of muscle-specific genes is the finding that precocious expression of Mef2d and activated MKK6 kinase (which activates p38 MAPK signaling) in fibroblasts undergoing MyoD-induced skeletal myogenesis leads to early expression of several MyoD target genes.Citation7 The fact that recruitment of Ash2L/MLL2 to muscle-specific promoters depends on p38α-mediated phosphorylation of Mef2d provides insight into the mechanism by which p38α increases transcriptional activity of Mef2 family membersCitation37 and is consistent with the critical role of this kinase in promoting myogenesis.Citation38–Citation40 Furthermore, it provides an explanation for the observation that Mef2d-mediated recruitment of RNA Pol II is independent of p38α MAPK signaling although it is necessary for transcriptional elongation at muscle genes.Citation7 Interestingly, while Ash2L/MLL2 interacts most strongly with phosphorylated Mef2d, this TrxG complex can also interact with phosphorylated Mef2c, but not phosphorylated Mef2a.Citation16 This variable ability of different Mef2 isotypes to interact with Ash2L/MLL2 suggests some degree of functional specificity between the three members of the Mef2 family of transcriptional activators expressed in muscle. Taken together, these findings have revealed several levels of regulation to ensure proper spatial and temporal patterning of muscle-specific gene expression.
Consistent with our studies demonstrating a cooperation between MyoD, Six4 and Mef2d in the activation of the myog gene,Citation16,Citation28 genome-wide studies by Cao et al.Citation11 identified enrichment of Mef2 binding sites at MyoD target genes undergoing transactivation, but not at genes whose expression remains unchanged during myogenesis. Similarly, ChIP-microarray studies have identified enrichment for both Mef2 and Six4 binding elements adjacent to muscle regulatory factor bound E-boxes during myogenesis.Citation10 These findings further support an important role for Mef2 and Six4 as key modulators of MyoD transcriptional activity, allowing this master regulator of myogenesis to overcome PcG-mediated transcriptional repression at specific loci to establish the muscle gene expression program.
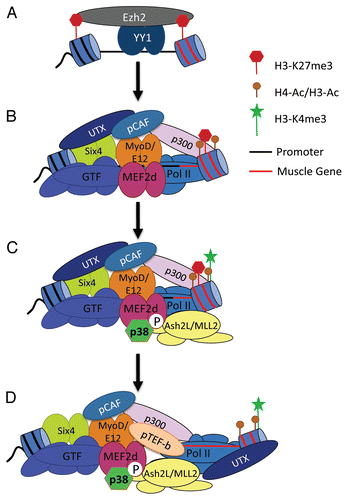
To explain the activation of MyoD-dependent transcription during myogenesis, we propose the following model (). In a first step the binding of MyoD, Six4 and Mef2d within the transcriptional regulatory region of muscle-specific genes leads to the formation of a poised promoter,Citation41 which is loaded with the general transcriptional machinery,Citation20,Citation21 RNA Pol II,Citation7 the acetyltransferases p300 and pCAFCitation15–Citation17 and the histone demethylase UTX.Citation28 The presence of p300 leads to acetylation within the nucleosomes adjacent to MyoD binding sitesCitation15–Citation17 while UTX mediates a localized demethylation of H3K27me3 within the promoter/enhancer region of the gene.Citation28 Meanwhile, the coding region of the gene remains enriched for the repressive H3K27me3 mark, preventing transcription.Citation28 Upon cell-cell contact, activation of the MAPK signaling pathway (possibly via CDO receptor mediated cascades)Citation42 results in p38α-directed phosphorylation of the transactivation domain of Mef2d.Citation7,Citation16 Phosphorylated Mef2d in turn recruits Ash2L/MLL2-containing methyltransferase complexes to the promoter, leading to formation of the transcriptionally permissive H3K4me3 mark at the 5′-end of the gene.Citation16 The transcriptionally permissive mark then allows engagement of the RNA Pol II and phosphorylation of its C-terminal domain (CTD) at Serine2 via MyoD-dependent recruitment of pTEF-b.Citation26 Upon phosphorylation of the CTD, UTX associates with the elongating polymeraseCitation43 and removes the repressive H3K27me3 mark across the coding region of the gene.Citation28 According to this model the ability of MyoD to activate gene expression is regulated by the antagonistic activities of PcG (Ezh2) and TrxG (Ash2L/MLL2 and UTX) through a mechanism mediated by the transcriptional activators Mef2d, Six4 and modulated by p38α MAPK signaling. It is interesting to note that recruitment of the TrxG protein complex SWI/SNF to the myog promoter is also modulated by p38α MAPK signaling.Citation17 The functional relationship between these two TrxG protein complexes (Ash2L/MLL2 and SWI/SNF) at the myog promoter remains to be determined.
Our finding that muscle-specific expression of myog is directed through a Mef2d-dependent mechanism is consistent with transgenic mouse studies demonstrating that mutation of the Mef2 site within the proximal myog promoter inhibits expression in limb buds and a subset of cells in the somite myotome at day 11.5 p.c.Citation33,Citation34 However, this promoter element is not required for expression of myog in somites anterior to somite 10 at this stage of development. Furthermore, by day 12.5 p.c. expression of the transgene is observed in the limb buds.Citation33,Citation34 There are two possible explanations for this finding. The first possibility is that the same Mef2 element is required for recruitment of the transcriptionally repressive PcG complex onto the muscle-specific promoter at specific stages of development. In that case, the lack of PcG protein recruitment may negate the requirement for TrxG to establish transcriptional competence.Citation44 However, since expression of myog was not observed outside the myogenic lineages, it is more likely that at different developmental stages, alternate transcriptional activators are responsible for recruiting the Ash2L/MLL2-containing methyltransferase complexes to activate the myog promoter in a spatially restricted manner. Indeed, several transcription factors can recruit the Ash2L/MLL2 complex to specific genes. For example, in activated satellite cells Ash2L/MLL2 is recruited to the Myf5 promoter by the transcriptional regulator Pax7.Citation45 Since the transgenic mouse studies showing muscle-specific reporter expression in the absence of the conserved Mef2 binding site used an extended myog promoter of 1,565 bp, it is currently unclear which additional DNA binding sites are involved in recruiting Ash2L/MLL2-containing methyltransferase complexes to the myog gene at alternate spatio-temporal stages. It is important to note that the use of alternate DNA sequences to promote expression of a muscle-specific gene in distinct regions of the embryo is not without precedence. Indeed, extensive mapping of the Myf5/Mrf4 locus during murine development has led to the identification of multiple enhancer elements which act to modulate expression of Myf5 within the muscle cell lineage at specific developmental stages.Citation46 Future studies looking at the recruitment of the Ash2L/MLL2 complex to myog promoter in muscle at different stages of development will be facilitated by advances in chromatin immunoprecipitation (ChIP) protocols which currently require less than 100 cells.Citation47
While we are beginning to understand how tissue-specific gene expression is regulated, it remains to be determined how a high level of gene expression is established at particular loci. One consideration is the cell-type specific nuclear reorganization that occurs during differentiation.Citation48,Citation49 Recent studies in C. elegans show that the muscle-specific Myo-3 promoter localizes to the nuclear envelope in non-muscle cells while shifting to the nuclear lumen where it is transcribed at high levels in cells of the muscle lineage.Citation50 While muscle-specific expression of the Myo-3 transgene is observed in the nuclear periphery at early stages of differentiation, transcriptional output from the locus is greatly increased upon transition to transcription factories located in the nuclear lumen.Citation50 Based on these observations, it is interesting to note that while the −133 to +18 bp region of the myog promoter is sufficient for muscle-specific gene expression, high level expression of the transgene is only observed when a region of −1,092 to +18 of the promoter is included in the construct.Citation33,Citation34 This suggests the existence of a transcriptional enhancer in the region upstream of the promoter, which may facilitate incorporation of the myog gene into the transcription factories of the nuclear lumen. The mechanism by which cells undergo extensive nuclear reorganization during myogenesis is not clear. An interesting possibility was suggested by a recent study demonstrating a critical role for the caspase activated nuclease (CAD) in the myogenic process whereby it generates transient DNA strand breaks.Citation51 It is enticing to propose that the role of CAD in myogenesis could be to facilitate reorganization of chromosomes in the context of the densely packed nucleus.
To conclude, we propose that while MyoD binds to a large number of sites throughout the genome, its ability to regulate transcription is restricted by the additional requirement for Six4 and Mef2d to overcome the repressive effects of PcG-mediated histone methylation. The need for MyoD, Six4 and Mef2d to interact at specific promoters suggests a mechanism by which spatial and temporal regulation of skeletal muscle gene expression program could be achieved. Furthermore, the compounding need for p38α MAPK signaling provides a fine-tuning mechanism that ensures a highly regulated spatio-temporal expression pattern for these genes. Thus our findings have revealed a previously unappreciated mechanism to control the potency and function of cell fate determinants. Interestingly, unlike MyoD whose expression is restricted to cells of the muscle lineage, Six4 and Mef2d are expressed in multiple tissue types. Similarly, p38α MAPK signaling is involved in directing differentiation of multiple cell types. Thus, it is interesting to speculate that in other tissues, alternate bHLH transcriptional activators might collaborate with Six4, Mef2d and p38α MAPK to regulate cell-specific gene expression through a similar mechanism.
Figures and Tables
Figure 1 Model for the coordinate activation of PcG-repressed muscle genes. (A) The transcriptional repressor YY1 targets Ezh2 to muscle-specific genes, establishing the repressive H3K27me3 mark across the locus. (B) MyoD binding at the promoter, in conjunction with Mef2d and Six4, establishes a transcriptionally poised promoter characterized by a localized demethylation of H3K27me3 (limited to the promoter) and the presence of acetylated histones. However, transcriptional competency is not achieved due to the presence of repressive H3K27me3 mark within the gene. (C) Phosphorylation of Mef2d by p38 MAPK allows the recruitment of Ash2L/MLL2 complex leading to H3K4me3 within the gene. (D) Phosphorylation of the CTD of RNA Pol II allows the transfer of UTX onto the elongating polymerase to mediate demethylation into the gene, permitting muscle-specific gene expression. See text for further details.
Acknowledgements
We thank Marjorie Brand, Lynn Megeney and Lawrence Puente for insightful discussions and comments on this manuscript. Work in the Dilworth laboratory is supported by the Canadian Institutes of Health Research (MOP-77778 and MOP-93777). F.J.D. is a Canadian Research Chair in the Epigenetic Regulation of Transcription.
References
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 2005; 132:38 - 40
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 1993; 75:1351 - 1359
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987; 51:987 - 1000
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA 1989; 86:5434 - 5438
- Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, et al. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 1991; 66:305 - 315
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell 2002; 9:587 - 600
- Penn BH, Bergstrom DA, Dilworth FJ, Bengal E, Tapscott SJ. A MyoD-generated feed forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev 2004; 18:2348 - 2353
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 1989; 58:537 - 544
- Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 1990; 250:1104 - 1110
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev 2005; 19:553 - 569
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell 2010; 18:662 - 674
- Davis RL, Weintraub H. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science 1992; 256:1027 - 1030
- Bengal E, Flores O, Rangarajan PN, Chen A, Weintraub H, Verma IM. Positive control mutations in the MyoD basic region fail to show cooperative DNA binding and transcriptional activation in vitro. Proc Natl Acad Sci USA 1994; 91:6221 - 6225
- Dilworth FJ, Seaver KJ, Fishburn AL, Htet SL, Tapscott SJ. In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation. Proc Natl Acad Sci USA 2004; 101:11593 - 11598
- Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell 1997; 1:35 - 45
- Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, et al. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol 2007; 14:1150 - 1156
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet 2004; 36:738 - 743
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, et al. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J 2006; 25:502 - 511
- Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, et al. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell 1999; 4:725 - 734
- Heller H, Bengal E. TFIID (TBP) stabilizes the binding of MyoD to its DNA site at the promoter and MyoD facilitates the association of TFIIB with the preinitiation complex. Nucleic Acids Res 1998; 26:2112 - 2119
- Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell 2008; 32:96 - 105
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, et al. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol 2005; 25:3997 - 4009
- Li ZY, Yang J, Gao X, Lu JY, Zhang Y, Wang K, et al. Sequential recruitment of PCAF and BRG1 contributes to myogenin activation in 12-O-tetradecanoylphorbol-13-acetate-induced early differentiation of rhabdomyosarcoma-derived cells. J Biol Chem 2007; 282:18872 - 18878
- Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol Cell Biol 2007; 27:384 - 394
- Simone C, Stiegler P, Bagella L, Pucci B, Bellan C, De Falco G, et al. Activation of MyoD-dependent transcription by cdk9/cyclin T2. Oncogene 2002; 21:4137 - 4148
- Giacinti C, Bagella L, Puri PL, Giordano A, Simone C. MyoD recruits the cdk9/cyclin T2 complex on myogenic-genes regulatory regions. J Cell Physiol 2006; 206:807 - 813
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 2004; 18:2627 - 2638
- Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, et al. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J 2010; 29:1401 - 1411
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 2007; 8:9 - 22
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell 2007; 128:735 - 745
- Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol 2008; 20:201 - 207
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 2006; 20:1123 - 1136
- Yee SP, Rigby PW. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev 1993; 7:1277 - 1289
- Cheng TC, Wallace MC, Merlie JP, Olson EN. Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science 1993; 261:215 - 218
- Spitz F, Demignon J, Porteu A, Kahn A, Concordet JP, Daegelen D, et al. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc Natl Acad Sci USA 1998; 95:14220 - 14225
- Liu Y, Chu A, Chakroun I, Islam U, Blais A. Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic Acids Res 2010; In press
- Han J, Jiang Y, Li Z, Kravchenko VV, Ulevitch RJ. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 1997; 386:296 - 299
- Zetser A, Gredinger E, Bengal E. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J Biol Chem 1999; 274:5193 - 5200
- de Angelis L, Zhao J, Andreucci JJ, Olson EN, Cossu G, McDermott JC. Regulation of vertebrate myotome development by the p38 MAP kinase-MEF2 signaling pathway. Dev Biol 2005; 283:171 - 179
- Perdiguero E, Ruiz-Bonilla V, Gresh L, Hui L, Ballestar E, Sousa-Victor P, et al. Genetic analysis of p38 MAP kinases in myogenesis: fundamental role of p38alpha in abrogating myoblast proliferation. EMBO J 2007; 26:1245 - 1256
- Brookes E, Pombo A. Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep 2009; 10:1213 - 1219
- Takaesu G, Kang JS, Bae GU, Yi MJ, Lee CM, Reddy EP, et al. Activation of p38alpha/beta MAPK in myogenesis via binding of the scaffold protein JLP to the cell surface protein Cdo. J Cell Biol 2006; 175:383 - 388
- Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, et al. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol 2008; 28:1041 - 1046
- Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep 2004; 5:373 - 377
- McKinnell IW, Ishibashi J, Le Grand F, Punch VG, Addicks GC, Greenblatt JF, et al. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol 2008; 10:77 - 84
- Carvajal JJ, Keith A, Rigby PW. Global transcriptional regulation of the locus encoding the skeletal muscle determination genes Mrf4 and Myf5. Genes Dev 2008; 22:265 - 276
- Dahl JA, Collas P. MicroChIP—a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res 2008; 36:15
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol 2006; 7:540 - 546
- Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev 2006; 20:1447 - 1457
- Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev 2010; 24:766 - 782
- Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci USA 2010; 107:4230 - 4235