Abstract
Abnormal methylation in gene promoters is a hallmark of the cancer genome; however, factors that may influence promoter methylation have not been well elucidated. As the one-carbon metabolism pathway provides the universal methyl donor for methylation reactions, perturbation of this pathway might influence DNA methylation and, ultimately, affect gene functions. Utilizing approximately 800 breast cancer tumor tissues from a large population-based study, we investigated the relationships between dietary and genetic factors involved in the one-carbon metabolism pathway and promoter methylation of a panel of 13 breast cancer-related genes. We found that CCND2, HIN1 and CHD1 were the most “dietary sensitive” genes, as methylation of their promoters was associated with intakes of at least two out of the eight dietary methyl factors examined. On the other hand, some micronutrients (i.e., B2 and B6) were more “epigenetically active” as their intake levels correlated with promoter methylation status in 3 out of the 13 breast cancer genes evaluated. Both positive (hypermethylation) and inverse (hypomethylation) associations with high micronutrient intake were observed. Unlike what we saw for dietary factors, we did not observe any clear patterns between one-carbon genetic polymorphisms and the promoter methylation status of the genes examined. Our results provide preliminary evidence that one-carbon metabolism may have the capacity to influence the breast cancer epigenome. Given that epigenetic alterations are thought to occur early in cancer development and are potentially reversible, dietary modifications may offer promising venues for cancer intervention and prevention.
Introduction
Promoter hypermethylation of cancer-related genes, accompanied by global hypomethylation, is a common feature of tumor cells.Citation1 While large numbers of studies have been performed to investigate the functional consequence of aberrant DNA methylation, little is known about what might influence the DNA methylation status of the genome. Since epigenetic processes are dynamic, reversible and susceptible to exogenous factors, identifying environmental or lifestyle factors that influence the epigenome may offer a unique opportunity for chemoprevention or diet-based interventions that target epigenetic pathways.
Increasing evidence suggests that certain constituents in food and dietary supplements have the capacity to influence the epigenome and, ultimately, an individual's risk of developing cancer.Citation2,Citation3 Among such compounds, micronutrients in folate-mediated one-carbon metabolism are of particular interest.Citation4 The universal methyl donor in the cellular reactions, S-adenosylmethionine (SAM), is generated in the one-carbon pathway. The key micronutrients in this pathway are folate, methionine and several B vitamins (i.e., B2, B6 and B12) that are essential co-factors for one-carbon transfer reactions. Other methyl donors, such as choline and betaine, can also affect SAM status, primarily through choline-mediated one-carbon metabolism, and ultimately impact DNA methylation. It has been shown that dietary methyl donors are capable of modulating methylation patterns in both animal models and humans.Citation5–Citation8 Furthermore, there are reports showing that functional polymorphisms in one-carbon-metabolizing genes can also influence DNA methylation status.Citation4,Citation9,Citation10 Nevertheless, human data on whether one-carbon metabolism influences DNA methylation are limited and inconsistent. This issue has been explored primarily in colon cancerCitation11,Citation12 and to a much lesser extent in breast cancer.
In this study, we chose a panel of 13 genes that had been shown to play an important role in breast carcinogenesis and tumor progression. For example, steroid hormone genes (e.g., ESR1 and PGR) and tumor suppressors (e.g., BRCA1, APC and p16INK4a) are well-established breast cancer-related genes. More importantly, promoters of these genes are frequently methylated in breast tumor tissues and this aberrant methylation has been associated with malignant phenotypes and survival rates of breast cancer cases.Citation13–Citation22 Thus, promoter methylation was proposed to be the underlying mechanism for the loss of gene function.Citation23,Citation24
We previously reported that one-carbon metabolism influenced breast cancer risk and survival in the population-based Long Island Breast Cancer Study Project (LIBCSP).Citation25–Citation27 Herein, we investigate the influence of one-carbon metabolism (i.e., dietary intake of co-factors and genetic polymorphisms of genes) on promoter methylation status of a panel of breast cancer-related genes.
Results
Promoter methylation of 13 breast cancer-related genes was assessed in up to 851 breast tumor samples from the LIBCSP, including 104 in situ and 747 invasive tumors. lists the genes examined in this study, with their established or proposed functions. Also shown in are number of tumor samples examined for each gene's methylation status. The main reason for missing methylation data was an insufficient amount of DNA yielded from the tumor blocks.
We have previously reported the relationship of promoter methylation of the same panel of genes with demographic and clinical-pathological characteristics in the population.Citation28,Citation29 This study focuses on the influence of diet on promoter methylation of the same panel of genes. Dietary methyl constituents examined in this study include folate, methionine, choline, betaine, B vitamins (B2, B6 and B12), and alcohol (a folate antagonist). Inter-relationships between dietary intake of these methyl constituents and 13 breast cancer genes are summarized in Table S1.
There are multiple gene-nutrient associations; however, the effects were moderate in size, 2-fold in general, when the high nutrient intake categories were compared to low ones (in tertiles). Although folate, methione and choline are considered key methyl donors in one-carbon metabolism, there is little evidence that these three micronutrients were associated with promoter methylation of any of the 13 breast cancer genes; the only exception is the inverse relationship between folate and CCND2 methylation (Table S1). Interesting patterns emerge from the gene-nutrient matrix (). Two co-factors for one-carbon metabolizing enzymes, B2 (for MTHFR) and B6 (for cSHMT), were associated with 3 of the 13 breast cancer genes () and two of these genes, HIN1 and CDH1, were both associated with B2 and B6. Furthermore, both positive and inverse associations between dietary intake and promoter methylation were observed (). Another interesting observation from the gene-nutrient matrix is that promoter methylation of several genes (CCND2, HIN1 and CHD1) appeared to be associated with multiple (at least two) dietary micronutrients (). While high intake of methyl constituents positively associated with HIN1 promoter methylation (with B2 and B6), inverse associations were observed for CCND2 (with folate and B6) and CDH1 (with B2, B6 and B12).
We then explored associations between 13 functional polymorphisms in one-carbon metabolizing genes and gene promoter methylation status (Table S2). Several suggestive dose-dependent relations were observed (in other words, if there was a corresponding increase in the magnitude of the odd ratios (ORs) with an increase in the number of variant alleles for a particular polymorphism, then that association was considered a positive dose-dependent relationship and, conversely, if the magnitude of the ORs decreased with the number of alleles, then that association was considered to be an inverse dose-dependent relationship): the TYMS 2R allele, ESR1 hypermethylation; the DHFR 19 bp deletion allele, CDH1 hypomethylation; CHDH rs12676 T allele, GSTP1 hypermethylation; and CHDH rs9001 C allele, GSTP1 hypomethylation. However, unlike the gene-nutrient matrix above, we did not observe any clear patterns between one-carbon polymorphism and promoter methylation.
Discussion
Changes in DNA methylation have been shown to play important roles in breast cancer development; these alterations are potentially reversible.Citation30 The panel of genes chosen in this study was based on their important role in breast carcinogenesis and tumor progression. These genes are frequently methylated in breast cancer and promoter methylation was proposed to be the underlying mechanism for the loss of function.Citation23,Citation24
To date, few studies have identified factors, especially modifiable lifestyle factors, that are capable of influencing the epigenome.Citation3 In this study, we focus on the one-carbon metabolism, in which potentially modifiable factors are involved. The goal of this study was to examine the potential influence of nutrient intake and genetic polymorphisms in the key enzymes involved in the one-carbon metabolic pathway on promoter methylation of breast cancer-related genes. Our study, based on a large population-based case-control study, is the first to systematically examine lifestyle factors that may influence the breast cancer epigenome, and may lead to the development of better preventive strategies for the disease.
Several interesting findings emerged from our study. First, from a panel of 13 breast cancer related genes, promoter methylation of CCND2, HIN1 and CHD1 appeared to be more “sensitive” to dietary modulation, as they were correlated with at least 2 out of 8 micronutrients examined in the study. In the meantime, two one-carbon micronutrients, vitamin B2 and B6, appeared to be more “epigenetically active” as they were associated with promoter methylation of 3 out of 13 genes examined. While vitamin B2 is co-factor for the rate-limiting enzyme in folate-mediated one-carbon metabolism, MTHFR, vitamin B6 is the co-factor of another critical enzyme, cSHMT, involved in regenerating the methyl-carrying folate in the pathway. These results suggest that intake of certain B vitamins may affect the supply of key co-factors required for maintaining proper functions of one-carbon metabolism in the cell, thus influencing the metabolic output of methyl donors for methylation reactions.
Second, associations between high intake of one-carbon micronutrients and promoter methylation are not unidirectional; both positive (hypermethylation) and inverse (hypomethylation) relationships were observed. For example, high B2 intake was associated with lower likelihood of having a methylated promoter for CDH1 but with higher likelihood of having a methylated promoter for TWIST1 and HIN1. These results may reflect the complex regulatory mechanisms in the cell. One-carbon metabolism provides the methyl-donor (i.e., SAM) for the methylation reaction; interruption of this pathway by suboptimal nutrients intake or by suboptimal enzyme activity caused by genetic polymorphism may influence methyl supply. However, the regulation of the methyltransferases (e.g., DNMT3) is not known, especially for gene specific methylation. Genomic structure, sequence dependence and genetic mutation could all play a role.
Third, we found suggestive evidence that functional polymorphisms in one-carbon metabolizing genes could influence promoter methylation. However, we did not pursue the analyses of gene-diet interactions due to limited power. There are few other reports studying gene promoter methylation status in relation to one-carbon metabolism. One study examined seven genes (RARβ2, CDH1, ESR1, BRCA1, CCND2, p16 and TWIST) among ∼200 tumors.Citation31 They found that patients who were homozygous for the TYMS double tandem repeat (2R/2R) showed a trend for more frequent promoter methylation (by methylated gene counts) in their breast cancers, and patients who were homozygous for the MTHFD1 G1958A polymorphism showed a significantly higher frequency of methylation. However, none of the individual methylated CpG sites showed a significant association with any of the polymorphisms. We observed an ESR1-TYMS association with the same trend in our study (Table S2). Another recent study examined promoter methylation of three genes (E-cadherin, p16 and RARβ2) among ∼800 breast tumors and found no evidence that MTHFR C677T, A1298C and MTR A2756G polymorphisms were associated methylation status.Citation32 We did not observe significant association for these three polymorphisms in our study, either.
There are other studies on the influence of one-carbon metabolism on gene promoter methylation with limited sample size. The majority of these published papers are on colorectal cancer, focusing on one or few of the nutrients (e.g., folate), SNPs (MTHFR C677T) and/or gene methylation status (p16, hMLH).Citation9,Citation33–Citation35 These results are far from conclusive. Variations in sample sizes, and the use of different assays to detect methylation status, could have contributed to the inconsistency.
Given the number or micronutrients and genetic polymorphisms examined in this study, one important issue that warrants careful consideration is that of “multiple comparison,” which results in increased likelihood of false positive findings. We examined eight micronutrients and 13 genetic polymorphisms in relation to the promoter methylation status of 13 genes. At the p < 0.05 level, there could be five significant results for micronutrients and eight for polymorphisms by chance alone. There is continuing debate on whether/when/how multiple comparisons should be taken into account.Citation36 When evaluating results of molecular epidemiology studies, in addition to the magnitude of the p value, statistical power and the priority of the tested hypothesis, are also need to take into account.Citation37 In this study, we opt to report the crude p values without adjusting for multiple comparisons for the following reasons. First, instead of randomly considering multiple genes without regarding of the biological relevance but only relying on statistical significance to interpret the study findings, we chose to focus only on genes and dietary/genetic factors that have functional relevance to breast cancer. Second, using the nutrient-gene matrix, our study focuses on the patterns of micronutrients-methylation associations, rather than of the magnitude of specific associations. Lastly, multiple comparison adjustment, such as Bonferroni method, may safeguard false positive findings, but it may increase Type II error (false negative) and reduce sensitivity,Citation38 thus being too conservative for exploratory purpose. Nevertheless, we need to be cautious and results from our study warrant replication in other population studies.
Several limitations of our study are discussed below. (1) In our study, tumor DNA was not available for all cases of the LIBCSP participants. Although there were some differences between those with and without tumor DNA available for our analyses (as described in the Methods section), the benefit of utilizing our population-based sample was that we were able to quantify the differences between the two groups. This valuable contrast aided in our interpretation of the generalizability of our study results to the general population. It is this type of information that is often unavailable from other study populations, such as those derived from a hospital-based case series. (2) The association between one-carbon metabolism and gene promoter methylation was examined only in tumor tissues; whether the same association exists in tissues that precede malignancy (i.e., normal adjacent tissues) is not known. This limits our ability to draw definitive causal-inference. Nevertheless, our results add knowledge in the breast cancer research field and provide a rationale for future mechanistic research. (3) The dietary factors may affect methylation of multiple CpG sites in the gene promoter region. The assays used in this study examined a limited number of CpG sites in the region, which may limit the sensitivity of detecting a diet-methylation relationship. (4) In our study, dietary intake values for one-carbon related micronutrients and compounds were assessed using a modified Block food frequency questionnaire (FFQ). There is continuing debate over the accuracy of this method to assess usual adult diet. Despite these potential limitations, the ranking of intakes (which was the approach used in our analyses) was unlikely to be invalid, given that the Block FFQ has been shown to be a valid and reliable dietary assessment tool for estimating usual food intake and ranking individuals into categories of intake of micronutrients.Citation39,Citation40
In summary, we examined the influence of one-carbon metabolism on gene promoter methylation in 13 breast-cancer related genes in a relatively large epidemiologic study. Our results provide preliminary evidence that dietary nutrients might influence the breast cancer epigenome. Given that epigenetic alterations are reversible and occur early in cancer development, dietary intervention may offer promising venues for cancer prevention.
Materials and Methods
Study population.
We utilized the resources of the Long Island Breast Cancer Study Project, a population-based study. The study participants included 1,504 women newly diagnosed with a first primary breast cancer in 1996–1997 and who participated in the original, parent case-control study. Details of the study design have been described in detail previously in references Citation41 and Citation42. Participant information used in this ancillary study was obtained as part of in-person interviews and medical record abstraction that were collected as part of the case-control parent study.Citation41 Questionnaires were administrated to assess the demographic characteristics and breast cancer-related factors. Tumor characteristics such as ER/PR status were extracted from the medical records. Among cases with this information available, 583 (58.9%) were ER+/PR+; 143 (14.4%) were ER+/PR−; 52 (5.3%) were ER−/PR+; and 212 (21.4%) were ER−/PR−.
Archived pathology blocks for the first primary breast cancer of the LIBCSP cases were requested from the 33 hospitals in the Long Island study area and successfully retrieved for 962 women.Citation43 After review and processing in the laboratory by a trained breast cancer pathologist (H. Hibshoosh), tumor tissues from 859 subjects (89.3%) were available for our study analyses. We compared the demographic and clinico/pathological features between cases with or without tumor blocks available for methylation analysis in our study. Although most characteristics are similar between these two groups, some factors were different. Case women who had tumor samples available for methylation analysis tended to be older (mean age 59.6 vs. 57.9 years; p = 0.005), to have an invasive tumor (87.8% vs. 80.1%; p < 0.001), and to be post-menopausal (70.7% vs. 64.6%; p = 0.01). The study protocol was approved by the Institutional Review Boards of the collaborating institutions.
Determining gene promoter methylation in tumor DNA.
Tumor block retrieval and DNA extraction were performed as previously described in reference Citation43 and Citation44. Tumor DNA first underwent bisulfite modification using the CpGnome DNA Modification Kit (Chemicon International, cat. no. S7820) following the protocol from the manufacturer. We selected a panel of 13 genes that have been implicated in breast carcinogenesis () for methylation analysis. Promoter methylation of ER, PR and BRCA1 was determined by methylation-specific PCR (MSP) as described previously in reference Citation44 and Citation45. The MethyLight assay was used for determining the methylation status of the rest of genes as described in reference Citation46 and Citation47. The percentage of methylation was calculated by the 2−ΔΔCT method, where ΔΔCT = (CT,Target − CT,Actin)sample − (CT,Target − CT,Actin)fully methylated DNACitation48 and multiplying by 100. Samples containing ≥4% fully methylated molecules were designated as methylated, whereas samples containing <4% were designated as unmethylated. The main reason for some missing methylation data was that there was not enough DNA as template for PCR reaction regardless of methods for assessing methylation status. The limiting amount of DNA is primarily due to the small size of the original tumor.
Genotyping and dietary assessment.
Genotyping was conducted on 13 functional polymorphisms in the one-carbon metabolism pathway using methods described in reference Citation25, Citation26 and Citation49. Dietary intake values for one-carbon related micronutrients and compounds were calculated based on data collected as part of the in-person case-control interview assessed using a modified Block food frequency questionnaire (FFQ), which was self-completed by the study participants.Citation50,Citation51
Statistical analysis.
The MSP-PCR assay for ER, PR and BRCA1 promoter methylation generated dichotomized outcomes, i.e., methylated and unmethylated status. MethyLight assay, on the other hand, yielded percentage of methylation for gene promoters that were then dichotomized into methylated or unmethylated cases using a cut-off of 4%, according to previous reports in reference Citation47, Citation52 and Citation53. Relationships of gene promoter methylation status with demographic and clinical-pathological characteristics were examined using chi-square test. Associations between methylation status and diet or genetic factors were analyzed in a case-case setting using unconditional logistic regression.Citation54 With this approach, the odds ratios (ORs) and 95% confidence interval (CI) represent the likelihood of a case possessing a methylated promoter for the gene of interest given certain nutrient intake levels or genotypes. Nutrient intakes in the year prior to the case interviews were divided into tertiles based on the distributions in cases with methylation data. Tests for linear trend of ORs were calculated by regressing on the median values of each tertile.Citation54 For genetic polymorphisms, an ordered categorical variable by assigning a score to each genotype [i.e., 0 (no variant allele), 1 (carrying one variant allele) and 2 (carrying two variant alleles)] was used as the variable to test for trend.
Potential confounders included age at diagnosis (entered as 5-year age groups), ER/PR IHC status [positive group: ER and PR both positive (ER+/PR+) and negative group: all others (ER+/PR−, ER−/PR+, ER−/PR−)], cancer type (in situ vs. invasive), menopausal status (pre- vs. post-), race and family history of breast cancer and benign breast. If eliminating a covariate from the full regression model changed the effect estimate by 10% or more, the covariate was considered a confounder and kept in the model.Citation54 None of the covariates tested met this criterion and, thus, only results from the age-adjusted model are presented. Because of the exploratory nature of this study, we choose to report the crude p values without adjusting for multiple comparison; the implications of this approach are discussed in the ‘Discussion’ section. All statistical analyses were performed using SAS statistical software version 9.1(SAS Institute, Cary, NC).
Figures and Tables
Figure 1 Example of epigenetically active micro-nutrients for association between gene promoter methylation status and dietary intake of one-carbon related nutrients. Odd ratios (ORs) and 95% confidence intervals are shown on the y-axis, comparing medium and high intake to the low intake group. p values are of the test for liner trend of ORs.
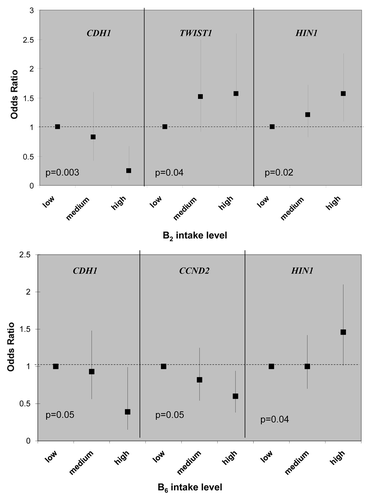
Figure 2 Example of dietary sensitive genes for association between gene promoter methylation status and dietary intake one-carbon related nutrients. Odds ratios and 95% confidence intervals were shown on the y-axis, comparing medium and high intake to the low intake group. p values are of the test for liner trend of ORs.
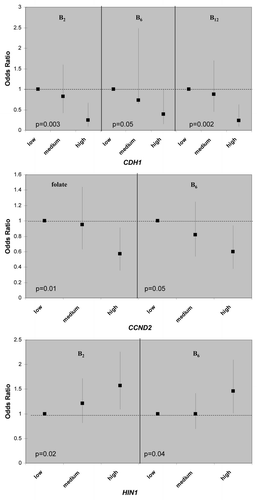
Table 1 Genes selected for methylation analysis and their putative functions
Table 2 Summary of significant trends between one-carbon micronutrient intake and gene promoter methylation status
Additional material
Download Zip (290.2 KB)Acknowledgments
This work was supported by grants from the National Cancer Institutes (R01CA109753 and 3R01CA109753-04S1) and in part by grants from Department of Defense (BC031746 and W81XWH-06-1-0298) and National Cancer Institute and National Institutes of Environmental Health and Sciences (UO1CA/ES66572, UO1CA66572, P30CA013696, P30ES009089 and P30ES10126).
References
- Esteller M. Epigenetics in cancer. N Engl J Med 2008; 358:1148 - 1159; http://dx.doi.org/10.1056/NEJMra072067
- Chen J, Xu X. Diet, epigenetic and cancer prevention. Adv Genet 2010; 71:237 - 255; PMID: 20933131; http://dx.doi.org/10.1016/B978-0-12-380864-6.00008-0
- Ulrich CM, Grady WM. Linking epidemiology to epigenomics—where are we today?. Cancer Prev Res 2010; 3:1505 - 1508; PMID: 21149325; http://dx.doi.org/10.1158/1940-6207.CAPR-10-0298
- Stern LL, Mason JB, Selhub J, Choi SW. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol Biomarkers Prev 2000; 9:849 - 853
- Christman JK, Sheikhnejad G, Dizik M, Abileah S, Wainfan E. Reversibility of changes in nucleic acid methylation and gene expression induced in rat liver by severe dietary methyl deficiency. Carcinogenesis 1993; 14:551 - 557; PMID: 8472313
- Fowler BM, Giuliano AR, Piyathilake C, Nour M, Hatch K. Hypomethylation in cervical tissue: Is there a correlation with folate status?. Cancer Epidemiol Biomarkers Prev 1998; 7:901 - 906; PMID: 9796635
- Rampersaud GC, Kauwell GP, Hutson AD, Cerda JJ, Bailey LB. Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. Am J Clin Nutr 2000; 72:998 - 1003
- Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, Miller BJ, et al. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr 1998; 128:1204 - 1212; PMID: 9649607
- Paz MF, Avila S, Fraga MF, Pollan M, Capella G, Peinado MA, et al. Germ-line variants in methyl-group metabolism genes and susceptibility to DNA methylation in normal tissues and human primary tumors. Cancer Res 2002; 62:4519 - 4524
- Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci USA 2002; 99:5606 - 5611; http://dx.doi.org/10.1073/pnas.062066299
- Kim YI. Folate and DNA methylation: A mechanistic link between folate deficiency and colorectal cancer?. Cancer Epidemiol Biomarkers Prev 2004; 13:511 - 519; PMID: 15066913
- Estécio MRH. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PloS one 2007; 2:399; PMID: 17476321
- Jin Z, Tamura G, Tsuchiya T, Sakata K, Kashiwaba M, Osakabe M, et al. Adenomatous polyposis coli (APC) gene promoter hypermethylation in primary breast cancers. Br J Cancer 2001; 85:69 - 73; PMID: 11437404; http://dx.doi.org/10.1054/bjoc.2001.1853
- Silva JM, Dominguez G, Villanueva MJ, Gonzalez R, Garcia JM, Corbacho C, et al. Aberrant DNA methylation of the p16INK4a gene in plasma DNA of breast cancer patients. Br J Cancer 1999; 80:1262 - 1264; PMID: 10376981; http://dx.doi.org/10.1038/sj.bjc.6690495
- Agathanggelou A, Honorio S, Macartney DP, Martinez A, Dallol A, Rader J, et al. Methylation associated inactivation of RASSF1A from region 3p21.3 in lung, breast and ovarian tumors. Oncogene 2001; 20:1509 - 1518; PMID: 11313894; http://dx.doi.org/10.1038/sj.onc.1204175
- Dulaimi E, Hillinck J, Ibanez de Caceres I, Al-Saleem T, Cairns P. Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients. Clin Cancer Res 2004; 10:6189 - 6193; PMID: 15448006; http://dx.doi.org/10.1158/1078-0432.CCR-04-0597
- Nass SJ, Herman JG, Gabrielson E, Iversen PW, Parl FF, Davidson NE, et al. Aberrant methylation of the estrogen receptor and E-cadherin 5′ CpG islands increases with malignant progression in human breast cancer. Cancer Res 2000; 60:4346 - 4348; PMID: 10969774
- Sirchia SM, Ferguson AT, Sironi E, Subramanyan S, Orlandi R, Sukumar S, et al. Evidence of epigenetic changes affecting the chromatin state of the retinoic acid receptor beta2 promoter in breast cancer cells. Oncogene 2000; 19:1556 - 1563; PMID: 10734315; http://dx.doi.org/10.1038/sj.onc.1203456
- Esteller M, Corn PG, Urena JM, Gabrielson E, Baylin SB, Herman JG. Inactivation of glutathione S-transferase P1 gene by promoter hypermethylation in human neoplasia. Cancer Res 1998; 58:4515 - 4518; PMID: 9788592
- Evron E, Umbricht CB, Korz D, Raman V, Loeb DM, Niranjan B, et al. Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res 2001; 61:2782 - 2787; PMID: 11289162
- Fackler MJ, McVeigh M, Mehrotra J, Blum MA, Lange J, Lapides A, et al. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res 2004; 64:4442 - 4452; PMID: 15231653; http://dx.doi.org/10.1158/0008-5472.CAN-03-3341
- Gort EH, Suijkerbuijk KP, Roothaan SM, Raman V, Vooijs M, van der Wall E, et al. Methylation of the TWIST1 promoter, TWIST1 mRNA levels and immunohistochemical expression of TWIST1 in breast cancer. Cancer Epidemiol Biomarkers Prev 2008; 17:3325 - 3330; PMID: 19064546; http://dx.doi.org/10.1158/1055-9965.EPI-08-0472
- Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene 2002; 21:5462 - 5482; http://dx.doi.org/10.1038/sj.onc.1205606
- Yang X, Yan L, Davidson NE. DNA methylation in breast cancer. Endocr Relat Cancer 2001; 8:115 - 127; PMID: 11446343
- Chen J, Gammon MD, Chan W, Palomeque C, Wetmur JG, Kabat GC, et al. One-carbon metabolism, MTHFR polymorphisms and risk of breast cancer. Cancer Res 2005; 65:1606 - 1614; http://dx.doi.org/10.1158/0008-5472.CAN-04-2630
- Xu X, Gammon MD, Zhang H, Wetmur JG, Rao M, Teitelbaum SL, et al. Polymorphisms of one-carbon metabolizing genes and risk of breast cancer in a population-based study. Carcinogenesis 2007; 28:1504 - 1509; PMID: 17372271; http://dx.doi.org/10.1093/carcin/bgm061
- Xu X, Gammon MD, Wetmur JG, Bradshaw PT, Teitelbaum SL, Neugut AI, et al. B-vitamin intake, one-carbon metabolism and survival in a population-based study of women with breast cancer. Cancer Epidemiol Biomarkers Prev 2008; 17:2109 - 2116; http://dx.doi.org/10.1158/1055-9965.EPI-07-2900
- Xu X, Gammon MD, Zhang Y, Cho YH, Wetmur JG, Bradshaw PT, et al. Gene promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Res Treat 2010; 121:685 - 692; PMID: 19921426; http://dx.doi.org/10.1007/s10549-009-0628-2
- Cho YH, Shen J, Gammon MD, Zhang YJ, Wang Q, Gonzalez K, et al. Prognostic significance of gene-specific promoter hypermethylation in breast cancer patients. Breast Cancer Res Treat In press
- Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet 2001; 10:687 - 692; PMID: 11257100
- Li SY, Rong M, Iacopetta B. Germ-line variants in methyl-group metabolism genes and susceptibility to DNA methylation in human breast cancer. Oncol Rep 2006; 15:221 - 225; PMID: 16328059
- Tao MH, Shields PG, Nie J, Marian C, Ambrosone CB, McCann SE, et al. DNA promoter methylation in breast tumors: No association with genetic polymorphisms in MTHFR and MTR. Cancer Epidemiol Biomarkers Prev 2009; 18:998 - 1002; PMID: 19240236; http://dx.doi.org/10.1158/1055-9965.EPI-08-0916
- van Engeland M, Weijenberg MP, Roemen GM, Brink M, de Bruine AP, Goldbohm RA, et al. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: The netherlands cohort study on diet and cancer. Cancer Res 2003; 63:3133 - 3137; PMID: 12810640
- Mokarram P, Naghibalhossaini F, Saberi Firoozi M, Hosseini SV, Izadpanah A, Salahi H, et al. Methylenetetrahydrofolate reductase C677T genotype affects promoter methylation of tumor-specific genes in sporadic colorectal cancer through an interaction with folate/vitamin B12 status. World J Gastroenterol 2008; 14:3662 - 3671; PMID: 18595133
- Stidley CA, Picchi MA, Leng S, Willink R, Crowell RE, Flores KG, et al. Multivitamins, folate and green vegetables protect against gene promoter methylation in the aerodigestive tract of smokers. Cancer Research 2010; 70:568 - 574; http://dx.doi.org/10.1158/0008-5472.CAN-09-3410
- Michels KB, Rosner BA. Data trawling: To fish or not to fish. The Lancet 1996; 348:1152 - 1153; http://dx.doi.org/10.1016/S0140-6736(96)05418-9
- Wacholder S, Chanock S, Garcia-Closas M, El ghormli L, Rothman N. Assessing the probability that a positive report is false: An approach for molecular epidemiology studies. Journal of the National Cancer Institute 2004; 96:434 - 442; http://dx.doi.org/10.1093/jnci/djh075
- Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology 1990; 1:43 - 46; PMID: 2081237
- Block G, Hartman AM, Dresser CM, Carroll MD, Gannon J, Gardner L. A data-based approach to diet questionnaire design and testing. Am J Epidemiol 1986; 124:453 - 469
- Potischman N, Swanson CA, Coates RJ, Weiss HA, Brogan DR, Stanford JL, et al. Dietary relationships with early onset (under age 45) breast cancer in a case-control study in the united states: Influence of chemotherapy treatment. Cancer Causes Control 1997; 8:713 - 721
- Gammon MD, Neugut AI, Santella RM, Teitelbaum SL, Britton JA, Terry MB, et al. The long island breast cancer study project: Description of a multi-institutional collaboration to identify environmental risk factors for breast cancer. Breast Cancer Res Treat 2002; 74:235 - 254
- Gammon MD, Santella RM, Neugut AI, Eng SM, Teitelbaum SL, Paykin A, et al. Environmental toxins and breast cancer on long island. I. polycyclic aromatic hydrocarbon DNA adducts. Cancer Epidemiol Biomarkers Prev 2002; 11:677 - 685
- Rossner P Jr, Gammon MD, Zhang YJ, Terry MB, Hibshoosh H, Memeo L, et al. Mutations in p53, p53 protein overexpression and breast cancer survival. J Cell Mol Med 2008; PMID: 19602056; http://dx.doi.org/10.1111/j.1582-4934.2008.00553.x
- Xu X, Gammon MD, Zhang Y, Bestor TH, Zeisel SH, Wetmur JG, et al. BRCA1 promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Res Treat 2009; 115:397 - 404; PMID: 18521744; http://dx.doi.org/10.1007/s10549-008-0075-5
- Liu Z, Maekawa M, Horii T, Morita M. The multiple promoter methylation profile of PR gene and ERα gene in tumor cell lines. Life Sci 2003; 73:1963 - 1972; http://dx.doi.org/10.1016/S0024-3205(03)00544-7
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res 1999; 59:2302 - 2306
- Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, et al. Fields of aberrant CpG island hypermethylation in barrett's esophagus and associated adenocarcinoma. Cancer Res 2000; 60:5021 - 5026
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001; 25:402 - 408; PMID: 11846609; http://dx.doi.org/10.1006/meth.2001.1262
- Xu X, Gammon MD, Zeisel SH, Lee YL, Wetmur JG, Teitelbaum SL, et al. Choline metabolism and risk of breast cancer in a population-based study. FASEB J 2008; 22:2045 - 2052; PMID: 10.1096/fj.07-101279
- Gaudet MM, Britton JA, Kabat GC, Steck-Scott S, Eng SM, Teitelbaum SL, et al. Fruits, vegetables and micronutrients in relation to breast cancer modified by menopause and hormone receptor status. Cancer Epidemiol Biomarkers Prev 2004; 13:1485 - 1494; DOI:13/9/1485 PMID: 15342450
- Zeisel SH, Mar MH, Howe JC, Holden JM. Concentrations of choline-containing compounds and betaine in common foods. J Nutr 2003; 133:1302 - 1307; PMID: 12730414
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, et al. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000; 28:32; PMID: 10734209
- Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn 2006; 8:209 - 217; PMID: 16645207; http://dx.doi.org/10.2353/jmoldx.2006.050135
- Rothman KJ, Greenland S. Modern Epidemiology. Ed 1998; Philadelphia, Pa Lippincott-Raven Publishers 2