Abstract
Benzo(a)pyrene (BaP), is an environmental pollutant present in tobacco smoke and a byproduct of fossil fuel combustion which likely contributes to the tumorigenic processes in human cancers including lung and esophageal. Long Interspersed Nuclear Element-1 (LINE-1) or L1 is a mobile element within the mammalian genome that propagates via a “copy-and-paste” mechanism using reverse transcriptase and RNA intermediates. L1 is strongly expressed during early embryogenesis and then silenced as cells initiate differentiation programming. Although the complex transcriptional control mechanisms of L1 are not well understood, L1 reactivation has been described in several human cancers and following exposure of mouse or human cells to BaP. In this study we investigated the molecular mechanisms and epigenetic events that regulate L1 reactivation following BaP exposure. We show that challenge of HeLa cells with BaP induces early enrichment of the transcriptionally-active chromatin markers histone H3 trimethylated at lysine 4 (H3K4Me3) and histone H3 acetylated at lysine 9 (H3K9Ac), and reduces association of DNA methyltransferase-1 (DNMT1) with the L1 promoter. These changes are followed by proteasome-dependent decreases in cellular DNMT1 expression and sustained reduction of cytosine methylation within the L1 promoter CpG island. Pharmacological inhibition of the proteasome signaling pathway with the inhibitor MG132 blocks degradation of DNMT1 and alters BaP-mediated histone epigenetic modifications. We conclude that genetic reactivation of L1 by BaP involves an ordered cascade of epigenetic events that begin with nucleosomal histone modifications and is completed with alterations in DNMT1 recruitment to the L1 promoter and reduced DNA methylation of CpG islands.
Background
Long interspersed element-1 (LINE-1 or L1) is the most active and abundant mammalian retrotransposon.Citation1,Citation2 A full-length human L1 element is 6–7 kb and contains a 1.0 Kb 5′ untranslated region (5′UTR) region with promoter activity, as well as two open reading frames (ORF1 and ORF2) encoding for proteins involved in reverse transcription and retrotransposition.Citation3,Citation4 Although many of the regulatory proteins and cofactors involved in transcriptional control of mammalian L1s remain unknown, chromatin remodeling and DNA methylation may participate in epigenetic control of retroelements.Citation5–Citation7 The nuclear transcription factor Yin Yang-1 (YY1) has been identified as an important regulator of L1 transcription,Citation8,Citation9 while runt-domain transcription factor 3 (RUNX3) regulates L1 activation and retrotransposition in culture.Citation10
To date, little is known about the complex molecular interactions involved in epigenetic control of L1. The transmission of heritable states of gene expression in the absence of changes in DNA sequence is regulated in part by methylation of DNA at the 5′ position of cytosines by DNA methyltransferases (DNMTs).Citation11 DNA methylation is also an important mechanism for epigenetic silencing of retroelements.Citation5–Citation7 L1 promoter hypomethylation along with increased expression have been described in several cancers, including testicular tumors,Citation12 urothelial bladder carcinoma,Citation13,Citation14 prostate carcinoma,Citation14 hepatocellular carcinoma,Citation15 chronic lymphocytic leukemiaCitation16 and chronic myeloid leukemia.Citation17 These findings suggest that loss of DNMT activity may contribute to L1 reactivation. In addition to L1-specific and global DNA hypomethylation, transformed cells show tumor suppressor specific-DNA hypermethylation.Citation18,Citation19 Other factors known to influence levels of DNA methylation in mammalian cells include histone tail modificationsCitation20 and diet and exogenous insults.Citation21,Citation22 Among these, histone tail modifications have received considerable attention with a focus on acetylation,Citation23 methylation,Citation20 citrullination,Citation24,Citation25 phosphorylationCitation26 and ADP ribosylation.Citation27 For instance, H3K4 trimethylation (H3K4Me3) and H3K9 acetylation (H3K9Ac) are both characteristic of transcriptionally-active chromatin,Citation28,Citation29 while H3K9 trimethylation (H3K9Me3) and H3K27 trimethylation (H3K27Me3) are primarily found in regions of transcriptionally-silent chromatin.Citation30,Citation31 Considerable crosstalk between histone and DNA methylation has been documented.Citation20 Cytosine methylation within promoter CpG repeats is recognized by methyl group-binding proteins (MBDs), which in turn recruit corepressor complexes with histone-modifying enzymatic activities, such as deacetylases (HDACs), to reinforce gene silencing.Citation32
Modulation of epigenetic control may play an important role in chemical carcinogenesis.Citation33 However, studies on the role of environmental insult on the reactivation of repetitive elements such as L1, or the epigenetic mechanisms that regulate tumorigenesis are limited. Previous studies in this laboratory have established that benzo(a)pyrene (BaP), a ubiquitous genotoxic carcinogen, inhibits DNA repair and reactivates L1 in mammalian cells by disruption of transcriptional control.Citation34–Citation38 More recently we have shown that epigenetic control of mammalian L1 is mediated partly by recruitment of E2F/Rb/HDAC complexes.Citation39 The degree to which recruitment of E2F/Rb/HDAC complexes modify DNA methylation and histone covalent modifications in BaP-treated cells remains unknown. This information is critical given the inconsistencies in epigenetic modification profiles described in the literature for carcinogen-treated mammalian cells. For example, in mouse embryonic fibroblasts (MEFs), extended exposures to BaP for seven days leads to overexpression of DNMT1, genetic instability at expanded tandem repeats and global DNA hypermethylation, as measured by cytosine extension assays (CEA).Citation40 Conversely, using a methyl acceptor assay (MAA), Sadikovic and RodenhiserCitation41 did not observe differences between control and BaP-treated human breast cancer cells. Similarly, mice exposed to complex mixtures of PAHs from urban/industrial origin showed increased global hypermethylation in sperm cells, as measured with both CEA and MAA assays,Citation42 while peripheral blood cells from humans exposed to environments rich in PAHs show either hypermethylationCitation43 or hypomethylationCitation44 of L1 retroelements. Thus, the extensive variability in epigenetic response of mammalian cells poses a challenge to our understanding the influence of environmental injury on global epigenetic control.
In this report, we investigated the molecular epigenetic events that regulate the reactivation of specific repetitive sequences within the mammalian genome following BaP exposure. Our studies focused on chromatin dynamics and DNA methylation profiles and their role in the reactivation of the L1 retrotransposon by BaP. We examined the effects of BaP on modulation of histone activation marks (H3K4Me3, H3K9Ac, total H3Ac and total H4Ac), histone silencing marks (H3K9Me3 and H3K27Me3) and transcription factors/corepressor recruitment (Rb, DNMT1, MBD2). Moreover, we studied global DNA methylation changes at six different CpG loci within the L1 5′UTR region. Carcinogen treatment of HeLa cells was associated with early recruitment of H3K4Me3 and H3K9Ac and reduced DNMT1 association with the L1 promoter, followed by proteasome-dependent reductions in DNMT1 protein levels and hypomethylation of L1. These findings establish functional linkages between genotoxic stress, chromatin remodeling and DNA methylation in the regulation of L1 retrotransposon in mammalian cells.
Results
L1 reactivation is associated with early increases in H3K4 trimethylation and H3K9 acetylation.
The first set of ChIP and real-time PCR experiments were conducted to examine histone marks within the L1 promoter (). ChIP-grade antibodies were used to immunoprecipitate H3K4Me3, H3K9Me3, H3K27Me3, H3K9Ac and total H3, while retinoblastoma protein (Rb), a negative regulator of E2F-dependent transcription, was used as a control for precipitation of L1 promoter DNA and IgG isotype antisera was used to control for nonspecific immunoprecipitation. Challenge of cells with BaP for 12 h increased L1 promoter enrichment for H3K4Me3 and H3K9Ac, but did not change accumulation of the repressive marks H3K9triMe or H3K27triMe. In keeping with previous findings, enrichment for L1 DNA using Rb antisera was observed.Citation39 Total histone H3 control was comparable for both treatments, while IgG background levels were undetectable, indicating that enrichment for L1 was specific. Real time PCR targeting the 3′UTR confirmed that chromatin modifications were promoter-specific (Sup. Fig. 1A). BaP treatment also reduced L1 promoter recruitment of DNMT1 (), while both IgG (shown in ) and 3′UTR amplification of the L1 promoter (Sup. Fig. 1B) yielded values close to background. These findings indicated that enrichment of L1 was specific for the antisera used and that carcinogen challenge involves downstream changes in L1 methylation. Thus, BaP likely activates L1 via signaling mechanisms that modulate chromatin dynamics and favor association of proteins that induce active chromatin conformations proximal to the L1 promoter, while inhibiting association of the DNA methylation machinery.
Hypomethylation of the L1 promoter in human and mouse cells by BaP.
The next set of studies focused on promoter methylation dynamics in BaP-treated cells. These experiments examined the L1 retinitis pigmentosa (L1RP) element, an active human retrotransposon containing 34 CpGs spread over a 371 bp CpG island within the promoter region, as predicted by MethPrimer™ CpG island softwareCitation45 (). HeLa cells were challenged with vehicle or 3 µM BaP for 96 h and DNA extracted, quantified and processed for DNA methylation studies. Several techniques for studying CpG methylation are available, including methylation specific PCR (MS-PCR) and pyrosequencing. We performed preliminary studies using MS-PCR primer sets for the human L1 promoter spanning two CpG sites (CpG 20 and 38) located on the CpG island, −887 and −869 base pairs upstream of the ORF1 start site, respectively. These CpG spots encompass the binding site for YY1,Citation9 (). Increased PCR amplification signal for unmethylated cytosine on the genomic template was obtained in cells treated with 3 µM BaP for 96 h compared to DMSO control (Sup. Fig. 2). Furthermore, using primary mouse aortic vascular smooth muscle cells (vSMC) in studies for the active L1MdA5 mouse retrotransposonCitation38 we identified a similar pattern to that of HeLa cells, suggesting that methylation of this L1 subtype in mouse primary cells is also subject to modulation of promoter DNA methylation status in a BaP-dependent manner (Sup. Fig. 2).
Because MS-PCR is limited in the number of CpG loci that can be analyzed at one time, and only provides a qualitative estimate of DNA methylation status, pyrosequencing was completed for multiple CpG sites ( and ). These CpGs included binding sites for, or proximal to, SP1, AP-1, CREB, ERα and NFκβ transcription factors (). In cytosine methylation analyses, C and T (C/T) are the input nucleotide at the locus being analyzed, and the output gives percentage amounts of incorporation of each added base at that locus. Quantitative analysis of L1 CpG islands in cells challenged for 96 h with vehicle or BaP showed reductions in methylation of CpG sites within the promoter ranging from 4–7% (). Collectively, these findings established that carcinogen treatment causes hypomethylation of specific CpG sites within the L1 promoter.
Given that changes in chromatin status precede changes in methylation, as exemplified by epigenetic inactivation of the RASSF1A tumor suppressor,Citation46 early chromatin modifications may set the stage for changes in DNA methylation at later time points. This hypothesis is consistent with previous findings showing that L1 activation kinetics by BaP display peaks at 12 and 72 h following carcinogen challenge.Citation47 To test this hypothesis, HeLa cells were treated with BaP for 12, 24, 48, 72 and 96 h, and processed for measurements of DNA methylation. We analyzed CpG 205 and CpG 252 as critical sites lying proximal to SP1, AP1, CREB, ERα and C/EBP putative binding sites. Significant hypomethylation occurred at both sites only at the 96 h time point, confirming that changes in DNA methylation exhibit delayed kinetics. A time-course pyrosequencing analysis for CpG 205 and 252 is shown in . In all, the data indicate that BaP exposure leads to decreased DNA methylation of retroelement promoters in both human epithelial tumor cells and mouse primary vascular cells. Likely, cells must undergo several replication cycles before significant DNA demethylation changes could be detected.
Depletion of DNA methyltransferase mediates L1 promoter DNA hypomethylation and modulates L1 expression.
To evaluate the effects of DNMTs on L1 promoter methylation, HeLa cells were transfected with siRNA targeting DNMT1, DNMT3A and DNMT3B, or off target siRNA. mRNA knockdown was confirmed by real-time PCR (). Since trace contaminants of DNA might lead to non-specific amplification of target sequences, an effect that could be more severe when detecting transcript levels of repetitive elements, RNA samples were subjected to DNase treatment before cDNA synthesis. Also, control reactions for cDNA synthesis in the absence of reverse transcriptase (no RT) were processed for real time PCR under similar conditions (Sup. Fig. 3A). The results indicate that the products obtained in the RT reactions are specific, and do not originate from exogenous DNA contamination. In addition, western blot assays for DNMT1, DNMT3a and GAPDH protein expression levels in HeLa cells following siRNA treatment were performed ( and C). A decreased level of cellular DNMT protein levels specific for the respective siRNA targets was observed, suggesting that concomitant with decreased message levels, siRNA treatment reflected diminished cellular availability of the target proteins, (). Genomic DNA was isolated, subjected to bisulphite treatment and analyzed for L1 promoter CpG methylation status via quantitative pyrosequencing. Following siRNA treatment, a significant reduction in L1 promoter methylation was observed across several CpG loci examined (), showing that disruption of DNMT1, DNMT3A and DNMT3B demethylates the L1 promoter. The response to siRNA was comparable to that induced by 5-aza-2′-deoxycytidine, a potent demethylating agent used as a positive control.Citation48 Interestingly, L1 CpGs 103 and 205 remained unaltered in cells lacking DNMT1. To determine if functional links exist between L1 promoter hypomethylation and L1 expression, HeLa cells were transfected with siRNAs targeting DNMT1, DNMT3A and DNMT3B for 48 h and processed for measurement of L1 mRNA by real-time PCR. Primers targeting ORF 1 and ORF 2 were used to amplify both L1 open reading frames. Silencing of DNMT1 significantly increased L1 ORF1 and ORF2 mRNA levels (). In contrast, siRNAs targeting DNMT3A and DNMT3B were without effect on L1. This was unexpected because siRNA targeting DNMT3A and DNMT3B decreased methylation across several CpG dinucleotides. To control for exogenous DNA contamination in the samples tested, no RT reactions were run under similar conditions (Sup. Fig. 3B).
BaP reduces DNA methyltransferase levels and promotes epigenetic changes on both L1 promoter DNA and histones.
Since mammalian cells silence retrotransposons primarily via promoter methylation, and BaP reduces L1 DNA methylation, we next determined the effects of BaP on the maintenance DNMT1 and de novo methyltransfersases, DNMT3A and DNMT3B, in HeLa cells. Cells were treated with vehicle or BaP, and protein extracted and processed for western blotting using antisera against DNA methyltransferases. Detectable signals were only seen for DNMT1 and DNMT3A. GAPDH was included as a loading control. Challenge of HeLa cells with 3 µM BaP reduced the levels of both DNMT1 and DNMT3A proteins, but not GAPDH (), suggesting that decreased methylation of the L1 CpG islands is mediated through disruption of the cellular DNA methylation machinery. Next, we determined if BaP targeted DNMT1 for degradation by the proteasome pathway. HeLa cells were pretreated with the proteasome inhibitor MG132 at 1 µM for 3 h followed by 3 µM BaP challenge for 48 h. As seen earlier, reduced levels of DNMT1 protein were seen in cells treated with BaP compared to vehicle alone. Cells co-treated with MG132 and BaP showed stabilization of DNMT1 protein levels (). Consistent with the notion that BaP targets AHR for proteasomal degradation,Citation49–Citation51 AHR protein levels decreased when cells were treated with BaP, and this response was inhibited by MG132. Parallel experiments showed that combined MG132 and BaP treatment increased methylation at several CpG sites (). However, treatment of cells with MG132 alone stabilized DNMT1 levels and induced demethylation at CpG 252. This finding is consistent with previous reports showing that MG132 modulates multiple histone and DNA modifying enzymes.Citation52 Collectively, these results indicate that protection of DNMT1 from degradation is translated into specific increases in DNA methylation at several CpG dinucleotides within the L1 promoter. Importantly, cigarette smoke leads to proteasomal-dependent degradation of HDAC2,Citation53 and HDAC1 plays a role in the proteosomal-mediated degradation of DNMT1 through modulation of HSP90 activity.Citation54 We therefore studied MG132 actions at the chromatin level following short-term exposure of HeLa cells to BaP. Cells were treated with 1 µM MG132 for 3 h followed by 12 h exposure to DMSO vehicle or 3 µM BaP. Since acetylation of histones is a key feature of transcriptionally-active chromatin, antisera against total histone H3 or total histone H4 was used. Also, antisera against histone H3 lysine 9 trimethyl repressive mark, total histone H3 and isotype-matched IgG were included to control for non-specific enrichment of L1 DNA. “Mock” samples containing protein A-agarose beads exposed to buffers were also examined. Our results showed that the isotype-matched IgG samples yielded negligible amounts of background amplification (). No amplification was seen in mock samples (not shown), indicating that DNA amplification was specific for enriched sequences. Treatment of cells with proteasome inhibitor diminished overall acetylation for histone H4, and this effect was not reversed by BaP treatment. Conversely, treatment of cells with MG132 decreased the silencing methylation mark H3K9triMe, but this effect was reversed upon addition of BaP. These data suggest that MG132 reverses and/or prevents epigenetic activation marks and promotes histone silencing marks in the presence of BaP. These data are consistent with the accumulation of DNMTs seen upon pharmacological inhibition of the proteasomal degradation pathway.
Discussion
During embryonic development the genome undergoes extensive reprogramming based upon global cytosine demethylation, later followed by specific patterns of DNA methylation established de novo during preimplantation.Citation55,Citation56 Maintenance methylation in somatic cells is essential, as aberrant changes in DNA methylation are linked to genomic instability, cell death and tumorigenesis.Citation17 Repetitive sequences are extensively silenced via DNA methylation, and L1 methylation status is often used as a proxy of global cellular methylation.Citation57,Citation58 Although literature focusing on environmental influences on repetitive sequences is scarce, genetically identical agouti mice have been found to develop markedly different phenotypes into adulthood which are accounted for by epigenetic changes during early embryogenesis.Citation59 Environmental factors including endocrine disruptors and diet have also been linked to epigenetic modulation. Bisphenol A, a widely publicized estrogenic endocrine disruptor, targets the Agouti gene and induces in utero hypomethylation of the inserted murine intracisternal A-particle (IAP) retrotransposon,Citation60 a retroelement located at the 5′ end of Agouti.Citation61 Here we show that challenge of HeLa cells with the carcinogen BaP reactivates L1RP retroelement by modulation of nucleosomal histone marks and global patterns of DNA hypomethylation. BaP treatment enriched the L1 promoter for H3K4Me3 and H3K9Ac, markers of transcriptionally-active chromatin. These findings are consistent with recent studies showing that L1 reactivation involves histone H3 acetylation at or near the 5′UTR regulatory region, a modification tightly controlled by E2F/Rb.Citation39 Early changes in covalent histone modification may contribute to sustained reactivation of L1 via inhibition of DNMT1 recruitment and activity. This interpretation is consistent with our finding that changes in DNA methylation were not seen with short term exposures, and work by others showing that changes in DNA methylation require at least one replication cycle, while chromatin modifications are observed within a few hours following recognition of the appropriate stimulus.Citation62
Pavanello et al.Citation43 examined the methylation status of L1 repetitive sequences in human peripheral blood lymphocytes isolated from coke-oven workers subjected to chronic exposure to polycyclic aromatic hydrocarbons (PAHs) in combination with metals. This study reported that both L1 and Alu sequences are hypermethylated in exposed workers compared to controls, and exhibited significantly higher levels of anti-B[a]PDE-DNA adducts and micronuclei, a marker of genetic injury. Using univariate linear regression analysis the authors also reported statistically significant correlations between LINE-1 and Alu hypermethylation and PAH exposure (1-pyrenol) or anti-B[a]PDE-DNA adducts. These observations are inconsistent with our findings of L1 hypomethylation in HeLa cells challenged with BaP. Although only one L1 family has been identified in humans, several subtypes of retroelements are known to exist within the mammalian genome. Our study focused on the L1RP, an element found within the retinitis pigmentosa-2 gene, and one of the few retroelements retaining a strong capacity for retrotransposition.Citation34,Citation63 As such, the difference in intrinsic activity between L1RP and other L1s may account for differences in patterns and extent of epigenetic regulation. Interestingly, the same primer set used by Pavanello et al. to cover a single, conserved CpG island located at position 252 of the 5′-UTR of a non-L1RP retroelement, was used by Bollati et al.Citation64 to study individuals exposed to low-dose airborne PAH (urban traffic officers and gas station attendants). Bollati et al. found hypomethylation of L1 sequences without changes in Alu elements, a finding that is in keeping with our finding of hypomethylation, but inconsistent with Pavanello et al.Citation43. Such discrepancies notwithstanding, differences in pattern or extent of DNA methylation between these studies may be accounted for by differences in length of chemical exposures, absence or presence of multiple chemicals, as well as contextual differences between different cell types.
In the present study, we examined six different CpG loci, four of which showed a consistent and statistically significant pattern of hypomethylation. The fidelity of the response in multiple experiments indicates that the BaP effect is highly reproducible and specific in this cell type. Using breast cancer cell lines, Sadikovic and RodenhiserCitation41 reported that BaP impairs DNA methylation and induces global DNA hypomethylation of SINEs and LTR retroelements. To determine if the hypomethylation response is tumor cell specific, we also examined the response of murine primary vSMC to BaP and found that L1MdA is also hypomethylated in these cells. These data are consistent with the ability of BaP challenge to reactivate L1 in both primary and tumor cell lines.Citation35 In addition, L1 elements in mouse and humans are subject to epigenetic silencing via E2F/Rb/HDAC1/HDAC2 protein complexes.Citation39 Since HDAC2 is downregulated by cigarette smokeCitation53 and HDAC1 promotes the degradation of DNMT1,Citation54 both transformed and primary cells may undergo L1 hypomethylation via proteasomal-mediated degradation of HDACsCitation53 and DNMTsCitation54,Citation65 and disruption of the repressor activity of E2F/Rb on the L1 promoter.Citation39,Citation66 Furthermore, Bradley et al.Citation67 used normal human mammary epithelial cells to examine the effects of carcinogens, including TCDD and BaP, on the global epigenetic histone H3 lysine 4 monomethyl (H3K4Me) mark and the di- and tri-methyl marks on H3K4. Two points regarding this study are important; first, treatment with different doses of BaP at different times did not alter H3K4Me and second, no detectable histone H3K4 tri-methylation was found in either treated or control cells, a finding that makes their results questionable given that H3K4Me3 is a widely recognized mark of transcriptional activation in the 5′ region of most active eukaryotic genes.Citation29 Sadikovic et al.Citation68 exposed MCF7 breast cancer cells to 0.5 µM BaP for four days. ChIP assays with antisera against histone H3 lysine 9 mono-acetylation (H3K9Ac) followed by microarray hybridization to the Affimetrix Human Promoter 1.0R platform (ChiP-on-Chip) employing repeat masker to exclude repetitive sequences was used to detect 1,456 gene promoters with increased H3K9 monoacetylation following BaP treatment. Because repetitive elements were excluded from this analysis, direct comparisons to our findings could not be made.
Extended treatment of HeLa cells with BaP decreased cellular levels of DNMT1 and DNMT3A, and elicited concomitant decreases in methylation at several CpG loci within the L1 promoter. Also, siRNAs targeting DNMTs decreased methylation across several CpG sites, confirming that changes in methylation status are linked to changes in DNMT activity. Endogenous regulation of L1 activity was selectively modulated by DNMT1, a regulator of maintenance DNA methylation.Citation69 Although DNMT3 may influence methylation of L1 CpG loci through cooperative interactions, this was not seen in our study, and may in fact be restricted to developmental periods when methylation patterns are being established.Citation70,Citation71 Hence, cell division following siRNA-mediated DNMT1 silencing would render hemimethylated L1 sequences prone to transcriptional reactivation and increasing number of methylation marks would be lost as replication cycles continue. Since other DNMTs cannot compensate for maintenance DNMT1 activity, this would explain the selective increase of L1 expression seen after DNMT1 siRNA treatment. The time lag between decreased levels of protein and effects on CpG loci may reflect the semiconservative nature of DNA replication requiring at least two complete rounds for removal of methyl groups from regulatory regions of DNA.
The absence of DNMT1 may play a role in disassembly of one or more repressor complex(es) involving CpG loci methylation and nucleosomal histone modifications. Of note was the finding that the two L1 promoter CpG cytosines unaffected by siRNA targeting DNMT1 were among the farthest away from the open reading frame, supporting the view that specific methylation of CpG cytosines plays a direct role in L1 silencing. The CpGs at positions 137, 205 and 351 contain proximal SP1 binding sites, while the CpG on position 205 flanks an NFκβ and ERα binding site, and CpG 252 contains a CREB binding site. These relationships are potentially relevant given that SP1, NFκβ and CREB are important nodes within the AHR regulatory network in mammalian cells,Citation72 and AHR in partnership with RB function as regulators of L1.Citation35,Citation39 Together, these findings suggest that BaP induces changes in nucleosomal histone modification and transcription factor recruitment that impact cytosine methylation. BaP has previously been shown to inhibit assembly of the DNA methylation machinery,Citation73,Citation74 and to dysregulate DNA methylation in breast cancer cells.Citation41 Interestingly, the CpG at position 103 with no proximal transcription factor binding sites was neither affected by BaP treatment nor DNMT1 silencing. Thus, differential recruitment of chromatin-modifying proteins by different CpG sites likely contributes to spatio-temporal regulation of the L1 promoter.
Evidence was also obtained that BaP targets DNMT1 for degradation via the proteasome pathway, and that pretreatment of cells with the proteasome inhibitor MG132 stabilized DNMT1 following challenge with BaP and increased DNA methylation of several CpG dinucleotides. These are important findings in that BaP may contribute to alterations of global DNA methylation via proteasome-dependent mechanisms that regulate DNMT1 and other proteins involved in maintenance of genomic imprints inherited via epigenetic mechanisms. The observation that MG132 alone reduced methylation on CpG 252 is consistent with the ability of MG132 to modulate multiple chromatin modifying enzymes, as well as DNA methyltransferases and demethylases.Citation52,Citation75 It is not yet clear why siRNA targeting DNMT3A and DNMT3B decreased methylation across several CpG dinucleotides, but did not regulate L1 activity. In this regard, the degree to which adduction of DNA by BaP intermediates influences the interaction of DNA binding proteins with the L1 promoter may be a significant factor. DNA binding of 7r,8t-dihydroxy-9t,10t-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (BPDE) alters the affinity of Sp1 for its canonical target by inducing recruitment of Sp1 protein to DNA in non-motif regions and abolishing binding to GC boxes.Citation76 Also, BPDE adducts force abnormal bending of the DNA double helix hence contributing to aberrant DNA-protein interactions, mutagenesis and cellular transformation. Remarkably, in vitro experiments with heterodimers of the transcription factors DP1 and E2F1 or DP1 and E2F4 show increased affinity for BPDE-modified DNA oligonucleotides carrying the canonical E2F binding sequence, as well the non-canonical BPDE-DNA sequences.Citation77 Thus, DNA adduction may influence patterns of DNMT binding and activity within the mammalian genome, and differentially modulate the interaction of DNMTs with proteins involved in chromatin remodeling, such as Rb and Rb-related proteins.Citation40 In agreement with this interpretation, short-term exposure to BaP in the presence of MG132 diminished total acetylation levels for histone H4 on the L1 promoter, and this effect could not be reversed by addition of BaP. Conversely, histone H3 lysine 9 trimethylation, a silencing mark, is augmented by BaP suggesting that activity of the silencing machinery is dependent on proteasomal pathways and that surveillance systems in the cells work to maintain L1 silenced through rapid histone modifications, including H3K9triMe.
Materials and Methods
Cell culture.
Mouse vascular smooth muscle cells (VSMCs) and mK4 cells were grown in M199, while HeLa cells were grown in DMEM, all supplemented with 10% FBS and 1% w/v antibiotic (penicillin-streptomycin).
Western blotting.
Total cellular proteins were isolated using Mammalian Protein Extraction Reagent (MPER, PIERCE) or RIPA buffer supplemented with 1X protease inhibitor cocktail (PIERCE). Twenty to forty micrograms of total protein were separated by 4–12% gradient SDS-PAGE (Invitrogen) and transferred to PVDF membranes overnight followed by blocking with 5% non-fat skim milk or BSA. PVDF membranes were probed with antibodies to DNMTs: anti-DNMT1 (IMGENEX, IMG 261A, lot AB 030707D-01) and anti-DNMT3A (Cell Signalling, Catalog # 2160S, lot #1), or GAPDH (Sigma-Aldrich, St. Louis, MO) in 1x Tris-buffered saline. Target antibodies were used at a dilution of 1:1,000 overnight in a cold chamber followed by HRP-conjugated secondary antibody at room temperature for 1 hour at 1:20,000 dilution. All washes were performed with TBS-0.05% Tween 20, 5× for 5 min each time. Blots were developed using West Dura chemiluminescent western blot detection reagent (PIERCE) and visualized using an x-ray film (KODAK). Membranes were exposed to film from 5 sec up to 3 min.
Quantitative real time PCR.
Total RNA was extracted using Trizol reagent (Invitrogen) in combination with RNeasy columns (Qiagen). RNA was then quantified and subjected to DNase treatment following manufacturer's instructions (Ambion). One µg of DNA-free RNA was used for cDNA synthesis using Invitrogen Superscript II reverse transcriptase system according to manufacturer's instructions (Invitrogen). Following reverse transcription, all real time amplifications were performed using SYBR Green (BIORAD). For each reaction, 25 µL of 2x SYBR green was mixed with 10 µM final concentration of forward and reverse primers. One µL of cDNA was added and volume brought up to 50 µL with DEPC water. Control reactions containing RNA subject to cDNA synthesis, but in the absence of reverse transcriptase (no RT) were used to control for exogenous DNA contamination. Cycling conditions were as follow: initial denaturation step at 95°C for 3 min, and 35–50 cycles at 95°C for 30 s, 55°C for 30 s and 72°C for 45 s. A real-time melting curve was done to ensure homogeneity of PCR products and data were analyzed based on the ΔΔCt approach. A minimum of three independent replicate samples was used for all experiments and each assay was repeated two or three times.
Methylation-specific PCR.
HeLa cells, mouse vascular smooth muscle cell (VSMC) and embryonic kidney (mK4) cells were grown to 50% confluence and challenged with 3 µM BaP for 96 h. DNA was extracted, quantified and 1 µg subjected to bisulfite treatment. Allele-specific PCR was used to differentiate between methylated alleles (M) versus unmethylated alleles (U). PCR primer sets for the human L1 promoter spanned three CpG sites located at 887, 873 and 869 base pairs upstream of the ORF1 start site, while those for the mouse promoter spanned two CpG sites located 487 and 483 base pairs upstream of the start site. Primers spanning multiple CpG spots were used to ensure high specificity in distinguishing methylated from non-methylated alleles.
DNA pyrosequencing.
HeLa cells at 50% confluence were treated with 3 µM BaP for 96 h and genomic DNA extracted using DNAzol (Invitrogen 10502-027). Quantitative pyrosequencing analyses were completed as described by Alexander et al.Citation80 to examine cytosine methylation at multiple CpG dinucleotidess spanning the L1 promoter following BaP treatment. The CpG island was identified using the MethPrimer™ software (www.urogene.org/methprimer/index1.html). Briefly, DNA was extracted and treated with bisulfate using the Qiagen Epitect® Bisulfite Kit (Cat #59104) according to the manufacturer's instructions. Following bisulfate treatment, the DNA sequence of the 420 fragment comprising the CpG island of L1RP was converted in silico using the “Snake Charmer™” software and the resulting sequence used to design primers for PCR. (The snake charmer can be found at insilico.ehu.es/restriction/two_seq/snake_charmer.html). The reverse PCR primer was biotinylated for pyrosequencing analyses experiments. PCR products were purified to remove excess primers using the Qiagen QIAquick® PCR Purification Kit (Cat #28104). For each sample a binding reaction comprising 35 µL of binding buffer® (Biotage 40-0033), 40 µL water, 2 µL Streptavidin (Amersham #17-5113-01) and 5 µL purified PCR reaction was placed in a 96-well plate and vortexed at high speed for 15 min. The biotinylated PCR product was trapped by the avidin beads. Using a vaccum preparation tool (# 600180) with the corresponding filter probes (impermeable to biotin-avidin tagged PCR products), biotinyl PCR product was temporarily immobilized on the probes by suction, and washed for 8 seconds in 70% ethanol, denatured in 0.2 M sodium hydroxide for 8 seconds and washed in 1X Biotage wash buffer (Biotage #40-005). The vacuum sucks off the denatured DNA, but traps the biotin-avidin DNA complexes. The denatured biotin-avidin strands were then transferred onto the annealing reaction prepared on Biotage PSQ 96 plates (# 40-0010). The annealing reaction in each well contained 8.4 µL annealing buffer (Biotage #40-0036) and 3.6 µL of 1 µM sequencing primer. Sequencing primers used are shown on . The plate was heated on a heat block at 94°C for 2 min and set at room temperature to cool. The samples were quantitatively sequenced using a Biotage Pyrosequencer HS 96A. Pyrosequencing begins with the release of pyrophosphate (PPi) as a result of dNTP incorporation by DNA polymerase. The released PPi is subsequently converted to ATP by ATP sulfurylase. ATP provides energy for the next step which is oxidation of luciferin by a luciferase enzyme. This reaction generates light, which is proportional to the number of dNTPs incorporated. Unincorporated nucleotides are degraded by a pyrase before addition of the next nucleotide, allowing predetermined addition of nucleotides based on the expected sequence (dispensation order). Since the dispensation order of nucleotides is known, the sequence of the template can be determined based on percent incorporation of sequentially added nucleotides.
Chromatin immunoprecipitation.
Recruitment of DNMTs and specific histone modifications of the L1 promoter following BaP treatment were assessed using chromatin immunoprecipitation (ChIP) assays, as described in reference Citation81, with minor modifications. HeLa cells were grown in 10 cm petri dishes to 90% confluence and fixed with 1% formaldehyde for 7–10 min at room temperature with gentle agitation. Formaldehyde quenching was performed by adding glycine to a final concentration of 0.125 M for 10 min. Plates were rinsed 3x with cold PBS and cells were lysed with modified RIPA buffer (50 mM Tris-Cl pH 7.4; 1 mM EDTA; 150 mM NaCl; 0.1% SDS; 1% TritonX-100; 0.1% sodium deoxycholate). Chromatin was sheared to fragment sizes below 1 Kb by sonication in a Sonifier® Model 450 ultrasonic cell disruptor (Branson Ultrasonics Corporation, Danbury, CT) with five to ten cycles (30 seconds each) at 40% amplitude depending on the amount of chromatin and fragment size after each cycle. To facilitate chromatin fragmentation, glass beads were added to the chromatin solution. Fragment sizes were checked after every three rounds of sonication. Before immunoprecipitation, chromatin samples were precleared for 2 h with protein A-agarose beads. Immunprecipitation reactions were performed using 25–40 µg of chromatin per sample. Four micrograms of antisera for histone H3K9Me3 (Upstate 07-442 lot DAM1411287), histone H3K4Me3 (Upstate 05-747 lot DAM1460170), H3K9Ac (Upstate 07-352 lot DAM1394804), H3K20Me3 (Upstate 07-449 lot DAM1421462), total Histone H3 (Upstaste 05-928 or 07-690 lot JBC 1353328 or 30374 respectively), E2F-1 (sc-193; C-20, lot G0505), histone H3 acetyl (Upstate biotechnology, 06-599 lot 29505), histone H4 acetyl (Upstate biotechnology, 06-598 lot 29867), histone H3K9 trimethyl (abcam, ab8898-100 lot 100455) or rabbit IgG (Santa Cruz Biotechnology, sc-2027 lot A170) were added and immunocomplexes allowed to form overnight at 4°C with gentle rocking in a volume no larger than 200 µL. To control for exogenous DNA contamination, mock samples containing protein A-agarose beads in 200 µL of modified RIPA buffer were run alongside with samples. Twenty microliters of a 50% slurry of protein A-agarose beads (Invitrogen Corporation, Carlsbad, CA) were used to capture immunocomplexes and extensive washes with modified RIPA buffer were performed to disrupt non-specific interactions. One final wash with LiCl buffer (50 mM Tris-Cl pH 7.4; 1 mM EDTA; 250 mM LiCl; 1% TritonX-100; 0.1% sodium deoxycholate) and two washes with TE were performed. Immunoprecipitated complexes and total input chromatin equivalent to 20% of the starting material were treated for 30 min with 10 µg RNAse A and then overnight with 20 µg proteinase K (Sigma-Aldrich, St. Louis, MO). Protein-DNA associations were reversed by incubation at 65°C for 6 h and the DNA purified using phenol-chloroform extraction and ethanol precipitation in the presence of glycogen. Precipitated DNA from both immunoprecipitated and input samples were resuspended in either 50 µL or 1,000 µL of TE buffer, respectively, and 0.5–2.0 µL of each sample was used in a PCR reaction to amplify the 5′UTR promoter region of the human L1RP retrotransposon. PCR primer sequences used were L1 5′UTR Fwd: 5′-TTG CTA GCA CAG CAG TCT GA-3′, L1 5′UTR Rv: 5′-AGC TGC AGG TCT GTT GGA AT-3′, L1 3′UTR Fwd: 5′-ACA CCG CAT ATT CTC ACT CAT AGG T-3′, L1 3′UTR Rv: 5′-TGT ATA CAT GTG CCA TGC TGG TGC-3′.
Measurement of the L1 promoter sequence enrichment within ChIP samples was performed according to the SABiosciences ChIP-qPCR Data Analysis manual (www.sabiosciences.com/chipqpcrresource.php) and plotted as a function of percent enrichment relative to input chromatin. DNA was extracted from 20% of the total amount of chromatin used for each immunoprecipitation, resuspended to an equivalent dilution of 1% and used as template for PCR. Briefly, the Ct value from each sample was subtracted from the input Ct and corrected for dilution corresponding to 1% input. To calculate the immunoprecipitation enrichment expressed as % input, 2 is raised to negative ΔCt and multiplied ×100.
siRNA transfection.
HeLa cells were seeded at 60% confluence without antibiotic and transfected with 50 nM siRNA (final concentration) targeting DNMTs. After 24 h the transfection medium was changed and cells were cultured for an additional 48 h and RNA or total cellular proteins extracted and processed for real time PCR or western blot assays as described above. Replicate samples were incubated an additional 48 h, DNA extracted and processed for pyrosequencing as described above.
Proteasome inhibitor assays.
HeLa cells were seeded at 60% confluence, and pre-treated with 1 µM MG132 for 3 h. The drug was removed followed by the addition of fresh medium containing vehicle DMSO or 3 µM BaP for an additional 48 h. Total protein lysates were obtained as described for “western blot procedures” and 40 µg of each sample resolved on a 4–12% gradient SDS PAGE followed by western blot assays using antisera against DNMT1, DNMT3A and GAPDH or β-actin as controls. For ChIP assays, MG132 pretreatment was followed by vehicle DMSO or B(a)P treatment for 12 h, as specified. Samples were then fixed and processed for ChIP assays as described.
Statistical analyses.
Analyses were done either by student's t test or ANOVA (p < 0.05) followed by post-hoc tests.
Conclusions
Our study is the first to examine global changes in several CpG islands within the human L1RP element and provides novel insights into specific epigenetic alterations of L1 induced by BaP in mammalian cells. We show that early reactivation of L1 retrotransposon by BaP involves complex epigenetic changes that favor early changes in chromatin assembly that culminate in long-term changes in DNA methylation and sustained reactivation. As such, L1 elements, once thought to be silenced via DNA methylation, are subject to complex regulatory control involving histone acetylation and methylation, and transcription factor recruitment, for silencing and/or reactivation. Overall, a novel mechanism for retrotransposon reactivation involving changes in nucleosomal histone code, DNMT availability and CpG hypomethylation can be proposed (). These relationships have profound implications for processes involving environmental influences on mammalian development, differentiation and malignant transformation.
Financial Support
This work was supported in part by National Institutes of Health (NIH, USA), Grant 5R01 ES004849 and ES014443 to K.S.R.
Authors' Contributions
I.T. carried out cell culture work, BaP-related molecular studies including ChIP assays, pyrosequencing studies, western blots, qPCR, performed the statistical analysis and drafted part of the manuscript.
D.E.M.D.: carried out cell culture work, BaP-related molecular studies including ChIP assays, western blots, qPCR, pharmacological treatments and participated in drafting the manuscript.
J.L.Q.: participated in transient transfection and western blot experiments.
M.E.L.: carried out BaP and pharmacological treatments and ChIP experiments.
K.S.R.: conceived the study, and participated in its design and coordination and helped to draft the manuscript.
All authors read and approved the final manuscript.
Abbreviations
| 5-aza-dC | = | 5-aza-2′-deoxycytidine |
| BPDE | = | 7r,8t-dihydroxy-9t,10t-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene |
| 5′UTR | = | 5′ untranslated region |
| AHR | = | aryl hydrocarbon receptor |
| BaP | = | benzo(a)pyrene |
| DNMT1 | = | DNA methyltransferase-1 |
| HDACs | = | histone deacetylases |
| H3K4Me3 | = | histone H3 trimethylated at lysine 4 |
| H3K9Ac | = | histone H3 acetylated at lysine 9 |
| H3K9Me3 | = | histone H3 trimethylated at lysine 9 |
| H3K27Me3 | = | histone H3 trimethylated at lysine 27 |
| LINE-1 or L1 | = | long interspersed nuclear element-1 |
| L1RP | = | L1 retinitis pigmentosa |
| MBDs | = | methyl group-binding proteins |
| IAP | = | murine intracisternal A-particle |
| ORF1 and ORF2 | = | open reading frames 1 and 2 |
| PAHs | = | polycyclic aromatic hydrocarbons |
| pRb | = | retinoblastoma protein |
| RUNX3 | = | runt-domain transcription factor 3 |
| YY1 | = | yin yang-1 |
Figures and Tables
Figure 1 BaP increases H3K4 trimethylation and H3K9 acetylation events and modulates recruitment of DNMT1 to the L1 promoter. HeLa cells treated with DMSO vehicle or 3 µM BaP for 12 hours were formaldehyde-cross-linked and chromatin processed for ChIP assays. Four µg of antisera were used for each immunoprecipitation with antibodies targeting different histone modifications or transcription factors as shown and purified DNA subjected to real time PCR. (A) Real time PCR was performed using primers targeting the L1 promoter (5′UTR). (B) Real time PCR for DNA samples from ChIP assays with antibodies shown was performed using primers targeting the L1 promoter (5′UTR). Isotype-matched IgG control for all reactions (A and B) is shown in (A). The results shown are representative of experiments performed in triplicate for each sample and repeated 3×. Calculations for L1 promoter sequence enrichment within ChIP samples was performed according to the SABiosciences ChIP-qPCR Data Analysis manual (www.sabiosciences.com/chipqpcrresource.php) and plotted as a function of percent enrichment relative to input (% input) chromatin. Statistical analyses done using ANOVA (* indicates statistically significant differences, p < 0.05).
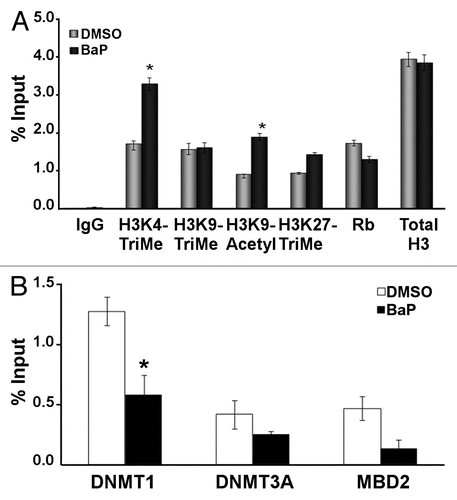
Figure 2 BaP causes hypomethylation of the human and mouse L1 promoters. (A) Schematic showing CpG island and the location of MS-PCR primers used for analyses in HeLa cells. The mouse primers targeted similar locations on the mouse L1MdA5 promoter (B). CpG positions analyzed on the human L1RP promoter and putative transcription factor binding sites as identified by MatInspector (www.genomatrix.de). Sequencing primers were chosen such that specific CpG spots could be analyzed via pyrosequencing. (C) HeLa cells were treated with DMSO vehicle or 3 µM BaP for 96 h, genomic DNA extracted, treated with bisulfite and pyrosequenced. Percent cytosine methylation for the indicated CpG loci was thus determined, and statistic analyses done using ANOVA (* indicates statistically significant differences, p < 0.05). The results shown are representative of three independent experiments. Each CpG locus was analyzed at least twice for each pyrosequencing run.
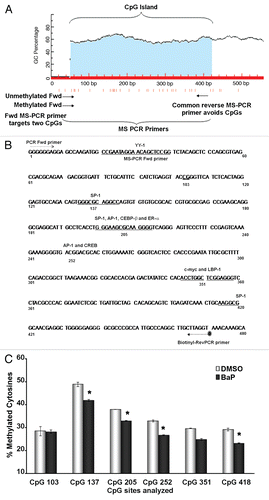
Figure 3 BaP-induced L1 DNA hypomethylation is observed at the 96 h time point. HeLa cells were treated with BaP for 12, 24, 48, 72 and 96 h, DNA harvested, bisulfite treated, PCR amplified (with a biotin-tagged reverse PCR primer) and pyrosequenced using sequencing primers specific for CpG loci 205 and 252. (A) shows a quantitative representation of pyrosequencing analyses for CpG 205. (B) shows the same for CpG252. Results are shown as percent cytosine methylation and statistic analyses done using ANOVA (* indicates statistically significant differences, p < 0.05). The data are representative of three independent experiments with each CpG locus analyzed at least twice for each pyrosequencing run.
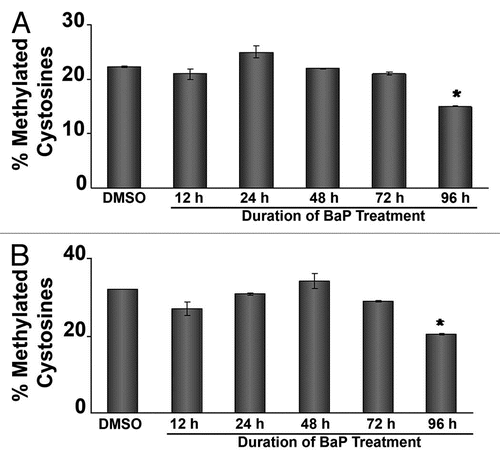
Figure 4 siRNA targeting DNMTs decreases L1 promoter methylation and increases L1 expression. HeLa cells were transfected with 50 nM siRNA targeting DNMT1, DNMT3A or DNMT3B, RNA was extracted and cDNA prepared for qPCR assays. (A) validation of DNMT mRNA downregulation via real time PCR. (B) Western blot experiments using 20 µg of total cellular protein lysate from HeLa cells transfected with siRNA against DNMT1. GAPDH was used as a loading control. (C) Western blot experiments using 20 µg of total cellular protein lysate from HeLa cells transfected with siRNA against DNMT3A. GAPDH was used as a loading control. (D) siRNA targeting DNMTs decreases L1 promoter methylation at several CpG sites. HeLa cells were transfected with 50 nM siRNA targeting DNMT1, DNMT3A or DNMT3B. As a control for DNA methylation status, the demethylating agent 5-aza-2′-deoxycytidine (5-AzaC) was used. DNA was isolated and pyrosequenced for quantitative analyses of DNA methylation using sequencing primers targeting six unique CpG dinucleotides. (E) siRNA targeting DNMT1 increases L1 transcripts. HeLa cells pretreated with 50 nM siRNA targeting DNMTs and 48 h later mRNA levels of LINE 1 ORF 1 and ORF 2 were measured by real time PCR. For all experiments involving real time PCR, relative quantitative analyses were done using the Livak method of ΔΔCt and statistic analyses were done using ANOVA (* indicates statistically significant differences, p < 0.05). Each experiment was run with three replicates and repeated at least 2×.
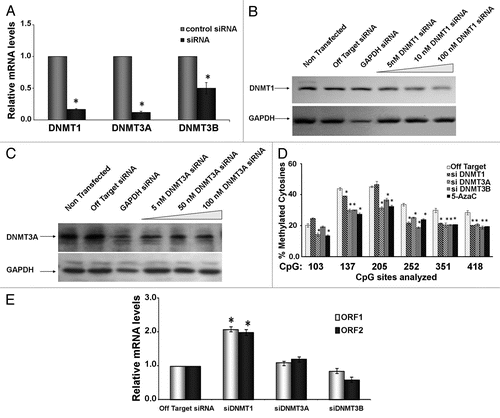
Figure 5 BaP targets DNMT1 for degradation via a proteasome-dependent pathway. (A) HeLa cells were treated with vehicle DMSO or 3 µM BaP for 48 h. Forty micrograms of total protein extract were resolved on a 4–12% gradient SDS PAGE. Membranes were probed for DNMT1 and DNMT3A. GAPDH was used as loading control. (B) MG132 inhibits degradation of DNMT1 following BaP treatment. HeLa cells pretreated with 1 µM MG132 for 3 h prior to DMSO vehicle or 3 µM BaP treatment for an additional 48 h were lysed and total protein used for western blot assays with the indicated antisera. AHR (control) is a protein targeted for degradation by BaP. (C) Inhibition of proteasomal degradation of DNMT1 results in reversal of BaP mediated hypomethylation at CpGs on the L1 promoter. Cells subjected to pharmacological treatment as in (B) were exposed to DMSO vehicle or 3 µM BaP for 96 h followed by DNA pyrosequencing of CpG loci 205, 252 and 351. Results are shown as percent cytosine methylation.
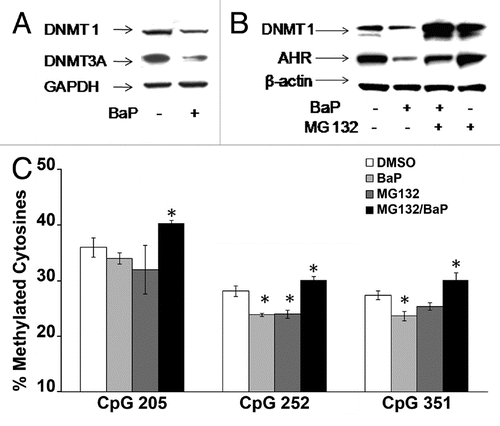
Figure 6 MG132 alters histone epigenetic modifications. HeLa cells were treated with 1 µM MG132 followed by a 12 h treatment with DMSO vehicle or 3 µM BaP. Cells were processed for ChIP assays using 25 µg of chromatin per immunoprecipitation. Real time PCR for DNA samples from ChIP assays with antibodies shown was performed using primers targeting the L1 promoter (5′UTR). Isotype-matched IgG control was used to control for non-specific enrichment of L1 DNA sequences. The results shown are representative of experiments with four replicate samples and repeated two independent times. Calculations for L1 promoter sequence enrichment within ChIP samples was performed according to the SABiosciences ChIP-qPCR Data Analysis manual (www.sabiosciences.com/chipqpcrresource.php) and plotted as a function of percent enrichment relative to input (% input) chromatin.
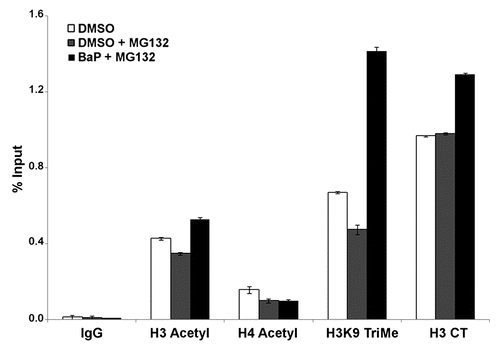
Figure 7 Proposed model for epigenetic crosstalk in the regulation of L1 expression. Early events in L1 expression involve histone modifications with enrichment for H3K4Me3 and H3K9Ac which are hallmarks of active gene expression. At this phase we observe reduced DNMT RNA and protein levels probably due to degradation. As cells divide with reduced levels of DNA methyltransferase, L1 promoter becomes hypomethylated.
Table 1 Pyrosequencing primers and dispensation order
Table 2 Transcription factor binding sites proximal to CpG dinucleotides analyzed
Additional material
Download Zip (537.3 KB)Acknowledgements
Thanks to Pasano Bojang Jr, Marlene C. Steffen and Vilius Stribinskis for editorial assistance and meaningful suggestions.
References
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA 2003; 100:5280 - 5285
- Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res 2008; 18:343 - 358
- Martin SL. The ORF1 protein encoded by LINE-1: Structure and function during L1 retrotransposition. xsJ Biomed Biotechnol 2006; 2006:45621
- Feng Q, Moran JV, Kazazian H Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 1996; 87:905 - 916
- Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene 1997; 189:227 - 234
- Steinhoff C, Schulz WA. Transcriptional regulation of the human LINE-1 retrotransposon L1.2B. Mol Genet Genomics 2003; 270:394 - 402
- Harony H, Bernes S, Siman-Tov R, Ankri S. DNA methylation and targeting of LINE retrotransposons in Entamoeba histolytica and Entamoeba invadens. Mol Biochem Parasitol 2006; 147:55 - 63
- Becker KG, Swergold GD, Ozato K, Thayer RE. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum Mol Genet 1993; 2:1697 - 1702
- Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acid Res 2004; 32:3846 - 3855
- Yang N, Zhang L, Zhang Y, Kazazian H Jr. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res 2003; 31:4929 - 4940
- Esteller M. Epigenetic lesions causing genetic lesions in human cancer: promoter hypermethylation of DNA repair genes. Eur J Cancer 2000; 36:2294 - 2300
- Bratthauer GL, Fanning TG. Active LINE-1 retrotransposons in human testicular cancer. Oncogene 1992; 7:507 - 510
- Florl AR, Lower R, Schmitz-Drager BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Brit J Canc 1999; 80:1312 - 1321
- Santourlidis S, Florl A, Ackermann R, Wirtz HC, Schulz WA. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. The Prostate 1999; 39:166 - 174
- Lin CH, Hsieh SY, Sheen IS, Lee WC, Chen TC, Shyu WC, et al. Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Res 2001; 61:4238 - 4243
- Dante R, Dante-Paire J, Rigal D, Roizes G. Methylation patterns of long interspersed repeated DNA and alphoid repetitive DNA from human cell lines and tumors. Anticancer Res 1992; 12:559 - 563
- Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene 2005; 24:7213 - 7223
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004; 4:143 - 153
- Morey Kinney SR, Smiraglia DJ, James SR, Moser MT, Foster BA, Karpf AR. Stage-specific alterations of DNA methyltransferase expression, DNA hypermethylation and DNA hypomethylation during prostate cancer progression in the transgenic adenocarcinoma of mouse prostate model. Mol Cancer Res 2008; 6:1365 - 1374
- Ben-Porath I, Cedar H. Epigenetic crosstalk. Mol Cell 2001; 8:933 - 935
- Zhou X, Sun H, Ellen TP, Chen H, Costa M. Arsenite alters global histone H3 methylation. Carcinogenesis 2008; 29:1831 - 1836
- Wichmann AE, Thomson NM, Peterson LA, Wattenberg EV. Genotoxic methylating agents modulate extracellular signal regulated kinase activity through MEK-dependent, glutathione- and DNA methylation-independent mechanisms in lung epithelial cells. Chem Res Toxicol 2003; 16:87 - 94
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem 2001; 70:81 - 120
- Mastronardi FG, Wood DD, Mei J, Raijmakers R, Tseveleki V, Dosch HM, et al. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J Neurosci 2006; 26:11387 - 11396
- Thompson PR, Fast W. Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock?. ACS Chem Biol 2006; 1:433 - 441
- Kim HG, Lee KW, Cho YY, Kang NJ, Oh SM, Bode AM, et al. Mitogen- and stress-activated kinase 1-mediated histone H3 phosphorylation is crucial for cell transformation. Cancer Res 2008; 68:2538 - 2547
- Realini CA, Althaus FR. Histone shuttling by poly(ADP-ribosylation). J Biol Chem 1992; 267:18858 - 18865
- Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, Turner BM. Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J Biol Chem 2007; 282:4408 - 4416
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 2007; 25:15 - 30
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 2007; 1:299 - 312
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 2008; 40:741 - 750
- Brown SE, Suderman MJ, Hallett M, Szyf M. DNA demethylation induced by the methyl-CpG-binding domain protein MBD3. Gene 2008; 420:99 - 106
- Klein CB, Costa M. DNA methylation, heterochromatin and epigenetic carcinogens. Mut Res/Rev Mut 1997; 386:163 - 180
- Stribinskis V, Ramos KS. Activation of human long interspersed nuclear element 1 retrotransposition by benzo(a)pyrene, an ubiquitous environmental carcinogen. Cancer Res 2006; 66:2616 - 2620
- Teneng I, Stribinskis V, Ramos KS. Context-specific regulation of LINE-1. Genes Cells 2007; 12:1101 - 1110
- Lu KP, Hallberg LM, Tomlinson J, Ramos KS. Benzo(a)pyrene activates L1Md retrotransposon and inhibits DNA repair in vascular smooth muscle cells. Mutation Research 2000; 454:35 - 44
- Lu KP, Ramos KS. Identification of genes differentially expressed in vascular smooth muscle cells following benzo(a)pyrene challenge: implications for chemical atherogenesis. Biochem Biophy Res Commun 1998; 253:828 - 833
- Lu KP, Ramos KS. Redox regulation of a novel L1Md-A2 retrotransposon in vascular smooth muscle cells. J Biol Chem 2003; 278:28201 - 28209
- Montoya-Durango DE, Liu Y, Teneng I, Kalbfleisch T, Lacy ME, Steffen MC, et al. Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutat Res 2009; 665:20 - 28
- Yauk CL, Polyzos A, Rowan-Carroll A, Kortubash I, Williams A, Kovalchuk O. Tandem repeat mutation, global DNA methylation and regulation of DNA methyltransferases in cultured mouse embryonic fibroblast cells chronically exposed to chemicals with different modes of action. Environ Mol Mutagen 2008; 49:26 - 35
- Sadikovic B, Rodenhiser DI. Benzopyrene exposure disrupts DNA methylation and growth dynamics in breast cancer cells. Toxicol Appl Pharmacol 2006; 216:458 - 466
- Yauk C, Polyzos A, Rowan-Carroll A, Somers CM, Godschalk RW, Van Schooten FJ, et al. Germ-line mutations, DNA damage and global hypermethylation in mice exposed to particulate air pollution in an urban/industrial location. Proc Natl Acad Sci USA 2008; 105:605 - 610
- Pavanello S, Bollati V, Pesatori AC, Kapka L, Bolognesi C, Bertazzi PA, et al. Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int J Cancer 2009; 125:1692 - 1697
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer research 2007; 67:876 - 880
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002; 18:1427 - 1431
- Strunnikova M, Schagdarsurengin U, Kehlen A, Garbe JC, Stampfer MR, Dammann R. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol Cell Biol 2005; 25:3923 - 3933
- Rempala GA, Ramos KS, Kalbfleisch T, Teneng I. Validation of a mathematical model of gene transcription in aggregated cellular systems: application to l1 retrotransposition. J Comput Biol 2007; 14:339 - 349
- Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 2002; 21:5483 - 5495
- Pollenz RS. The mechanism of AH receptor protein downregulation (degradation) and its impact on AH receptor-mediated gene regulation. Chem Biol Interact 2002; 141:41 - 61
- Pollenz RS, Buggy C. Ligand-dependent and -independent degradation of the human aryl hydrocarbon receptor (hAHR) in cell culture models. Chem Biol Interact 2006; 164:49 - 59
- Song Z, Pollenz RS. Ligand-dependent and independent modulation of aryl hydrocarbon receptor localization, degradation and gene regulation. Mol Pharmacol 2002; 62:806 - 816
- Kinyamu HK, Collins JB, Grissom SF, Hebbar PB, Archer TK. Genome wide transcriptional profiling in breast cancer cells reveals distinct changes in hormone receptor target genes and chromatin modifying enzymes after proteasome inhibition. Mol Carcinog 2008; 47:845 - 885
- Adenuga D, Yao H, March TH, Seagrave J, Rahman I. Histone Deacetylase 2 is phosphorylated, Uuiquitinated and degraded by cigarette smoke. Am J Respir Cell Mol Biol 2009; 40:464 - 473
- Zhou Q, Agoston AT, Atadja P, Nelson WG, Davidson NE. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol Canc Res 2008; 6:873 - 883
- Arányi T, Páldi A. The constant variation: DNA methylation changes during preimplantation development. FEBS Lett 2006; 580:6521 - 6526
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 2002; 241:172 - 182
- Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene 2004; 23:8841 - 8846
- Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 2007; 16:108 - 114
- Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet 2002; 18:348 - 351
- Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA 2007; 104:13056 - 13061
- Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nat Genet 1994; 8:59 - 65
- Bui HT, Van Thuan N, Kishigami S, Wakayama S, Hikichi T, Ohta H, et al. Regulation of chromatin and chromosome morphology by histone H3 modifications in pig oocytes. Reproduction 2007; 133:371 - 382
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian H Jr. High frequency retrotransposition in cultured mammalian cells. Cell 1996; 87:917 - 927
- Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci USA 2006; 103:9590 - 9594
- Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, et al. 5-Aza-Deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain and nuclear localization signal. Mol Cell Biol 2005; 25:4727 - 4741
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet 2000; 25:338 - 342
- Bradley C, van der Meer R, Roodi N, Yan H, Chandrasekharan MB, Sun ZW, et al. Carcinogen-induced histone alteration in normal human mammary epithelial cells. Carcinogenesis 2007; 28:2184 - 2192
- Sadikovic B, Andrews J, Carter D, Robinson J, Rodenhiser DI. Genome-wide H3K9 histone acetylation profiles are altered in benzopyrene-treated MCF7 breast cancer cells. J Biol Chem 2008; 283:4051 - 4060
- Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet 2000; 9:2395 - 2402
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 2004; 431:96 - 99
- Bestor TH, Bourc'his D. Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb Symp Quant Biol 2004; 69:381 - 387
- Reymann S, Borlak J. Transcriptome profiling of human hepatocytes treated with Aroclor 1,254 reveals transcription factor regulatory networks and clusters of regulated genes. BMC Genomics 2006; 7:217
- Zhang N, Lin C, Huang X, Kolbanovskiy A, Hingerty BE, Amin S, et al. Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement.. J Mol Biol 2005; 346:951 - 965
- Weisenberger DJ, Romano LJ. Cytosine methylation in a CpG sequence leads to enhanced reactivity with Benzo[a]pyrene diol epoxide that correlates with a conformational change. J Biol Chem 1999; 274:23948 - 23955
- Kinyamu HK, Jefferson WN, Archer TK. Intersection of nuclear receptors and the proteasome on the epigenetic landscape. Environ Mol Mutagen 2008; 49:83 - 95
- MacLeod MC, Powell KL, Kuzmin VA, Kolbanovskiy A, Geacintov NE. Interference of benzo[a]pyrene diol epoxide-deoxyguanosine adducts in a GC box with binding of the transcription factor Sp1. Mol Carcinog 1996; 16:44 - 52
- Johnson DG, Coleman A, Powell KL, MacLeod MC. High-affinity binding of the cell cycle-regulated transcription factors E2F1 and E2F4 to benzo[a]pyrene diol epoxide-DNA adducts. Mol Carcinog 1997; 20:216 - 223
- Liu T, Xu J, Tsao H, Li B, Xu R, Yang C, et al. Base sequence-dependent bends in site-specific benzo[a]pyrene diol epoxide-modified oligonucleotide duplexes. Chem Res Toxicol 1996; 9:255 - 261
- Huber HE, Goodhart PJ, Huang PS. Retinoblastoma protein reverses DNA bending by transcription factor E2F. J Biol Chem 1994; 269:6999 - 7005
- Alexander AM, Pecoraro C, Styche A, Rudert WA, Benos PV, Ringquist S, et al. SOP3: a web-based tool for selection of oligonucleotide primers for single nucleotide polymorphism analysis by Pyrosequencing. Biotechniques 2005; 38:87 - 94
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 1997; 11:205 - 214