Abstract
Lower levels of LINE-1 methylation in peripheral blood have been previously associated with risk of developing non-communicable conditions, the most well-explored of these being cancer, although recent research has begun to link altered LINE-1 methylation and cardiovascular disease. We examined the relationship between LINE-1 methylation and factors associated with metabolic and cardiovascular diseases through quantitative bisulfite pyrosequencing in DNA from peripheral blood samples from participants of the Samoan Family Study of Overweight and Diabetes (2002-03). The sample included 355 adult Samoans (88 men and 267 women) from both American Samoa and Samoa. In a model including all sample participants, men had significantly higher LINE-1 methylation levels than women (p=0.04), and lower levels of LINE-1 methylation were associated with higher levels of fasting LDL (p=0.02) and lower levels of fasting HDL (p=0.009). The findings from this study confirm that DNA "global" hypomethylation, (as measured by methylation at LINE-1 repeats) observed previously in cardiovascular disease is associated with altered levels of LDL and HDL in peripheral blood. Additionally, these findings strongly argue the need for further research, particularly including prospective studies, in order to understand the relationship between LINE-1 DNA methylation measured in blood and risk factors for cardiovascular disease.
Introduction
The Samoan islands, composed of the independent nation of Samoa and the US territory of American Samoa, are geographical neighbors and are currently experiencing economic development and its associated nutritional transition at different rates, although both populations are characterized by alarmingly high prevalence of obesity.Citation1 American Samoa has a higher prevalence of obesity compared with Samoa, with approximately 71% of women and 59% of men defined as obese (using Polynesian standards of BMI >32), relative to 53% of women and 29% of men in Samoa.Citation1 These high prevalences of obesity has lead to rapid rises in obesity-related diseases such as cardiovascular disease (CVD) and type 2 diabetes.Citation2
The rapid temporal rise of Samoan obesity and obesity-related diseases has been attributed to modernization and its associated nutritional transition, in which these developing nations are consuming more calorie-rich foods and expending less energy.Citation1 Although these behavioral factors are linked to obesity and obesity-related diseases, genetic factors have also been shown to play an important role in Samoan obesity and obesity-related risk factors.Citation3–Citation6
Both Samoan islands were settled by Polynesians approximately 3,000 y ago.Citation7 Genetic evidence suggests that these island nations were originally settled by small groups of voyagers.Citation7,Citation8 These original settlers may have endured food shortages and cold night-time open-ocean temperatures, perhaps favoring those with the ability to store body fat and those with efficient energy metabolism, thus suggesting a role for a thrifty genotype.Citation9 Although certain genetic loci have been associated with obesity phenotypes in Samoans,Citation3–Citation6 studies examining potential links of epigenetic alterations with these extreme metabolic phenotypes are currently lacking in this population.
Epigenetic alterations are DNA modifications that do not involve changes to the sequence, yet alter gene expression.Citation10 Epigenetic regulation of gene expression is based upon complex alterations in histone proteins, affecting chromatin conformation. These changes have been associated with coordinate changes in DNA methylation of cytosine bases in the context of CG dinucleotide pairs that often reside in gene promoter regions (CpG islands) or in DNA repeat regions.Citation11 DNA methylation is heritable, in that it may be passed from mother to daughter cell and stable, due to the fact that it is not easily reversed.Citation12 The detailed evaluation of the profile of DNA methylation in affected tissues, as well as in the peripheral blood, has recently become an important tool in cancer research, and the potential for epigenetic change to affect a multitude of other diseases has lead to an interest in evaluating the association of changes in DNA methylation with risk factors for other conditions.Citation13 Altered levels of DNA methylation in DNA sequence repeat regions in separate tissues, as well as in peripheral blood, have been associated with cancer and cancer risk factors.Citation14,Citation15
It is estimated that over half of the DNA within the human genome is made up of repetitive sequences such as transposons, retrotransposons and endogenous retroviruses.Citation16 These sequences are generally non-transcribed due to their constant heterochromatic state maintained by hypermethylation. Examination of methylation at these repeat sequences has become an important tool in disease research and is often referred to as “global” methylation. There are several types of repeat sequences in the human genome that are measured in research. Long Interspersed Nuclear Elements (LINE-1) are retrotransposon sequences that make up approximately 17% of the human genome.Citation16 Measuring methylation at LINE-1 sequences in lymphocyte-derived DNA is a well-recognized method for examining what often has been termed genome-wide or “global” methylation.Citation15,Citation17,Citation18
Preliminary studies have suggested that DNA methylation, including LINE-1 methylation, may play a role in chronic diseases other than cancer; specifically, cardiovascular disease and diabetes.Citation19–Citation24 There is also evidence to suggest that environmental exposures are related to DNA repeat sequence hypomethylation at these repeat regions.Citation25 The methyl groups that are substrates for DNA methylation are provided by diet and, therefore, dietary factors are candidates for playing an essential role in maintenance and regulation of DNA methylation.Citation14,Citation22
Here, we investigated several risk factors for metabolic and cardiovascular diseases by assessing their relationship with LINE-1 DNA methylation in women selected for a study of menstrual patterns and their spouses from a larger sample from American Samoa and Samoa.
Results
Socio-demographic characteristics of the study sample are shown in . The subjects were made up of slightly more American Samoans than Samoans (55.8% and 44.2%, respectively). The mean age among American Samoans was 31.6 y (±7.1) and 31.2 y (±7.3) among Samoans. Men were significantly older than women in both American Samoa (p < 0.0001) and Samoa (p < 0.0001). Among both men and women, American Samoans were significantly more likely to have at least a secondary education, higher BMIs, higher levels of fasting insulin and lower levels of fasting HDL. There was a higher proportion of men in American Samoa reporting alcohol consumption than in Samoa (p = 0.02), as well as higher levels of DBP among men in American Samoa than in Samoa (p < 0.0001). Samoan men were significantly older than American Samoan men (p = 0.01).
In both locations, women had higher BMIs than men, though this finding was only significant in Samoa (p = 0.046). Men were significantly more likely to be current alcohol users than women (p = 0.02 American Samoa; p = 0.0005 Samoa) or current cigarette smokers (p = 0.02 American Samoa; p = 0.0001 Samoa). American Samoan men had significantly lower levels of HDL, higher levels of fasting LDL, and higher levels of fasting glucose than American Samoan women (p = 0.002, p = 0.003, and p = 0.003, respectively). American Samoan men also had higher SBP (p < 0.0001) and DBP (p < 0.0001) levels than American Samoan women. Samoan women had higher levels of fasting insulin than Samoan men (p = 0.0005).
The overall range of DNA LINE-1 methylation in American Samoans and Samoans combined was 75.9% to 89.1% with a mean (SD) of 83.2% (1.8) and 82.8% (2.0) for men and women, respectively. LINE-1 methylation was significantly higher in men than in women among the entire sample (p = 0.04); however, there was no significant difference in LINE-1 methylation between American Samoans and Samoans (data not shown). Among the entire study sample, there was a significant positive association between BMI and LINE-1 methylation (p = 0.007), and HDL and LINE-1 methylation (p = 0.009), as well as a significant negative association between LDL and LINE-1 methylation (p = 0.02) in a multivariate model containing all of these variables, while controlling for gender and age (). Adding location to the model did not alter the relationship of LINE-1 methylation with sex, lipid profile or BMI (data not shown). When men and women were examined separately using the same multivariate model, LDL and HDL relationships with LINE-1 methylation were only significant among men (p = 0.007 for both; ), whereas the association between BMI and LINE-1 methylation was only significant among women (p = 0.03; ), although the direction of the effect estimates for all of the associations were consistent with the overall model. shows the unadjusted plots, stratified by sex, of HDL and LDL with LINE-1 DNA methylation level.
When LDL and HDL were dichotomized by clinically relevant levels, elevated levels of LDL (≥100 mg/dL) were significantly associated with lower levels of LINE-1 methylation among women (p = 0.04; ).
To determine if multiple lipoprotein risk factors acted additively in their association with LINE-1 methylation, individuals were grouped based on their LDL and HDL dichotomized levels. Individuals with both high LDL and low HDL were given a risk score of 2, those with either high LDL and high HDL, or low HDL and low LDL a risk score of 1, and those with both low LDL and high HDL a risk score of 0. In both men and women, a positive association was observed for increasing number of risk factors and decreasing levels of LINE-1 methylation, however this association was only significant among women (p = 0.13 in men and p = 0.04 in women; ).
These same associations in all models were observed when identical models were run in subsample populations in which siblings were removed (data not shown).
Discussion
The present study examined one ethnic group, Samoans, characterized by high prevalence of obesity and cardiovascular and metabolic diseases risk factors residing in two different nations with different patterns of economic development and changes in way of life. Men were found to have significantly higher levels of LINE-1 methylation than women, consistent with the literature, motivating our exploratory stratified analyses by gender. Higher levels of LINE-1 methylation in men than in women have been previously reported in reference Citation14, Citation15, Citation18 and Citation30. It has been suggested that this difference may be due to X-chromosome inactivation, but our analysis cannot address this directly.Citation31 Hormonal differences between men and women might also contribute to this difference in LINE-1 methylation.
In the combined sample of both men and women, lipoproteins (LDL and HDL) were found to be associated with levels of LINE-1 methylation, with lower levels of LINE-1 methylation associated with higher LDL and lower HDL levels. This is consistent with a recent prospective study that, at baseline, showed that healthy men with lower levels of LINE-1 methylation were more likely to develop ischemic heart disease.Citation20 It remains to be determined if altered LINE-1 DNA methylation is a cause or a consequence of cardiovascular anomalies. Homocysteine levels have long been known to be associated with cardiovascular risk and homocysteine is also involved in the methyl group donor pool, in which higher levels of homocysteine are associated with lower levels of available methyl groups.Citation32 Accordingly, DNA hypomethylation has been found to be associated with hyperhomocysteinaemia,Citation33,Citation34 although the role of hyperhomocysteinaemia in cardiovascular disease remains unknown.Citation32 It is possible that hyperhomocysteinaemia somehow creates cardiovascular risk through DNA hypomethylation in a substantial fraction of the genome, or perhaps these are simply markers for atherosclerotic inflammation.
Cardiovascular disease is characterized by atherosclerotic plaques, resulting in various cardiovascular morbidities and mortalities.Citation35 Elevated levels of peripheral LDL, as well as decreased levels of peripheral HDL are known risk factors for atherosclerosis.Citation36 DNA methylation in a substantial fraction of the genome has been related to atherosclerosis and has been found to precede the formation of atherosclerosis in Apoe-/- mice.Citation37,Citation38 Since Apoe-/- mice have higher LDL and lower HDL levelsCitation39 it is possible that lipoproteins in peripheral blood influence DNA methylation, including LINE-1 methylation, potentially contributing to DNA hypomethylation that has been observed in atherosclerotic tissues.Citation40–Citation42
A prominent feature of atherogenesis is an inflammatory response, in which inflammatory cells, including lymphocytes, become upregulated.Citation43 Previous work has shown that different cell types have unique patterns of DNA methylation.Citation44 Specifically, different inflammatory cells from peripheral blood samples have been shown to have varied levels of LINE-1 methylation; specifically, lymphocytes were associated with lowered levels of LINE-1 methylation.Citation18 Work by the same group also found that serum levels of VCAM-1, which are responsible for adhesion of inflammatory cells onto vascular endothelium, aiding in atherosclerosis, were associated with LINE-1 hypomethylation.Citation45 Our work, along with previous studies, suggests that the DNA hypomethylation that is observed in cardiovascular disease may in fact be a consequence of an inflammatory response, where an influx of inflammatory cells within peripheral blood alters the levels of DNA methylation at repeat regions as a result of a change in cellular profile within whole blood.
Finally, we found a significant positive association between LINE-1 methylation and BMI that was particularly prominent among women. Although the mechanism responsible for this relationship is unclear, it is of interest due to the fact that increased BMI is a risk factor for cardiovascular disease. Prior work in substantially less obese populations has not observed a significant association of BMI with LINE-1 methylation, although the direction of the (non-significant) associations was the same as we observed in Samoa and American Samoa.Citation18,Citation46,Citation47 Prospective studies will be needed in order to disentangle the apparently complex relationships between BMI, lipoprotein levels, cardiovascular disease and LINE-1 methylation.
Strengths of this study included the population-based study design, quantitative pyrosequencing to determine LINE-1 methylation and comprehensively collected data on a wide variety of population characteristics. The limitation of this study was set by the fact that a small portion of this sample was related, which could possibly affect LINE-1 levels, although our analysis did not suggest that this was a significant source of variability. Sensitivity analyses showed very similar model results between the full study sample and a smaller sample removing one or more members of the sibling pairs or triplets. The true heritability of LINE-1 DNA methylation remains unknown.
In conclusion, we have shown that LINE-1 methylation gender differences exist in this Samoan population, and that serum levels of fasting LDL and HDL are important correlates of LINE-1 methylation, in which risk levels of LDL and HDL are associated with lowered levels of LINE-1 methylation. The temporal sequence of LINE-1 hypomethylation and atherosclerosis is unclear and further longitudinal studies of human groups, including Samoans, are needed. Homocysteine levels were not measured in this population, so we could not determine whether or not homocysteine was associated with LINE-1 methylation in this population. We also did not look at the subcellular components of the peripheral blood that was collected and, therefore, could not explore the relationships between inflammatory cells and LINE-1 methylation. It would be interesting to conduct longitudinal analyses in order to more clearly understand the relationships between homocysteine, inflammation and DNA methylation in the development of cardiovascular disease; this is critically important due to the fact that cardiovascular disease is currently the top cause of death globally.Citation48
Materials and Methods
Subjects.
Subjects in this study were part of large pedigrees who participated in the Samoan Family Study of Overweight and Diabetes, with data collected in 2002–03, and described previously in reference Citation3, Citation4 and Citation6. Briefly, recruitment began in American Samoa based on random selection of probands who participated in the 1990–94 cohort study and had at least two adult siblings alive and residing in American Samoa. Recruitment in Samoa began in 2003 and first involved participants who were members of American Samoan families involved in the 2002 recruitment. Further villages were then selected throughout the nation to achieve geographic and economic diversity and families were chosen based on maximum number of available adult siblings. Protocols for this study were approved by the Brown University Institutional Review Board, the Government of Samoa, the Samoan Ministry of Health and the Samoan Health Research committee. Written informed consent was obtained from all participants.
The women included in this analysis were part of a previously defined subsample, derived from the Samoan Family Study of Overweight and Diabetes, which was designed to investigate patterns of menstrual irregularity reported by Samoan women and examine their relationship to adiposity and hormone levels.Citation26 This subsample included all non-pregnant women between 18–39 y old who did not report hysterectomy, ovariectomy or other unspecified pelvic surgery. Subjects were further excluded if their serum samples could not be located, or if menstrual data were missing. One additional woman was removed due to her mullerian inhibiting substance (MIS) value being an extreme outlier (45 ng/ml) suggestive of an ovarian tumor or other possible pathologic process. The study subsample totaled n = 322 women.
We further excluded women who did not have DNA available for analysis (n = 40). Men included in this analysis were spouses of the female subsample with available DNA (n = 97) so as to minimize overall relatedness among participants. Two men and two women were excluded due to parental relationships with women in the subsample in order to control for direct relationships within the sample, as heritable factors may potentially play a role in DNA methylation profiles.Citation27 Individuals on medication for high blood pressure and/or diabetes (n = 7 men and n = 13 women) were removed to control for confounding factors, although similar results were obtained when the full sample was included in the models; the models presented are for the reduced sample. Therefore, from the original 71 pedigrees containing 1,164 genotyped adults (at least 18 years old, 534 men and 630 women) we included 355 in the present sample (88 men and 267 women). Of the subjects included in the sample, 72 (20.3%) had at least one sibling relationship within the sample.
Data collection and measurements.
An in-person interview with each eligible study subject was conducted by a trained Samoan field worker using a structured questionnaire. The questionnaire gathered information on subject demographics, history of tobacco and alcohol use, medical history, physical activity, occupational history and dietary intake. Standard anthropometric techniques were used to measure height, weight and body circumferences, and to calculate body mass index (BMI) by dividing weight (kg) by height squared (m). Blood pressure was measured 3 times after participants were seated for 5 min. The mean of 3 measurements was used for analyses. Fasting blood specimens were drawn following a 10-h minimum overnight fast, and serum was separated by centrifugation in the field and stored at −40°C until shipped on dry ice. The following assays of sera were completed: serum leptin by radioimmunoassay (RIA) using a kit from ALPCO (Windham, NH); serum insulin using standard RIA kits from Diagnostic Products Inc.; serum glucose using an automatic analyzer, Beckman CX4; serum adiponectin using RIA kits form Linco Inc., (St. Charles, MI). Total cholesterol and triglycerides were measured by enzymatic assays on Gilford Impact 400 computer directed analyzer. HDL cholesterol was measured after precipitation of VLDL and LDL with heparin-Mn2+ reagent. Buffy coats were prepared from 10 ml of ethylenediamine-tetraacetic acid blood samples in the field, kept at −40°C, then shipped to Cincinnati, OH. Genomic DNA was isolated using the Puregene Kit (Gentra Systems, Inc., Minneapolis, MN) and quantified, and shipped to Providence, RI for DNA methylation analysis.
One microgram of peripheral lymphocyte DNA was sodium bisulfite modified using the EZ DNA Methylation Kit according to manufacturer's protocol (Zymo Research, Orange, CA). LINE-1 region methylation extent was quantified using quantitative bisulfite pyrosequencingCitation28 as previously described in reference Citation17, which examines the cytosine methylation status at 4 CpG sites in the LINE-1 region. All PCR reactions were performed using Qiagen Hot Star Taq polymerase, and each batch included a no template control, unmodified DNA control and a standardized methylation control. The PCR primers were as follows: TTT TGA GTT AGG TGT GGG ATA TA (forward) and AAA ATC AAA AAA TTC CCT TTC (reverse). Each sample was run in triplicate, with each pyrosequencing reaction using 20 µl of PCR product, and was run according to instrument/manufacturer's protocols on a PyroMark™ MD System (Qiagen). The standard error of the averaged individual repeats was found to be the same as the standard error for each replicate, so the average measure (percentage) of LINE-1 methylation across the 4 CpG sites for each replicate was used to calculate an average of the replicates for each sample.
Statistical methods.
The Chi-square test was used to examine the differences in the distributions of categorical variables and t-test for the differences in means of continuous variables between Samoans and American Samoans as well as between men and women. Variable distributions were analyzed using categorical values.
Education was dichotomized into those who did and did not complete a secondary education. Cigarette smoking and alcohol consumption were assessed based upon participant's response to their current status (yes/no). Fasting high-density lipoprotein (HDL) cholesterol was dichotomized at <50 mg/dL for women and <40 mg/dL for men, the American Heart Association's definition of low HDL cholesterol, which is associated with elevated risk for heart disease.Citation29 Fasting low-density lipoprotein (LDL) cholesterol was dichotomized at ≥100 mg/dL, the American Heart Association's definition for above optimal LDL cholesterol which is associated with elevated risk for heart disease.Citation29 In , these same cutoffs for LDL and HDL were used to examine lipoprotein risk factor clustering. Risk factor clustering and LINE-1 methylation was examined by grouping individuals based on high LDL (0 = no, 1 = yes) and low HDL (0 = no, 1 = yes) levels, and using their sum, which ranged from 0–2.
In order to control for sample plate variability bias, mixed linear models were used to assess the relationship between LINE-1 methylation level and selected variables. Age, BMI, fasting glucose, fasting insulin, SBP, DBP and fasting HDL and LDL cholesterol were modeled as continuous variables. Due to non-normal distributions of fasting glucose and insulin levels, these values were log transformed to normalize the data. Location, gender, education and current drinking and smoking status were modeled as categorical values. Bivariate models were used to calculate unadjusted p values, whereas multivariate mixed models were used to calculate adjusted p values. Mean LINE-1 values presented in – were derived from multivariate mixed linear models. All models included a random effect to account for plate variability of LINE-1 measurement. Intra-plate variability was controlled for by including a fixed regional effect and regional x plate (random) interaction coefficient within all models to account for small position effects of LINE-1 measurement.
Statistical analyses were performed using SAS version 9.2 statistical software package (SAS Institute, Cary, NC). All p values are two sided; p values lower than 0.05 were considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures and Tables
Figure 1 Scatter plots of LINE-1% methylation (y-axis) and (x-axis). (A) HDL among men, (B) LDL among men, (C) HDL among women and (D) HDL among women.
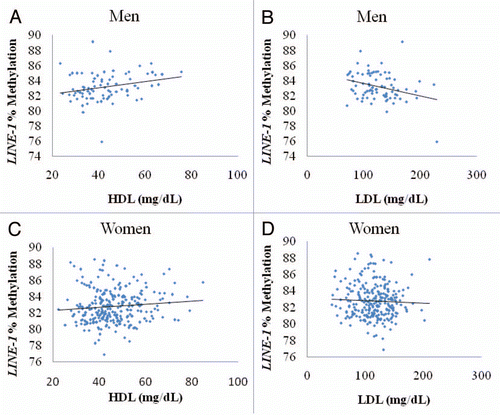
Table 1 Selected characteristicsTable Footnote* of study sample participants
Table 2 Association of selected characteristics and LINE-1 methylation among men and women
Table 3 Association of HDL and LDL clinical categories and LINE-1 methylation among men and women separately
Table 4 Association of LDL/HDL risk clustering and LINE-1 methylation among men and women separately
Acknowledgments
We thank Devin Koestler for statistical assistance, Graham Poage for helpful discussions and Dr. Ranjan Deka for preparation and shipment of DNA samples. We also thank the participants of the Samoan Family Study of Overweight and Diabetes. This study was funded by the NIH (NIH DK59642, HL093093, CA65726 and CA121147-04S1).
References
- Keighley ED, McGarvey ST, Quested C, McCuddin C, Viali S, Maga UA. Stanley J, Ulijaszek RO. Nutrition and health in modernizing Samoans: temporal trends and adaptive perspectives. Health Change in the Asia-Pacific Region 2007; Cambridge 147 - 191
- DiBello JR, McGarvey ST, Kraft P, Goldberg R, Campos H, Quested C, et al. Dietary patterns are associated with metabolic syndrome in adult Samoans. J Nutr 2009; 139:1933 - 1943; PMID: 19710163; http://dx.doi.org/10.3945/jn.109.107888
- Dai F, Keighley ED, Sun G, Indugula SR, Roberts ST, Aberg K, et al. Genome-wide scan for adiposity-related phenotypes in adults from American Samoa. Int J Obes (Lond) 2007; 31:1832 - 1842; PMID: 17621312; http://dx.doi.org/10.1038/sj.ijo.0803675
- Dai F, Sun G, Aberg K, Keighley ED, Indugula SR, Roberts ST, et al. A whole genome linkage scan identifies multiple chromosomal regions influencing adiposity-related traits among Samoans. Ann Hum Genet 2008; 72:780 - 792; PMID: 18616661; http://dx.doi.org/10.1111/j.1469-809.2008.00462.x
- Aberg K, Dai F, Sun G, Keighley E, Indugula SR, Bausserman L, et al. A genome-wide linkage scan identifies multiple chromosomal regions influencing serum lipid levels in the population on the Samoan islands. J Lipid Res 2008; 49:2169 - 2178; PMID: 18594117; http://dx.doi.org/10.1194/jlr.M800194-JLR200
- Åberg K, Dai F, Sun G, Keighley ED, Indugula SR, Roberts ST, et al. Susceptibility loci for adiposity phenotypes on 8p, 9p and 16q in American Samoa and Samoa. Obesity (Silver Spring) 2009; 17:518 - 524; PMID: 19238140; http://dx.doi.org/10.1038/oby.2008.558
- Kirch P. On the Road of the Winds: An Archaeological History of the Pacific Islands before European Contact 2000; Berkeley University of California Press
- Tsai HJ, Sun G, Smelser D, Viali S, Tufa J, Jin L, et al. Distribution of genome-wide linkage disequilibrium based on microsatellite loci in the Samoan population. Hum Genomics 2004; 1:327 - 334; PMID: 15588493
- McGarvey ST, Bindon J, Crews D, Schendel D. Little JDH. Modernization and adiposity: causes and consequences. Human Population Biology: A Trans-diciplinary Science 1989; New York Academic Press 263 - 279
- Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet 2000; 1:11 - 19; PMID: 11262868; http://dx.doi.org/10.1038/35049533
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002; 16:6 - 21; PMID: 11782440; http://dx.doi.org/10.1101/gad.947102
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature 2007; 447:433 - 440; PMID: 17522677; http://dx.doi.org/10.1038/nature05919
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3:415 - 428; PMID: 12042769
- Cash HL, Tao L, Yuan JM, Marsit CJ, Houseman EA, Xiang YB, et al. LINE-1 hypomethylation is associated with bladder cancer risk among non-smoking Chinese. Int J Cancer 2011; In press PMID: 21445976; http://dx.doi.org/10.1002/ijc.26098
- Wilhelm CS, Kelsey KT, Butler R, Plaza S, Gagne L, Zens MS, et al. Implications of LINE1 methylation for bladder cancer risk in women. Clin Cancer Res 2010; 16:1682 - 1689; PMID: 2017921; http://dx.doi.org/10.1158/1078-0432.CCR-09-2983
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet 2009; 10:691 - 703; PMID: 19763152; http://dx.doi.org/10.1038/nrg2640
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res 2007; 67:876 - 880; PMID: 17283117; http://dx.doi.org/10.1158/0008-5472.CAN-06-2995
- Zhu ZZ, Hou L, Bollati V, Tarantini L, Marinelli B, Cantone L, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol 2010;
- Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. DNA methylation as a biomarker for cardiovascular disease risk. PLoS ONE 2010; 5:9692; PMID: 20300621; http://dx.doi.org/10.1371/journal.pone.0009692
- Baccarelli A, Wright R, Bollati V, Litonjua A, Zanobetti A, Tarantini L, et al. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology 2010; 21:819 - 828; PMID: 20805753; http://dx.doi.org/10.1097/EDE.0b013e3181f20457
- Sharma P, Kumar J, Garg G, Kumar A, Patowary A, Karthikeyan G, et al. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol 2008; 27:357 - 365; PMID: 18613790; http://dx.doi.org/10.1089/dna.2007.0694
- Chiang EP, Wang YC, Chen WW, Tang FY. Effects of insulin and glucose on cellular metabolic fluxes in homocysteine transsulfuration, remethylation, S-adenosylmethionine synthesis and global deoxyribonucleic acid methylation. J Clin Endocrinol Metab 2008; 94:1017 - 1025; PMID: 19088160; http://dx.doi.org/10.1210/jc.2008-38
- Maier S, Olek A. Diabetes: a candidate disease for efficient DNA methylation profiling. J Nutr 2002; 132:2440 - 2443; PMID: 12163708
- Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, et al. Impact of inflammation on epigenetic DNA methylation—a novel risk factor for cardiovascular disease?. J Intern Med 2007; 261:488 - 499; PMID: 17444888; http://dx.doi.org/10.1111/j.1365-2796.2007.01777.x
- Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect 2008; 116:1547 - 1552; PMID: 19057709; http://dx.doi.org/10.1289/ehp.11338
- Lambert-Messerlian G, Roberts MB, Urlacher S, Ah-Ching J, Viali S, Urbanek M, et al. Assessment of reproductive status in women from Samoa
- Kile ML, Baccarelli A, Tarantini L, Hoffman E, Wright RO, Christiani DC. Correlation of global and gene-specific DNA methylation in maternal-infant pairs. PLoS ONE 2010; 5:13730; PMID: 21060777; http://dx.doi.org/10.1371/journal.pone.0013730
- England R, Pettersson M. Pyro Q-CpG™ quantitative analysis of methylation in multiple CpG sites by Pyrosequencing®. Nat Methods 2005; 2:2
- Grundy SM, Balady GJ, Criqui MH, Fletcher G, Greenland P, Hiratzka LF, et al. Primary prevention of coronary heart disease: guidance from Framingham: a statement for healthcare professionals from the AHA Task Force on Risk Reduction. American Heart Association. Circulation 1998; 97:1876 - 1887; PMID: 9603549
- El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, et al. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet 2007; 122:505 - 514; PMID: 17851693; http://dx.doi.org/10.1007/s00439-007-0430-3
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 2010; 141:956 - 969; PMID: 20550932; http://dx.doi.org/10.1016/j.cell.2010.04.042
- Cacciapuoti F. Hyper-homocysteinemia: a novel risk factor or a powerful marker for cardiovascular diseases? Pathogenetic and therapeutical uncertainties. J Thromb Thrombolysis 2011;
- Ingrosso D, Cimmino A, Perna AF, Masella L, De Santo NG, De Bonis ML, et al. Folate treatment and unbalanced methylation and changes of allelic expression induced by hyperhomocysteinaemia in patients with uraemia. Lancet 2003; 361:1693 - 1699; PMID: 12767735; http://dx.doi.org/10.1016/S0140-6736(03)13372-7
- Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, James SJ. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem 2000; 275:29318 - 29323; PMID: 10884384; http://dx.doi.org/10.1074/jbc.M002725200
- Lusis AJ. Atherosclerosis. Nature 2000; 407:233 - 241; PMID: 11001066; http://dx.doi.org/10.1038/35025203
- Krauss RM. Lipoprotein subfractions and cardiovascular disease risk. Curr Opin Lipidol 2010; 21:305 - 311; PMID: 20531184; http://dx.doi.org/10.1097/MOL.0b013e32833b7756
- Turunen MP, Aavik E, Yla-Herttuala S. Epigenetics and atherosclerosis. Biochim Biophys Acta 2009; 1790:886 - 891
- Lund G, Andersson L, Lauria M, Lindholm M, Fraga MF, Villar-Garea A, et al. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem 2004; 279:29147 - 29154; PMID: 15131116; http://dx.doi.org/10.1074/jbc.M403618200
- O'Neill TP. Apolipoprotein E-deficient mouse model of human atherosclerosis. Toxicol Pathol 1997; 25:20 - 21; PMID: 9061846; http://dx.doi.org/10.1177/019262339702500104
- Yideng J, Jianzhong Z, Ying H, Juan S, Jinge Z, Shenglan W, et al. Homocysteine-mediated expression of SAHH, DNMTs, MBD2 and DNA hypomethylation potential pathogenic mechanism in VSMCs. DNA Cell Biol 2007; 26:603 - 611; PMID: 17688412; http://dx.doi.org/10.1089/dna.2007.0584
- Hiltunen MO, Turunen MP, Hakkinen TP, Rutanen J, Hedman M, Makinen K, et al. DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc Med 2002; 7:5 - 11; PMID: 12083735; http://dx.doi.org/10.1191/1358863x02vm418oa
- Hiltunen MO, Yla-Herttuala S. DNA methylation, smooth muscle cells and atherogenesis. Arterioscler Thromb Vasc Biol 2003; 23:1750 - 1753; PMID: 12947012; http://dx.doi.org/10.1161/01.ATV.0000092871.30563.41
- Epstein R. Atherosclerosis—an inflammatory disease. N Engl J Med 1999; 340:115 - 126; PMID: 9887164; http://dx.doi.org/10.1056/NEJM199901143400207
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 2006; 38:1378 - 1385; PMID: 17072317; http://dx.doi.org/10.1038/ng1909
- Baccarelli A, Tarantini L, Wright RO, Bollati V, Litonjua AA, Zanobetti A, et al. Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA normative aging study. Epigenetics 2010; 5:5; PMID: 20305373
- Wu HC, John EM, Ferris JS, Keegan TH, Chung WK, Andrulis I, et al. Global DNA methylation levels in girls with and without a family history of breast cancer. Epigenetics 2011; 6:29 - 33; PMID: 20930546; http://dx.doi.org/10.4161/epi.6.1.13393
- Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: Association with risk factors in epidemiologic studies. Epigenetics 2011; 6:828 - 837; PMID: 21636973; http://dx.doi.org/10.4161/epi.6.7.16500
- AHA. Statistics ICD. 2007 update 2007;