Abstract
Tumor suppressor genes have antiproliferative and antimetastatic functions, and thus, they negatively affect tumor progression. Reactivating specific tumor suppressor genes would offer an important therapeutic strategy to block tumor progression. Mammary Serine Protease Inhibitor (MASPIN) is a tumor suppressor gene that is not mutated or rearranged in tumor cells, but is silenced during metastatic progression by transcriptional and epigenetic mechanisms. In this work, we have investigated the ability of Artificial Transcription Factors (ATFs) to reactivate MASPIN expression and to reduce tumor growth and metastatic dissemination in Non-Small Cell Lung Carcinoma (NSCLC) cell lines carrying a hypermethylated MASPIN promoter. We found that the ATFs linked to transactivator domains were able to demethylate the MASPIN promoter. Consistently, we observed that co-treatment of ATF-transduced cells with methyltransferase inhibitors enhanced MASPIN expression as well as induction of tumor cell apoptosis. In addition to tumor suppressive functions, restoration of endogenous MASPIN expression was accompanied by inhibition of metastatic dissemination in nude mice. ATF-mediated reactivation of MASPIN lead to changes in cell motility and to induction of E-CADHERIN. These data suggest that ATFs are able to reprogram aggressive lung tumor cells towards a more epithelial, differentiated phenotype, and thus, represent novel therapeutic agents for metastatic lung cancers.
Introduction
Tumor suppressors play a pivotal role in controlling unscheduled cell proliferation and they act as gatekeepers that block neoplastic progression. The expression of tumor suppressors is downregulated in tumor cells by means of genetic and epigenetic mechanisms, while recent evidence points to their regulation also by microRNAs.Citation1–Citation3 Epigenetic silencing modifications, such as DNA and histone methylation, impact chromatin structure and restrict the physical access of the nuclear factors to the underlying DNA. At the promoter level, this epigenetic state results in inhibition of tumor suppressor gene transcription.Citation2,Citation4,Citation5 Reversal and modification of the tumor suppressor epigenetic state can be achieved by blocking DNA and histone methyltransferases as well as histone deacetylases.Citation6,Citation7 While drugs can be used to achieve the inhibition of these enzymes in an untargeted manner, the precise mechanisms and the genetic cascades whereby these drugs induce biological changes are currently not well understood. Moreover, given that tumor suppressors sustain anti-proliferative and anti-metastatic functions, it is essential to target the native tumor suppressor function in order to develop more effective therapies.
MASPIN, or SERPINB5, is a class II tumor suppressor gene that is not found mutated or rearranged in tumor cells, but is silenced during neoplastic progression.Citation8MASPIN silencing involves transcriptional regulation [via p53,Citation9 ETS,Citation10 AP-1Citation10 and hormone receptorCitation11 (HR) transcription factor], aberrant promoter methylationCitation12,Citation13 and microRNAs (miR-21Citation3). High levels of MASPIN are associated with reduction of tumor growth,Citation14 decreased angiogenesis,Citation15 cell motility and invasion,Citation16,Citation17 as well as metastatic dissemination.Citation18–Citation21 The clinical relevance of MASPIN has been demonstrated in several types of human cancers including breast,Citation22,Citation23 prostate,Citation24 colon,Citation25 lungCitation26 and melanomaCitation27 malignancies.
Lung cancer is associated with high mortality and poor patient survival, mainly due to the lack of effective therapies. NSCLC, which are characterized by slow tumor cell growth and dissemination, are particularly refractory to chemotherapy and chest radiotherapy. In these carcinomas, MASPIN expression is a prognosis marker hence tumor specimens that are MASPIN positive are associated with better patient survival than tumor samples that lack MASPIN expression.Citation26,Citation28,Citation29 However, little is known about the precise mechanism and biological function of MASPIN in NSCLC. MASPIN silencing is correlated with lung cancer progression, metastasis and invasiveness.Citation30 Silencing of MASPIN in highly aggressive NSCLC has been associated with epigenetic regulation in which DNA methylation appears to be a key mechanism for silencing.Citation31 Since methylation is a reversible event, efforts have been made to restore MASPIN expression in cancer cells thereby inducing its tumor and metastasis suppressive functions.Citation32,Citation33 Ectopic expression of MASPIN-encoded cDNA or reactivation of endogenous MASPIN in breast cancer cells perturb multiple cellular networks, such as apoptosis and motility.Citation33–Citation35 Furthermore, ectopic expression of MASPIN in tumor cells results in reprogramming of the tumor cell proteosome, which affect multiple processes during tumor progression.Citation36,Citation37
In previous work we have described a strategy that combines chromatin-remodeling drugs and ATFs to overcome the epigenetic repression of MASPIN in breast cancer cells.Citation33 ATFs composed of modular zinc finger (ZF) domains were designed to bind unique 18-base pair (bp) sites in the MASPIN proximal promoter. The ATFs were constructed by linkage of six sequence-specific zinc finger domains with the VP64 transactivator domain, which is a tetramer of the herpex simplex virus VP16 activator domain.Citation38 Each ZF is a compact 30-amino acid domain composed of a recognition α-helix packed with two antiparallel β-strands coordinated with a zinc ion. The α-helix of each zinc finger specifically recognizes three bp in the DNA. Three MASPIN specific ATFs were engineered with recognition α-helices specific for their corresponding target sites in the MASPIN promoter using previously characterized ZF domains.Citation33
Among the three designed ATFs, ATF-126 was able to partially reactivate MASPIN in several breast cancer cell lines carrying a methylated MASPIN promoter.Citation33,Citation39 Interestingly, the degree of MASPIN reactivation by ATFs appeared to be cell line dependent. This suggests that the structure of the chromatin in the target promoter influences the regulatory potential of ATFs. Moreover, little is known as to how ATFs crosstalk with their chromatin microenvironments. Most importantly, it is presently unknown if ATFs are indeed able to modify individual epigenetic marks such as DNA methylation. To address these questions, we took advantage of the MASPIN specific ATFs to investigate if they could influence hypermethylated promoter microenvironments in highly aggressive NSCLC cell lines. Since NSCLC are known to silence MASPIN by DNA methylation,Citation31 we hypothesized that our ATFs could reactivate MASPIN by modifying the methylation status of the promoter. This should result in specific reactivation of MASPIN tumor suppressive functions as well as inhibition of metastatic behavior, which is a hallmark of NSCLC. Such reactivation of MASPIN may provide a powerful therapeutic tool for NSCLC as well as novel insights on how MASPIN mediates its functions in metastatic lung cancers.
In this paper, we have examined the effect of ATFs in combination with chromatin modifiers to upregulate MASPIN in a panel of NSCLC cell lines. We found that reactivation of the endogenous MASPIN in low-MASPIN expressing cells treated with a combination of ATFs and chromatin modifiers induced apoptosis and suppressed metastasis in immunodeficient mice. Furthermore, restoration of endogenous MASPIN expression also resulted in upregulation of E-CADHERIN thereby inducing a more differentiated epithelial phenotype in lung cancer cells. In addition, we show that ATFs linked to transactivator domains resulted in the demethylation of the MASPIN promoter. Overall, these experiments indicate that ATFs are able to epigenetically reprogram aggressive lung tumor cells towards a more epithelial, differentiated phenotype and, thus, represent novel therapeutic agents for metastatic lung cancers.
Results
MASPIN expression in NSCLC cell lines.
We first performed quantitative real-time PCR (qRT-PCR) to evaluate MASPIN mRNA levels. As shown in , these experiments revealed two categories of NSCLC cell lines, those with high- and low-MASPIN expressing cells. Interestingly, no significant differences in MASPIN expression were observed between the non-transformed immortalized lung epithelial cell line BEAS-2B, which expresses MASPIN, and the high-MASPIN expressing cells. In contrast, significant differences in MASPIN expression levels were detected between the high- and low-MASPIN expressing cell lines. The MASPIN mRNA levels in the high- and low-MASPIN expressing cell lines correlated with the amount of MASPIN protein as shown by western blot ().
Silencing of MASPIN in cancer cell lines has been previously associated with increased cellular invasiveness.Citation17,Citation40 We subsequently investigated the in vitro invasion ability in a panel of NSCLC cells by performing Matrigel invasion assays. As shown in , a reverse correlation between MASPIN expression and invasion ability was observed, corroborating previously published findings in breast and lung cancer lines.Citation16,Citation17,Citation30,Citation33 Overall our results show that high-MASPIN expressing cells were significantly less invasive than low-MASPIN expressing cells.
Next, we analyzed MASPIN protein expression and cellular localization in the NSCLC cell lines by immunofluorescence. As shown in , the immortalized lung epithelial cell line (BEAS-2B) and the cancer cell line NCI-460 localized MASPIN both in the nucleus and in the cytoplasm, whereas that in other NSCLC cell lines, such as NCI-H2009 and NCI-H2122, MASPIN was expressed mainly in the cytoplasm. As expected, we did not detect any MASPIN protein by immunofluorescence in the Low-MASPIN expressing cell line panel (including the NCI-H157 cell line). Overall these results suggest that MASPIN is silenced in a group of NSCLC and as expected, loss of MASPIN expression correlated with invasion ability.Citation30
ATF s reactivate MASPIN in NSCLC cell lines.
Previously, we have described the construction of three ATFs designed to bind 18-bp sites on the MASPIN proximal promoter.Citation28 The ATFs were constructed by linking six sequence-specific ZF domains with the VP64 activator domain. Among all ATFs, only ATF-126 was able to reactivate MASPIN in breast cancer cell lines.Citation33 In this study, we investigated if the same ATFs were able to reactivate MASPIN in the NSCLC cell panel (). The ATFs were delivered into NSCLC cell lines using the retroviral vector PMX-IRES-GFP, which allowed the tracking of transduced cells by flow cytometry using the GFP as a marker.Citation38 Seventy-two hours post-transduction cells were collected and processed for qRT-PCR analysis. First, we transduced two high-MASPIN expressing cell lines, NCI-H358 and NCI-H460. As shown in , two ATFs (ATF-97 and ATF-126) targeting the MASPIN proximal promoter (positions-97 and -126 upstream of the translation start site, respectively) were able to upregulate MASPIN in both cell lines as compared to control (cells transduced with an empty retroviral vector). In these cell lines the ATFs-97 and -126 upregulated MASPIN by 1.5- and 2-fold relative to controls. As illustrated in , both ATFs, ATF-97 and -126, were able to upregulate MASPIN by 25- to 125-fold relative to controls in lung cells having a silenced MASPIN promoter. Similarly to what we reported in breast cancer lines,Citation33,Citation39 ATF-452 had no significant activity in NSCLC lines. Effective ATF expression was also verified by qRT-PCR, using ATFs-specific primers (Sup. Fig. 1). Subsequently we focused on the NCI-H157 cell line as an archetype of an aggressive, low-MASPIN line, to study the functional and phenotypic properties of MASPIN reactivation by ATFs.
ATF-97 and ATF-126 induced apoptosis and suppressed cell invasion in NCI -H157 cells.
We first examined if expression of the ATFs induced the apoptotic function of MASPIN in NCI-H157 cells. We quantified the fraction of early apoptotic cells by Annexin V staining 96 h post-transduction. Non-transduced and transduced cells were stained and analyzed by flow cytometry. As illustrated in , ATF-97 and ATF-126 were able to induce apoptosis in NCI-H157 cells (44 and 45%, respectively) relative to control cells (cells transduced with an empty retroviral vector). As expected, ATF-452, which had no impact on MASPIN expression, had no significant effect in inducing apoptosis. These results differ from the breast cancer cell line models, in which only ATF-126 was able to induce apoptosis.Citation39 In lung cancer cell line models both ATFs induced MASPIN re-expression at similar levels, suggesting that cell type specific chromatin structure strongly influences the potential of ATFs to transactivate target promoters.
Next, we examined the capability of the ATFs to decrease the invasion ability of the highly invasive NCI-H157 cell line. For this purpose, we performed matrigel invasion assays with non-transduced cells as well as cells transduced with ATFs and controls. As we expected from their capability to regulate MASPIN expression, ATF-97 and ATF-126 were both able to downregulate cell invasion relative to controls ().
We subsequently investigated if the ATFs were able to suppress tumorigenicity in vitro. We performed an in vitro colony formation assay in soft agar, which measure anchorage-independent growth, and is considered the most stringent assay for detecting malignant transformation of cells in vitro. ATF-transduced cells and controls were seeded in soft agar for 28 days. Following this incubation period, colonies were analyzed morphologically and quantified. As shown in , cells transduced with ATF-97 and ATF-126 generated fewer number of colonies as compared to controls. Furthermore, control colonies exhibited a scattered amorphous shape with a complete loss of polarity, indicative of invasive properties. In contrast, both ATF-97- and ATF-126-transduced colonies appeared to have round and compact structure, suggesting the acquisition of epithelial-like properties. These results suggested that ATF-97 and ATF-126 were able to induce a more epithelial and less invasive phenotype in the highly aggressive NCI-H157 cell line.
To further examine the effects of ATF-97 and ATF-126 on cell polarity, we performed a three-dimensional matrigel culture assay. As shown in , ATF-97 and ATF-126 had a dramatic effect on the morphologic architecture of the NCI-H157 cells cultured in a three-dimensional matrix. Non-transduced cells and controls formed a disorganized aggregation of cells whereas that ATF-97 and ATF-126 generated highly organized spheroid structures (). These data suggest that MASPIN re-expression in NCI-H157 cells induced the acquisition of a more differentiated epithelial phenotype. To further explore this phenotype, we investigated the expression of E-CADHERIN, which plays a critical role for maintenance of normal epithelial function and architecture of epithelial cells. We analyzed E-CADHERIN mRNA levels in non-transduced, transduced and control NCI-H157 cells. We observed that in ATF-transduced cells E-CADHERIN mRNA levels were upregulated 50-fold relative to controls () at 72 h post-transduction. These results suggested that MASPIN re-expression could regulate in vitro growth and invasive abilities of NCI-H157 cells including re-expression of E-CADHERIN and the acquisition of a more differentiated epithelial phenotype. Overall these results demonstrate that re-activation of endogenous MASPIN by ATFs resulted in suppression of tumor cell growth and invasion in NSCLC cell lines.
ATF-126 abolishes experimental metastasis development in animal models.
The results described above demonstrated that both, ATF-97 and ATF-126 induced similar phenotypes in knocking-down cell invasion in vitro. We next focused on ATF-126 to study the ability of this ATF to decrease lung metastasis formation in a mouse model using non-invasive, bioluminescence imaging (BLI). The injection of cancer cells into the left cardiac ventricle of immunodeficient mice has been extensively used as animal model of bone and brain metastasis.Citation41 The NCI-H157-luc cells (stably expressing the firefly luciferase gene, Luc) were transduced with either a control (empty retroviral vector) or ATF-126-expressing retroviral vector (pMX-IRES-ATF-126-GFP). Twenty-four hours post-transduction 1 × 105-transduced cells were injected into the left ventricle of SCID mice (N = 5 per group). Animals were imaged 15 min after injection by monitoring luciferase activity. Four of the control animals and five of the ATF-126 animals exhibited a diffuse photon accumulation over the entire body, suggesting effective injection. Upon intracardiac injection, one control animal showed signs of distress and died within an hour post-injection. The surviving animals were imaged every week for a total of 12 weeks. Among the three control animals, only two developed metastasis after three weeks post-injection. The BLI in these control animals revealed distinct areas of photon accumulation, indicative of metastatic tumor growth, over the ventral projection of the thoracic vertebrae, the maxillofacial region and dorsal projection of the spine (). Control animals were euthanized at week six due to the large size of the metastatic lesions. These results illustrate the aggressive potential of the NCI-H157 lung cancer cell line. Noteworthy, the animals injected with ATF-126-transduced cells did not develop metastasis during the 12 weeks of imaging (), suggesting that ATF-126 was able to abolish metastasis formation in the highly aggressive NCI-H157 lung cell line.
ATF-97 and ATF-126 demethylate the MASPIN proximal promoter.
As a class II tumor suppressor gene, the MASPIN gene has not been found mutated or deleted in cancer cells. In contrast, it is well known that MASPIN is regulated by epigenetic mechanisms.Citation13 To investigate whether the MASPIN promoter was methylated at DNA level in NCH-H157 cells, we performed a sodium bisulfite sequencing of the MASPIN promoter after 72 h post-transduction. As shown in , the MASPIN promoter was found methylated with frequencies close to 100%. The ATF-97- and ATF-126-binding sites comprised one and two CpG dinucleotides, respectively. However, neither the ATF-452 nor the p53 binding sites contained CpG dinucleotides. To investigate if ATF-mediated upregulation of MASPIN resulted in changes in the MASPIN promoter methylation, we performed sodium bisulfate sequencing of ATF-97 and ATF-126 transduced NCI-H157 cells. As shown in , the ATFs decreased the DNA methylation levels of the MASPIN promoter. Interestingly, the ATF-126 decreased the DNA methylation frequencies in the MASPIN promoter upstream the -126 binding site. In contrast, ATF-97 affected the DNA methylation frequencies in the promoter region downstream the ATF-97 binding site.
ATF-97 synergizes with 5-Aza-2′-dC to reactivate MASPIN expression.
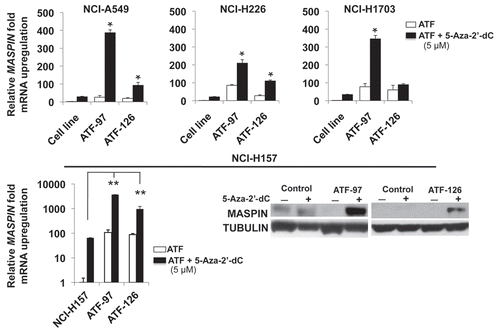
To further investigate the influence of DNA methylation in the re-expression of MASPIN, we challenged ATF-expressing cells with the methyltransferase inhibitor 5-Aza-2′-dC. We transduced four low-MASPIN expressing NSCLC cells lines with ATF-97, ATF-126 as well as controls and MASPIN mRNA levels were analyzed by real-time PCR 72 h post-transduction. As shown in top, the combination of 5-Aza-2′-dC with ATF-97 upregulated MASPIN in a synergistic manner in all the four different cell lines analyzed; hence, the upregulation of the combination treatment was higher than the addition of the effects of the single treatments. In addition, for ATF-126 we observed synergistic interactions with the methyltransferase inhibitor in some cell lines, including NCI-H157 (, bottom). Moreover, in NCI-H157 cells the ATFs strongly synergized with 5-Aza-2′-dC to upregulate MASPIN expression as assessed by real-time quantification and western blot (, bottom). In these cells, the combination treatment of the ATFs with 5-Aza-2′-dC resulted in a concomitant synergistic effect in inducing apoptosis, as assessed by Annexin V staining (Sup. Fig. 2). The differences in the levels of MASPIN reactivation in response to the methyltransferase inhibitor in combination with the two different ATFs could be due to local differences in chromatin structure among the cell lines examined.
Discussion
In this work we have investigated the ability of ATFs to reactivate MASPIN expression and to reduce tumor growth and metastatic dissemination in NSCLC cell lines carrying a hypermethylated promoter. We found that the ATFs were able to demethylate the MASPIN promoter and reactivate the endogenous gene. This was associated with tumor- and metastasis-suppressive functions as well as a phenotypic reprogramming of the aggressive lung tumor cells towards a more differentiated, epithelial phenotype.
Lung cancers are particularly aggressive and associated with high mortality due to their ability to colonize and quickly metastasize. Therefore, there is a clear need to develop novel targeting strategies for metastatic lung cancers. Extensive research in the field has revealed that high MASPIN expression is associated with improved prognosis and patient survival.Citation26,Citation28,Citation29,Citation42,Citation43 In lung carcinomas the MASPIN gene is not mutated or deleted but is silenced by a variety of molecular mechanisms, including transcription factors (for example, p53Citation44 and p63Citation30) and epigenetic regulation (DNA methylationCitation31). Importantly, the level of MASPIN promoter methylation correlates with tumor's aggressiveness.Citation45 Given that methylation is a reversible event, the specific restoration of endogenous MASPIN expression represents a promising therapeutic strategy to block lung cancer progression.
In previous work we have described the generation of ATFs as novel molecular tools to restore the endogenous expression of MASPIN in tumor cells.Citation33 We hypothesized that ATFs could provide a unique opportunity to specifically alter the epigenetic silencing of MASPIN in hyper-methylated promoter microenvironments.Citation39 The results of this work show that ATFs designed to bind methylated DNA sequences in the MASPIN promoter are able to reactivate the endogenous gene in a panel of NSCLC cell lines. Importantly, the ATFs induced MASPIN expression in the NCH-H157 cell line, which contains a hypermethylated promoter with methylation frequencies close to 100%. The reactivation of MASPIN appeared to be specific, since transduction of ATFs lacking the MASPIN-specific DNA-binding domains had no impact on MASPIN expression. Furthermore, truncated versions of ATF-126 and ATF-97 lacking the VP64 domain were not able to reactivate the gene.
It is presently unknown how the ATFs gain access to a hypermethylated MASPIN promoter. Both DNA- and histone methylation are epigenetic silencing marks associated with a compacted chromatin structure, which restricts the physical access of Transcription Factors and Pol II complex to the DNA.Citation4 Our results suggest that DNA-methylation represents indeed a partial blockade for gene reactivation, hence 5-Aza-2′-dC, a DNA-methyltransferase inhibitor, highly enhanced the transactivation capabilities of the ATFs. Future experiments are required to address if indeed the ATFs alter chromatin accessibility patters by inducing localized relaxation of the chromatin, for example by DNAse footprint assays. It is also presently not known if the ATFs are able to locally remodel the chromatin by altering the binding to the DNA of endogenous proteins, such as histones and methyl-binding proteins.
Importantly, MASPIN reactivation upon ATF transduction was associated with site-specific promoter demethylation. Both ATF-97 and ATF-126 significantly decreased the DNA methylation levels in the MASPIN proximal promoter region in the NCH-H157 cell line. ATF-126 decreased the frequency of DNA methylation by up to ∼70%. Interestingly, the DNA sequences exhibiting decreased methylation frequencies were comprised in a 400 bp promoter region upstream the ATF-126 recognition site. In contrast, ATF-97 reduced methylation frequencies by up to ∼50% in a promoter region localized downstream its binding site. Even though these DNA methylation changes are relatively modest, they are associated with a strong degree of reactivation of the endogenous gene, as evidenced by the 100-fold increase in MASPIN transcript. These results are consistent with previous reports in breast cancer cell lines where the frequency of DNA promoter methylation strongly correlated with the degree of MASPIN mRNA expression.Citation13 The localized changes in DNA demethylation induced by the two ATFs in the close proximity of their binding sites suggest that enhanced transcription could be associated with increases accessibility of the ATFs and Pol II complex.
The directionality of the demethylation pattern associated with ATF-126 and ATF-97 might be explained by the differential orientation of the activator domain of each ATF in complex with the target DNA. ATF-126 was designed to bind the “coding” DNA strand in the MASPIN promoter gene, thereby positioning the activator domain upstream the ATF binding site. In contrast, ATF-97 was designed to bind the complementary DNA strand, anchoring the activator domain downstream the ATF-binding site. It will be interesting to extend the mapping of the CpG dinucleotides in the MASPIN promoter further upstream and downstream the ATF-binding sites, which will allow us to have a better understanding of the ATF's demethylation potential. Nevertheless, in the best of our knowledge this is the first report of ATFs that is able to effectively demethylate their targeted promoter resulting in a reactivation of an otherwise silenced gene.
The mechanism by which the ATFs DNA demethylate the MASPIN promoter remains elusive. Recent studies suggest that cross-talk occurs between different epigenetic processes, such as histone acetylation, histone and DNA methylation and nucleosome remodelers.Citation46 Significantly, our results suggest that DNA demethylation and reactivation of the MASPIN promoter by ATFs require a VP64 activator domain properly anchored in the promoter by a specific DNA-binding domain. The VP64 effector domain a tetramer of VP16 (Herpes virus simplex VP16 activator domain) has been proposed to interact with MEDIATOR to recruit the RNA Pol II transcriptional complex.Citation47 As integral members of this complex, histone acetyltransferases (HAT) as well as chromatin remodelers (SWN/SNF)Citation48–Citation50 are able to modify the combination of histone acetylation marks within the promoter.Citation47 This allows a more open and accessible chromatin conformation, resulting in enhanced gene transcription. In this regard, demethylation of the MASPIN promoter represents an indirect or passive mechanism consequential from recruitment of the RNA Pol II complex into the MASPIN promoter. In future experiments it will be important to investigate if ATFs similarly alter other epigenetic marks in the MASPIN promoter, such histone H3 lysine9 methylation, which has been previously associated with the silencing of MASPIN and other tumor suppressors in breast cancer.Citation51
Alternatively to a passive mechanism of demethylation, it is also possible that DNA demethylases recruited to the RNA Pol II complex would enzymatically demethylate the promoter in a more direct manner. However, regardless of the mechanism of demethylation, our results suggest that ATFs can be used to target DNA demethylation into specific promoters. Our future engineering of ATFs will include the linkage of specific DNA-binding domains along with protein domains that actively demethylate DNA or histone substrates. The construction of sequence-specific ATFs could be used by investigators to address important questions such as the impact of demethylation in the generation and maintenance of other epigenetic marks, promoter accessibility and gene transcription.
The ATF-mediated reactivation of MASPIN in lung cancer cell lines resulted in complex functional outcomes, including induction of apoptosis, suppression of cell invasion and experimental metastasis formation in nude mice. This is consistent with the role of MASPIN as a multifaceted tumor suppressor involved in many cellular processes. In breast and prostate cancers, upregulation of MASPIN is associated with induction of apoptosisCitation52,Citation54 suppression of cell invasion,Citation16,Citation17 and suppression of angiogenesis.Citation15 ATFs that reactivated MASPIN in lung cancer cell lines induced approximately 40% apoptosis relative to control-transduced cells. In addition, when the same cells were treated with 5-Aza-2′-dC an increase on MASPIN expression and an additional 20% apoptosis was observed. More recently, MASPIN has been shown to physically associate with HDAC1 to inhibit histone acetylation.Citation55 As a putative HDAC inhibitor, MASPIN could bind and reactivate multiple tumor suppressor genes that are found repressed by HDAC1, including E-CADHERIN. In NCI-H157 cells the E-CADHERIN promoter is found repressed by transcription factor SNAIL.Citation55,Citation57 Consistently, treatment of NCI-H157 cells with HDAC inhibitors, and/or transduction with MASPIN-specific ATFs, also lead to reactivation of E-CADHERIN mRNA levels up to 50-fold relative to controls. Our results show that only a percentage of the transfected spheroids reactivate E-CADHERIN and the morphology of these spheroids exhibited more epithelial phenotypic features. It is possible that a percentage of ATF-transduced cells induced apoptosis as expected from the re-activation of tumor suppressive functions. The survival population could reactivate other tumor suppressor genes such as E-CADHERIN and E2F,Citation58 and thereby these cells would exhibit a more differentiated phenotype.
Overall, our work suggests that ATFs designed to upregulate MASPIN have therapeutic potential to suppress apoptosis and metastatic dissemination in lung cancers. Thus, reactivation of MASPIN by ATFs offers a unique therapeutic strategy to target multiple steps in the neoplastic progression. However, in order to be applied in the clinic, the ATFs need to be specifically delivered in tumor cells. The delivery of ATFs into tumor and metastasis is presently a critical limitation for its use as therapeutic agents in a clinical setting. However, novel nanoparticle targeted systems are being developed, witch will enable the encapsulation of the ATF-encoded DNA or the ATF's polypeptide specifically into tumor cells.Citation59 In addition, novel adenoviral delivery systems are also in development possessing tumor-cell tropism.Citation60,Citation61 ATFs made of three zinc finger modules designed to regulate the Vascular endothelial growth factor (VEGF) gene are presently in clinical trials, outlining the potential of the ATFs as therapeutic agents.Citation62
Materials and Methods
Cell lines.
The human bronchial epithelial cells (BEAS-2B; ATCC CRL#9609) were cultured in BEGM plus additives (BEGM Kit Catalog No. CC-3170, Lonza/Clonetics Corporation). The human lung cancer cell lines NCH-H1648, NCI-H211, NCH-H385, NCI-H460, NCI-A549, NCI-H226, NCI-H520, NCI-H157, NCI-H1703 and NCI-H661 were kindly provided by Dr. Robert Winn (University of Colorado Health Science Center). All the lung cancer cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and grown at 37°C in a humidified 5% CO2 incubator.
ATF retroviral transduction.
ATF-126, ATF-97 and ATF-452 were cloned into the retroviral vector pMX-IRES-GFP as described.Citation33 Retroviral vectors and a plasmid expressing the vesicular stomatitis virus envelope protein (pMDG.1) were first cotransfected into 293TGag-Pol cells to produce retroviral particles. Transfection was done using Lipofectamine according to the manufacture's instructions (Invitrogen). The viral supernatant was used to infect the host cell lines, and the infection efficiency was assessed by flow cytometry (FACSCalibur, BD Biosciences) using green fluorescent protein as a marker.
Stable NCI-H157-Luc cells.
The retroviral vector (pLNX-Luc) and the envelope plasmid expressing the vesicular stomatitis virus envelope protein (pMDG.1) were cotransfected into 293TGag-Pol cells to produce retroviral particles. The viral supernatant was used to infect the NCI-H157 cells. After two days, cells were placed under selection with Geneticin at 800 µg/ml for two weeks. Single colonies were isolated, expanded and used for animal experiments.
Real-time gene expression assays.
Total RNA from 1 × 106 non-transduced, ATF-transduced cells and control cells were extracted using the RNeasy extraction kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. For reverse transcription, 2.5 µg of total RNA was used to prepare cDNA using the High Capacity cDNA Archive Kit (Applied Biosystems). Quantification of MASPIN, E-CADHERIN and GAPDH (Human GAPDH Endogenous Control) transcript expression was measured by real-time PCR using hydrolytic probes (Taqman, Applied BioSystems). All PCR reactions were performed in triplicate and under PCR previously published conditionsCitation39 using independently prepared biological samples. Fold increase in gene expression for each sample and experimental condition was calculated as 2Ct(control) − Ct(sample) ± SD. Differences between groups were analyzed using the Student's t-test or ANOVA. Significance level was set-up at p < 0.05 (indicated as *) and p < 0.01 (indicated as **).
Western blotting.
The NSCLC cell lines were collected in RIPA buffer. Total protein was purified and the concentration measured by a Bio-Rad DC protein Assay Kit (Bio-Rad Laboratories). Equal amounts of protein were subjected to a 10% SDS-PAGE gel (Bio-Rad) and subsequently transferred to a PVDF membrane (Amersham Biosciences) by electroblotting. The membrane was washed twice in 0.05% TBST for 10 min, and then blocked with 5% milk in TBST for 1 hour at room temperature. The primary monoclonal antibody against MASPIN (anti-human MASPIN antibody, clone G167-70; Pharmingen International) was diluted 1:1,000 in 1% milk/TBST. The membrane was incubated overnight at room temperature and washed as described above. The secondary antibody, anti-human Tubulin (Amersham Biosciences), was diluted 1:5,000 in 1% milk/TBST for 45 min. MASPIN and TUBULIN proteins were detected with ECL Plus (Amersham Biosciences).
In vitro invasion assay.
The non-transformed immortalized bronchial epithelial cell line BEAS-2B and all the NSCLC cell lines were seeded in a 24-well matrigel-coated invasion chambers (BD Biosciences). The lower chambers were filled with 0.5 ml of RPMI-1640 medium containing 10% fetal bovine serum (FBS). Cells were starved overnight and 5 × 105 cells in 0.5 ml serum-free medium were added to the upper chamber. Cells were incubated for 18 h at 37°C in a humidified incubator with 5% CO2. The non-invading cells that remained on the upper surface of the membranes were removed by scraping. The invasive cells attached to the lower surface of the membranes were fixed in methanol and stained with Hematoxylin/Eosin (Diff Quick stain Kit, IMEB INC.). The average number of invasive cells was counted under a microscope from four different fields of view from each well and a total number of three wells. Each experiment was performed three times. Differences between groups were analyzed by the Student's t-test. Significance level was set-up at p < 0.05 (indicated as *) and p < 0.01 (indicated as **).
Immunofluorescence assay.
Lung cell lines were seeded in fibronectin-coated slides (5 × 104 cells) and grown for 24 h. Cells were washed with PBS (phosphate buffered saline pH 8.0) and fixed in paraformaldehyde for 20 min. Cells were washed and permeabilized with 0.5% Triton-X100 in PBS followed by blocking with 5% BSA for 2 h. These cells were then incubated with primary antibody (anti-MASPIN, BD Pharmingen) at 4°C overnight. Expression of MASPIN was determined by FITC labeled secondary antibody (Invitrogen).
Quantification of apoptotic cells.
Quantification of apoptosis was determined by flow cytometry using the Annexin-V staining kit (BD Biosciences) following the instructions of the manufacturer. Apoptosis was measured 96 h post-transduction. A positive control was induced with 10 µM camptothecin (Sigma) for 24 h prior analysis to calibrate the flow cytometry settings. Apoptosis was measured by flow cytometry using a FACS Calibur cytometer and CellQuest software (BD Biosciences). The percentage of apoptotic cells was calculated by comparing each sample with the non-transduced sample. Differences between groups were analyzed the Student's t-test. Significance level was set-up at p < 0.05 (indicated as *) and p < 0.01 (indicated as **).
Colony formation assay.
Non-transduced cells, control transduced cells and cells transduced with ATFs, were seeded in triplicate at a density of 3 × 103 cells per well in a six-well plate. The wells were pre-coated with 0.6% agarose in growth medium, and then coated with an additional layer containing 0.3% agarose in RPMI-1640 medium supplemented with 10% fetal bovine serum. After incubation at 37°C for three weeks, the cells were stained with 0.005% crystal violet. Colonies, which were defined as groups of a minimum of 50 cells, were counted using phase contrast microscopy. Colony formation was assessed in three independent experiments. Differences between groups were analyzed with the Student's t-test. Significance level was set-up at p < 0.05 (indicated as *) and p < 0.01 (indicated as **).
Three-dimensional cell culture.
Cells were grown in 3D basement membrane cultures as describedCitation49 with the following modifications. Growth factor-reduced Matrigel (BD Biosciences) was combined in a 1:1 ratio with complete serum medium (RPMI 1640 medium containing 10% fetal bovine serum). 100 µl were added to each well of an eight-well glass slide chamber and allowed to solidify for 2 h in at 37°C incubator. Cells were trypsinized, counted and diluted to 25,000 cells/ml. The cell suspension was combined in a 1:1 ratio with a 10% Matrigel solution and 200 µl of this mixture was added to each well for a final concentration of 5,000 cells/well in 5% Matrigel. Cells were fed with complete serum medium containing 5% Matrigel every three days for a period of three weeks.
Genomic sequencing of the MASPIN promoter.
The methylation status of 19 CpG dinucleotides within the MASPIN gene promoter region was examined in bisulfite-modified DNA as described.Citation12 Genomic DNA was isolated using the DNeasy Tissue Kit (Qiagen) and 1.5 µg was modified using EZ DNA Methylation-Gold kit (Zymo Research). Samples were then purified using Wizard DNA purification resin (Promega), treated with NaOH, precipitated with ethanol and resuspended in water. The MASPIN gene promoter was amplified from bisulfite-modified DNA by two rounds of PCR using nested primers specific to the bisulfite-modified sequence of the MASPIN gene as previously described.Citation13,Citation39 The resulting PCR products were cloned into a TA cloning vector according to the manufacturer's instructions (Invitrogen). Ten positive recombinants were isolated, sequenced and the methylation status of individual CpG dinucleotides was determined by comparison of the sequence obtained with the known MASPIN promoter sequence.Citation10 To obtain a percentage methylation value for each site, we divided the number of methylated CpGs at a specific site by the number of clones analyzed.
Assay of spontaneous metastasis.
Female SCID mice (age four weeks) were purchased from Taconic Farms, and housed under pathogen-free conditions. Each experiment group included five animals. Animals were injected with 1 × 105 NCI-H157-Luc cells transduced with ATF-126 and control vector (pMX-IRES-GFP empty retroviral vector) in 100 µl sterile PBS into the left ventricle of the heart by nonsurgical procedures. Bioluminescent imaging was performed with a highly sensitive, cooled CCD camera mounted in a light-tight specimen box (IVIS™, Xenogen). The signal quantification was controlled with the analysis software Living Image® (Xenogen). For in vivo imaging, animals were administered with the substrate D-luciferin by intraperitoneal injection at 150 mg/kg in DPBS and anesthetized (1–3% isoflurane). Animals were imaged from both dorsal and ventral views about 10 min after intraperitoneal injection of D-luciferin. Assessment of metastasis was monitored in vivo once a week for up to 12 weeks. Regions of interest (ROI) from displayed images were identified around the tumor sites. The bioluminescent signal was quantified as total photons per second (photons/s) by measuring the amount of highlighted pixels in each ROI with the aid of the Living Image® software (Xenogen). The total photons/s in ROI in each mouse were used to evaluate the differences between the two groups using the Wilcoxon rank sum test with the level of significance of p ≤ 0.1.
Figures and Tables
Figure 1 MASPIN expression in a panel of NSC LC cell lines. (A) MASPIN mRNA expression levels by quantitative real-time PCR in a panel of NSCLC cell lines. MASPIN mRNA levels were normalized to the immortalized bronchial epithelial cell line (BEAS-2B). Data represent the mean ± SD of three independent experiments. (B) MASPIN protein expression in high- and low-MASPIN expressing cell lines. (C) Loss of MASPIN expression correlates with invasion abilities in NSC LC cell lines. (D) Immunofluorescence staining of MASPIN in BEAS-2B and in the NSCLC cell lines. ↑ Indicates nuclear and ➭ indicates cytoplasmic staining, respectively.
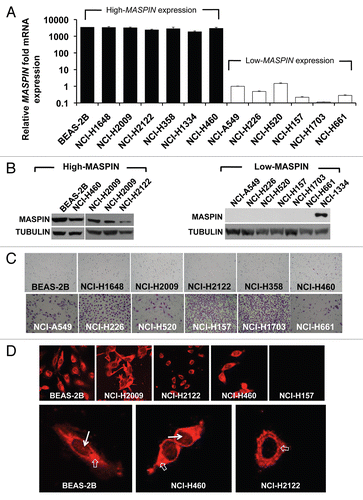
Figure 2 ATFs regulate MASPIN in high- and low-MASPIN expressing NSC LC cell lines. (A) Relative MASPIN mRNA upregulation in the high-MASPIN expressing cell lines NCI-H358 and NCI-H460. (B) Relative MASPIN expression in the low-MASPIN expressing cell lines NCI-A549, NCI-H157, NCI-H226, NCI-H520, NCI-H1703 and NCI-H661. MASPIN mRNA expression levels were measured by quantitative real-time PC R and data was normalized to the untransduced cell lines. Experiments were run in triplicate and data represent the mean ± SD of three independent biological replicates (*p < 0.05, as determined by ANOVA). Cell line refers to untransduced NSC LC cell lines; control, cells transduced with an empty retroviral vector; ATF-97, ATF-126 and ATF-452, cells transduced with the corresponding ATFs; ATF-126NOVP is a control ATF-126 lacking the VP64 activator domain; PMXVP64-SS is a control vector expressing the VP64 activator domain only.
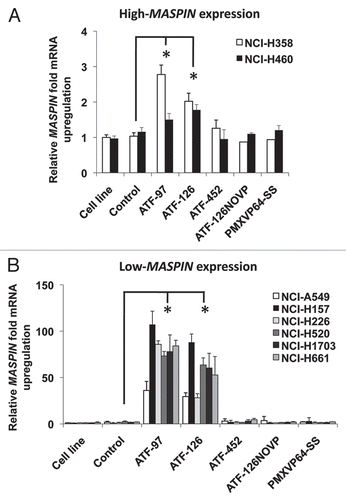
Figure 3 ATFs induce apoptosis and reduce cell invasion in the NCI-H157 NSCLC cell line. (A) ATF-97 and ATF-126 induce apoptosis in NCI-H157 cells. The percentage of apoptosis was quantitatively analyzed in untransduced cells, control cells and in ATF-transduced cells using an Annexin-V staining. Apoptosis was measured 96 h post-transduction. Data represents an average of three independent experiments (**p < 0.01, as determined by the Student's t-test). (B) ATFs downregulate cell invasion in NCI-H157 cells in matrigel invasion assays. Left panel shows a representative picture of invasive cells. Right panel shows the quantification of invasive cells. Experiments were run in triplicate and data represents an average of three different biological replicates (**p < 0.01, as determined by the Student's t-test). (C) In vitro colony formation assay in soft agar. The morphology of colonies of control and ATF-transduced cells are indicated. (D) 3D Matrigel culture assay illustrating that ATF-97- and ATF-126-transduced cells form colonies with an organized spheroid structure. (E) Expression of E-CADHERIN by qRT-PC R in control and ATF-transduced cells. Data was normalized to the untransduced cell line. Experiments were run in triplicate and data represent the mean ± SD three independent experiments (**p < 0.01, as determined by Student's t-test). NCI-H157 refers to untransduced cells; control, cells transduced with an empty retroviral vector; ATF-97, ATF-126 and ATF-452, cells transduced with the corresponding ATFs; ATF-126NOVP is a control ATF-126 lacking the VP64 activator domain.
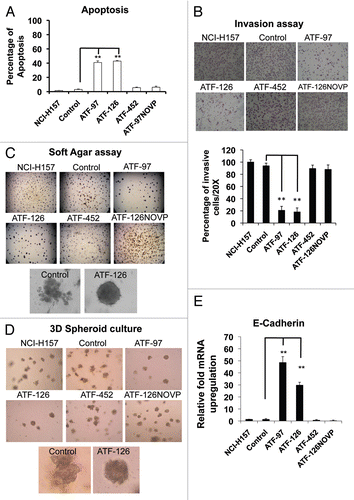
Figure 4 ATF-126 inhibits metastatic dissemination in the NSCLC NCI-H157 cell line stably expressing a luciferase reporter (Luc) in vivo. Top panel; in vivo Bioluminescence Imaging of control mice showing metastasis in the brain and spine at week six after receiving an intracardiac injection of 1 × 105 NCI-H157 cells transduced with control vector (empty retroviral vector). Bottom panel; Bioluminescence Imaging of ATF-126-injected mice twelve weeks after receiving an intracardiac injection of 1 × 105 NCI-H157 cells transduced with ATF-126 retroviral vector. Differences between the two groups were calculated using the Wilcoxon rank sum test with the level of significance of p ≤ 0.1.
Figure 5 ATFs demethylate the MASPIN promoter in the NCI-H157 cell line. (A) Methylation status of the MASPIN promoter in untransduced NCI-H157 cells. The X-axis represent the nucleotide position relative to the first methionine and Y-axis the percentage of methylation along the MASPIN proximal promoter region (495 to +134). The frequencies of 5-Methylcytosine were obtained by sodium bisulfite genomic sequencing of the MASPIN promoterCitation33 Known transcription factor binding sites were included (p53-binding sites as well as ATF-binding sites). Red nucleotides indicate the methylated cytosines in the ATF-binding sites. (B) Schematic representation of ATF-126 and ATF-97 illustrating the positioning of the six zinc finger (ZF-97 and ZF-126) DNA binding domains and the VP64 effector domain (blue circles) in the promoter. (C) Methylation status of the MASPIN promoter in ATF-97- and ATF-126-transduced cells.
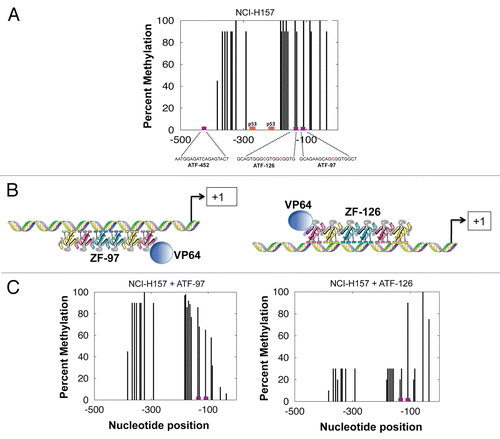
Figure 6 ATFs synergize with 5-Aza-2′-dC to reactivate MASPIN in NSCLC Low-MASPIN expressing cell lines. Non-transduced and transduced cells were treated with 5 µM of 5-Aza-2′-dC for 48 h. MASPIN mRNA expression levels were measured by quantitative real-time PCR. MASPIN protein expression in NCI-H157 cells was determined by western blot. Experiments were run in triplicate and data represent the mean ± SD three independent experiments (*p < 0.05, as determined by Student's t-test). NCH-A549, NCH-H226, NCH-H1703 and NCH-H157 are referred to as “cell line;” ATF-97, ATF-126 and ATF-452, represent cells transduced with the corresponding ATFs.
Additional material
Download Zip (110.9 KB)Acknowledgements
This work was supported from NCI/NIH grants 1R01CA125273, 3R01CA125273-03S1 and DoD W81XWH-10-1-0265 (P.B.). The authors thank Dr. Paul Lizardi (Yale University School of Medicine) for his assistance with the DNA bisulfate sequencing and M.D. Robert A. Winn (University of Colorado at Denver and Health Sciences Center) for kindly providing the NSCLC cell lines.
References
- Zardo G, Fazi F, Travaglini L, Nervi C. Dynamic and reversibility of heterochromatic gene silencing in human disease. Cell Res 2005; 15:679 - 690
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction?. Nat Rev Cancer 2006; 6:107 - 116
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 2008; 18:350 - 359
- Collingwood TN, Urnov FD, Wolffe AP. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J Mol Endocrinol 1999; 23:255 - 275
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 2007; 39:237 - 242
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the reexpression of genes silenced in cancer. Nat Genet 1999; 21:103 - 107
- Oshiro MM, Watts GS, Wozniak RJ, Junk DJ, Munoz-Rodriguez JL, Domann FE, et al. Mutant p53 and aberrant cytosine methylation cooperate to silence gene expression. Oncogene 2003; 22:3624 - 3634
- Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science 1994; 263:526 - 529
- Zou Z, Gao C, Nagaich AK, Connell T, Saito S, Moul JW, et al. p53 regulates the expression of the tumor suppressor gene maspin. J Biol Chem 2000; 275:6051 - 6054
- Zhang M, Magit D, Sager R. Expression of maspin in prostate cells is regulated by a positive ets element and a negative hormonal responsive element site recognized by androgen receptor. Proc Natl Acad Sci USA 1997; 94:5673 - 5678
- Khalkhali-Ellis Z, Christian AL, Kirschmann DA, Edwards EM, Rezaie-Thompson M, Vasef MA, et al. Regulating the tumor suppressor gene maspin in breast cancer cells: a potential mechanism for the anticancer properties of tamoxifen. Clin Cancer Res 2004; 10:449 - 454
- Domann FE, Rice JC, Hendrix MJ, Futscher BW. Epigenetic silencing of maspin gene expression in human breast cancers. Int J Cancer 2000; 85:805 - 810
- Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, et al. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet 2002; 31:175 - 179
- Sager R, Sheng S, Pemberton P, Hendrix MJ. Maspin. A tumor suppressing serpin. Adv Exp Med Biol 1997; 425:77 - 88
- Zhang M, Volpert O, Shi YH, Bouck N. Maspin is an angiogenesis inhibitor. Nat Med 2000; 6:196 - 199
- Seftor RE, Seftor EA, Sheng S, Pemberton PA, Sager R, Hendrix MJ. Maspin suppresses the invasive phenotype of human breast carcinoma. Cancer Res 1998; 58:5681 - 5685
- Sheng S, Carey J, Seftor EA, Dias L, Hendrix MJ, Sager R. Maspin acts at cell membrane to inhibit invasion and motility of mammary and prostatic cancer cells. Proc Natl Acad Sci USA 1996; 93:11669 - 11674
- Zhang M, Shi Y, Magit D, Furth PA, Sager R. Reduced mammary tumor progression in WAP-TAg/WAP-maspin bitransgenic mice. Oncogene 2000; 19:6053 - 6058
- Shi HY, Liang R, Templeton NS, Zhang M. Inhibition of breast tumor progression by systemic delivery of the maspin gene in a syngeneic tumor model. Mol Ther 2002; 5:755 - 761
- Cher ML, Biliran HR, Bhagat S, Meng Y, Che M, Lockett J, et al. Maspin expression inhibits osteolysis, tumor growth and angiogenesis in a model of prostate cancer bone metastasis. Proc Natl Acad Sci USA 2003; 100:7847 - 7852
- Watanabe M, Nasu Y, Kashiwakura Y, Kusumi N, Tamayose K, Nagai A, et al. Adeno-associated virus 2-mediated intratumoral prostate cancer gene therapy: long-term maspin expression efficiently suppresses tumor growth. Hum Gene Ther 2005; 16:699 - 710
- Sharma G, Mirza S, Parshad R, Srivastava A, Gupta SD, Pandya P, et al. Clinical significance of Maspin promoter methylation and loss of its protein expression in invasive ductal breast carcinoma: correlation with VEGF-A and MTA1 expression. Tumour Biol 2010;
- Maass N, Ho jo T, Rosel F, Ikeda T, Jonat W, Nagasaki K. Downregulation of the tumor suppressor gene maspin in breast carcinoma is associated with a higher risk of distant metastasis. Clin Biochem 2001; 34:303 - 307
- Machtens S, Serth J, Bokemeyer C, Bathke W, Minssen A, Kollmannsberger C, et al. Expression of the p53 and Maspin protein in primary prostate cancer: correlation with clinical features. Int J Cancer 2001; 95:337 - 342
- Bettstetter M, Woenckhaus M, Wild PJ, Rummele P, Blaszyk H, Hartmann A, et al. Elevated nuclear maspin expression is associated with microsatellite instability and high tumour grade in colorectal cancer. J Pathol 2005; 205:606 - 614
- Nakagawa M, Katakura H, Adachi M, Takenaka K, Yanagihara K, Otake Y, et al. Maspin expression and its clinical significance in non-small cell lung cancer. Ann Surg Oncol 2006; 13:1517 - 1523
- Vereecken P, Reynaert S, Lalmand MC, Zouaoui-Boudjeltia K, Heenen M, Van Den Heule B, et al. Decreased immunoreactive maspin expression in intermediate thickness and thick primary melanoma lesions. J Int Med Res 2006; 34:52 - 57
- Katakura H, Takenaka K, Nakagawa M, Sonobe M, Adachi M, Ito S, et al. Maspin gene expression is a significant prognostic factor in resected non-small cell lung cancer (NSCLC). Maspin in NSCLC. Lung Cancer 2006; 51:323 - 328
- Takanami I, Abiko T, Koizumi S. Expression of maspin in non-small-cell lung cancer: correlation with clinical features. Clin Lung Cancer 2008; 9:361 - 366
- Kim S, Han J, Kim J, Park C. Maspin expression is transactivated by p63 and is critical for the modulation of lung cancer progression. Cancer Res 2004; 64:6900 - 6905
- Yatabe Y, Mitsudomi T, Takahashi T. Maspin expression in normal lung and non-small-cell lung cancers: cellular property-associated expression under the control of promoter DNA methylation. Oncogene 2004; 23:4041 - 4049
- Hurtubise A, Momparler RL. Evaluation of antineo-plastic action of 5-aza-2′- deoxycytidine (Dacogen) and docetaxel (Taxotere) on human breast, lung and prostate carcinoma cell lines. Anticancer Drugs 2004; 15:161 - 167
- Beltran A, Parikh S, Liu Y, Cuevas BD, Johnson GL, Futscher BW, et al. Re-activation of a dormant tumor suppressor gene maspin by designed transcription factors. Oncogene 2007; 26:2791 - 2798
- Zhang W, Shi HY, Zhang M. Maspin overexpression modulates tumor cell apoptosis through the regulation of Bcl-2 family proteins. BMC Cancer 2005; 5:50
- Nam E, Park C. Maspin suppresses survival of lung cancer cells through modulation of Akt pathway. Cancer Res Treat 2010; 42:42 - 47
- Chen EI, Florens L, Axelrod FT, Monosov E, Barbas CF, Yates JR, et al. Maspin alters the carcinoma proteome. FASEB J 2005; 19:1123 - 1124
- Bailey CM, Hendrix MJ. IRF6 in development and disease: a mediator of quiescence and differentiation. Cell Cycle 2008; 7:1925 - 1930
- Beerli RR, Segal DJ, Dreier B, Barbas CF. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci USA 1998; 95:14628 - 14633
- Beltran AS, Sun X, Lizardi PM, Blancafort P. Reprogramming epigenetic silencing: artificial transcription factors synergize with chromatin remodeling drugs to reactivate the tumor suppressor mammary serine protease inhibitor. Mol Cancer Ther 2008; 7:1080 - 1090
- Bailey CM, Khalkhali-Ellis Z, Seftor EA, Hendrix MJ. Biological functions of maspin. J Cell Physiol 2006; 209:617 - 624
- Wetterwald A, van der Pluijm G, Que I, Sijmons B, Buijs J, Karperien M, et al. Optical imaging of cancer metastasis to bone marrow: a mouse model of minimal residual disease. Am J Pathol 2002; 160:1143 - 1153
- Nakashima M, Ohike N, Nagasaki K, Adachi M, Morohoshi T. Prognostic significance of the maspin tumor suppressor gene in pulmonary adenocarcinoma. J Cancer Res Clin Oncol 2004; 130:475 - 479
- Hirai K, Koizumi K, Haraguchi S, Hirata T, Mikami I, Fukushima M, et al. Prognostic significance of the tumor suppressor gene maspin in non-small cell lung cancer. Ann Thorac Surg 2005; 79:248 - 253
- Bircan A, Bircan S, Kapucuoglu N, Songur N, Ozturk O, Akkaya A. Maspin, VEGF and p53 expression in small biopsies of primary advanced lung cancer and relationship with clinicopathologic parameters. Pathol Oncol Res 2010;
- Woenckhaus M, Bubendorf L, Dalquen P, Foerster J, Blaszyk H, Mirlacher M, et al. Nuclear and cytoplasmic Maspin expression in primary non-small cell lung cancer. J Clin Pathol 2007; 60:483 - 486
- Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res 2008; 659:40 - 48
- Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol 1999; 145:1341 - 1354
- Herrera FJ, Triezenberg SJ. VP16-dependent association of chromatin-modifying coactivators and under-representation of histones at immediate-early gene promoters during herpes simplex virus infection. J Virol 2004; 78:9689 - 9696
- Mittler G, Stuhler T, Santolin L, Uhlmann T, Kremmer E, Lottspeich F, et al. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J 2003; 22:6494 - 6504
- Yang F, DeBeaumont R, Zhou S, Naar AM. The activator-recruited co-factor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci USA 2004; 101:2339 - 2344
- Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW. 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3K9 dimethylation levels are linked to tumor suppressor gene reactivation. Oncogene 2007; 26:77 - 90
- Jiang N, Meng Y, Zhang S, Mensah-Osman E, Sheng S. Maspin sensitizes breast carcinoma cells to induced apoptosis. Oncogene 2002; 21:4089 - 4098
- Liu J, Yin S, Reddy N, Spencer C, Sheng S. Bax mediates the apoptosis-sensitizing effect of maspin. Cancer Res 2004; 64:1703 - 1711
- Li Z, Shi HY, Zhang M. Targeted expression of maspin in tumor vasculatures induces endothelial cell apoptosis. Oncogene 2005; 24:2008 - 2019
- Li X, Yin S, Meng Y, Sakr W, Sheng S. Endogenous inhibition of histone deacetylase 1 by tumor-suppressive maspin. Cancer Res 2006; 66:9323 - 9329
- Kakihana M, Ohira T, Chan D, Webster RB, Kato H, Drabkin HA, et al. Induction of E-cadherin in lung cancer and interaction with growth suppression by histone deacetylase inhibition. J Thorac Oncol 2009; 4:1455 - 1465
- Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol 2004; 24:306 - 319
- Ben Shachar B, Feldstein O, Hacohen D, Ginsberg D. The tumor suppressor maspin mediates E2F1-induced sensitivity of cancer cells to chemotherapy. Mol Cancer Res 2010; 8:363 - 372
- Torchilin VP. Cell penetrating peptide-modified pharmaceutical nanocarriers for intracellular drug and gene delivery. Biopolymers 2008; 90:604 - 610
- Henning P, Andersson KM, Frykholm K, Ali A, Magnusson MK, Nygren PA, et al. Tumor cell targeted gene delivery by adenovirus 5 vectors carrying knobless fibers with antibody-binding domains. Gene Ther 2005; 12:211 - 224
- Harvey TJ, Burdon D, Steele L, Ingram N, Hall GD, Selby PJ, et al. Retargeted adenoviral cancer gene therapy for tumour cells overexpressing epidermal growth factor receptor or urokinase-type plasminogen activator receptor. Gene Ther 2010; 17:1000 - 1010
- Klug A. Towards therapeutic applications of engineered zinc finger proteins. FEBS Lett 2005; 579:892 - 894