Abstract
Malignant pleural mesothelioma (MPM) remains a cancer of poor prognosis. It is hoped that implementation of effective screening biomarkers will lead to earlier diagnoses and improved outcomes. Serum-measured soluble mesothelin-related peptide (SMRP) has been demonstrated to have excellent specificity for MPM, but poor sensitivity precludes its use as a screening biomarker. Using a case series of MPM patients from the International Mesothelioma Program at the Brigham and Women's hospital, we sought to determine whether epigenetic change at the MSLN gene in patient tumors is responsible for the poor sensitivity of SMRP. We identified three potential target regions for CpG methylation silencing in the MSLN promoter, one of which was amenable to bisulfite pyrosequencing and located 214 bp upstream of the transcription start site. MSLN promoter methylation was significantly higher in normal pleura than tumor tissue (P < 6.0x10-9). Next, we compared cases according to serum SMRP status and observed that MSLN methylation was significantly higher among tumors from patients testing negative for SMRP (<1.5nM) versus those that were SMRP positive (P < 0.03). These results demonstrate that MSLN is normally methylated in the pleura, and that methylation is lost in most tumors. However, in a subset of tumors methylation is retained, and this mechanism explains the poor sensitivity of the SMRP assay. These results may lead to additional biomarker targets that will resolve the poor sensitivity of the SMRP assay and allow implementation of screening among exposed populations.
Introduction
Malignant Pleural Mesothelioma (MPM) is a rapidly fatal tumor attributable to commercial use of asbestos.Citation1 The incidence of MPM has been steadily climbing, and is expected to level off in the US in the next decade with approximately 2,500–3,000 cases annually.Citation1 This trend is likely a reflection of OSHA regulations set in place in the mid 1970s and 80s that reduced occupational exposures, and the 40 year latency of MPM. However, worldwide MPM rates are expected to rise due to the uncontrolled use of asbestos in less industrialized countries.Citation2–Citation4 The expected cost for mesothelioma-related compensation within the US alone has been estimated at $200 billion over the next four decades.Citation5
Often, MPM is diagnosed at advanced stages making successful treatment unlikely. It is hoped that early diagnosis will improve clinical treatment and outcome. Therefore, the development of a screening tool for at-risk populations, namely those exposed to asbestos, has the promise of improving mesothelioma mortality. Serum soluble mesothelin-related peptide (SMRP), derived from the mesothelin protein, is of particular interest as a candidate biomarker for mesothelioma screening. Mesothelin is a 40 kDa cell surface glycoprotein and is likely involved in cell adhesion, although its true biologic function has yet to be identified. This protein originates from a 69 kDa precursor and is commonly overexpressed in mesothelioma, pancreatic and ovarian cancers.Citation6,Citation7 The protein is cleaved to a 31 kDa fragment, referred to as SMRP, and can be detected in sera via sandwich ELISA.Citation7,Citation8 Many groups have investigated the potential of SMRP as a diagnostic biomarker of mesothelioma,Citation7,Citation9–Citation14 and while specificity is high, the low sensitivity observed in many studies (a high false-negative rate), precludes its use as a screening biomarker.
Given the high specificity of SMRP for mesothelioma, it is imperative to improve the sensitivity of this biomarker so it may be implemented as a screening test. Poor SMRP sensitivity may be attributable to tumor alterations that change its expression. Epigenetic alterations, such as promoter hypermethylation of cancer-related genes contribute to mesothelioma and clinical outcome,Citation15–Citation17 through the control of expression of these genes.Citation18 We hypothesized that epigenetic changes at the MSLN gene in MPM tumors are responsible for the poor sensitivity of SMRP for MPM.
Results
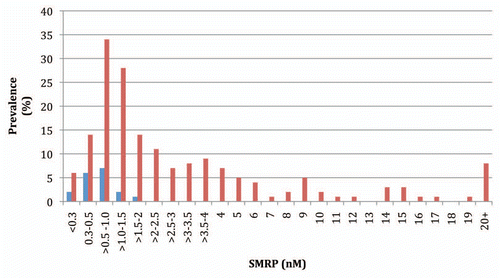
Demographic, asbestos exposure, histology and SMRP data for cases are presented in . To establish the sensitivity and specificity of SMRP in our case group we measured serum SMRP in 179 cases and 18 non-diseased controls. The SMRP values for the non-diseased serum controls ranged from <0.30–1.79 nM with a median of 0.56 nM, while cases ranged from <0.30–85.8 nM with a median of 1.71 nM (). Cases were significantly more likely to have SMRP values above the 1.5 nM cutoff (p < 0.0004). SMRP was 94.4% specific and 52.5% sensitive for MPM using the recommended assay cut-off value of 1.5 nM.
Three separate target CpG regions within the MSLN promoter were identified for methylation measurement by bisulfite pyrosequencing. Of these, one region containing three CpG sites ∼200 bp 5′ of the transcription start site was amenable to assay design and bisulfite pyrosequencing. This region does not meet the formal criteria of a CpG island. Among ten pathologically normal pleura samples the average methylation value at these loci was 67.9%, compared to 21.4% among the 36 case tumors available for study ( and p < 6.0 × 10−9). There was a statistically significant association of tumor MSLN methylation and the presence of serum SMRP. Among those positive for serum SMRP, the average tumor methylation at this locus was 14.3%, while those negative for SMRP had an average tumor methylation of 23.3% (p < 0.03, ).
Previously, we had reported on methylation status at ∼800 genes in mesothelioma (not including MSLN).Citation15 The broad aim of that work was to establish methylation profiles across known cancer genes, and we identified seven distinct classes of mesothelioma tumors based on a panel of 1,500 CpG's. Here, we tested whether the previously identified methylation classes are associated with serum SMRP (). There is a significant association of methylation class with serum SMRP (permutation p < 0.0001), with SMRP levels higher among methylation classes 5 and 7.
Discussion
We have tested the hypothesis that low serum SMRP values oberved in a subset of mesothelioma cases—one of the limiting factors in its utility as a population screening tool—are associated with CpG methylation status at the MSLN gene within tumors. Consistent with this hypothesis, our data show a statistically significant association between promoter methylation at the MSLN gene and high serum SMRP values.
Further, consistent with previous reports, we found that serum SMRP was highly specific to MPM patients compared with non-diseased controls, but with poor sensitivity.Citation8,Citation9,Citation11,Citation12,Citation20 The high specificity of serum SMRP fuels its promise as a non-invasive screening biomarker for mesothelioma. However, it is imperative that this is accompanied by a high degree of sensitivity. In the current study, we demonstrate that the normal state of the MSLN gene within pleura is methylated, and this likely explains the low levels of SMRP observed in non-diseased individuals. Further, on average, the MSLN gene is hypomethylated in mesothelioma tumors; it is assumed that this hypomethylation results in gene expression and thus, the elevated serum SMRP values observed in MPM cases. Despite observing a statistically significant association between SMRP and MSLN methylation, the data were not completely concordant; possibly because we have not captured the CpGs at MSLN whose methylation status has the highest correlation with expression of MSLN mRNA. Alternatively, the lack of complete concordance between MSLN methylation and SMRP values could be due to other factors that impact serum SMRP levels, such as post-transcriptional regulation of MSLN transcript by miRNA(s) or post-translational modifications of mesothelin itself that alter the quantification of the peptide by ELISA.
We acknowledge that we have not shown a direct relationship between methylation of MSLN and gene expression. However, among cases whose tumors have MSLN methylation, we observe that serum SMRP is low. In fact, it is under the condition of low SMRP that sensitivity of the SMRP assay is lost and this low sensitivity limits the assay's utility (and hence its application) as a population screening tool. In future work, it may be possible to improve the sensitivity of SMRP for detecting MPM by combining the assay results with those from a second biomarker. Including a second biomarker that measures epigenetic states in serum that are known to be MPM-specific could complement results from the SMRP assay and be robust enough to apply in a population screening context (to individuals with known asbestos exposure). However, additional study is required to identify MPM-specific epigenetic markers that can be detected in patient serum samples.
Previously, we have investigated DNA methylation changes in a broad array of cancer genes,Citation15 and observed that methylation classes are associated with asbestos exposure (methylation class 5) and poor survival (methylation class 7). In our present analysis we observe that these same two methylation classes are associated with elevated serum SMRP (and by inference loss of methylation at the MSLN gene).
Other groups have investigated how the status of the MSLN gene relates to mesothelin expression.Citation21–Citation23 In a study of pancreatic cancer Sato et al. showed that MSLN is methylated in normal pancreatic tissue, that 5-azacytidine treatment (inhibitor of methylation) of cell lines with methylated MSLN results in MSLN re-expression, and that the majority of primary pancreatic tumors are hypomethylated at MSLN.Citation23 Obulhasim et al. evaluated mesothelin expression in ovarian tumors (assessed by IHC) and its association with methylation at 20 CpG sites in the MSLN gene. In this instance, there was wide variation in MSLN methylation but, on average, only moderate correlation between the median MSLN methylation in ovarian tumors and tumor mesothelin expression (28% methylation in mesothelin positive tumors and 22% in mesothelin negative). These data are not highly consistent with our observations, though we measured the cleaved product of mesothelin in serum, not mesothelin protein expression in tumor tissue. Additionally, compared to our measurement at ∼200 bp 5′ of the transcription start site, Obulhasim et al. measured MSLN methylation more than 1.5 kb 5′ of the transcription start site and inconsistencies in results between studies could certainly be attributable to the difference in where MSLN methylation was measured. Most recently, promoter methylation of MSLN was investigated in mesothelioma by Tan et al.Citation21 In this case, MSLN methylation was associated with gene expression in epitheloid tumors. This is highly consistent with our work, which extends these observations to the screening marker SMRP.
As in all studies, we had limitations to our data and our analysis. We included a small number of controls; however, this was with intent as the sensitivity and specificity of SMRP as a screening biomarker has already been extensively studied. Rather, we capitalized on the availability of patient tumor tissue to investigate the underlying cause for the poor sensitivity of SMRP as a screening biomarker. This too had limitations, as not all subjects had available tumor tissue for further study. However, our study has identified the likely cause of poor sensitivity of SMRP for MPM and can inform future work in biomarker development for this disease.
Materials and Methods
Study population.
Incident cases of mesothelioma were identified through the International Mesothelioma Program at the Brigham and Women's Hospital beginning in 2005. The case participation rate was 85% and all patients provided informed consent under the approval of the appropriate Institutional Review Boards. Clinical information, including histologic diagnosis, was obtained from pathology reports. Each patient was assessed for history of exposure to asbestos as well as additional demographic and exposure data by obtaining their medical and occupational history with an in-person questionnaire or interview. Control serum samples were obtained from subjects (disease-free controls) without asbestos exposure who were participating in a study of head and neck cancer.Citation19 De-identified control pleural tissue samples were obtained from the BWH tissue bank from patients without mesothelioma, with no known asbestos exposure and no evidence of pleural disease. These samples were from patients in the lung transplant program (both donors and recipients) or patients undergoing lung surgery for other disease processes that did not involve the pleura. In all control pleural samples, there was no gross or microscopic evidence of pleural pathology.
SMRP measurement.
Serum mesothelin related protein was measured in serum from mesothelioma cases and non-diseased controls with the MESOMARK assay (Fujirebio Diagnostics) at the Division of Prenatal and Special Testing for Women and Infants Hospital of Rhode Island as described by reference Citation7. Samples were tested without operator knowledge of pathology. Inter-assay coefficents of variation were 4.0% at 4.5 nM and 8.0% at 13.5 nM. The assays sensitivity was 0.3 nM.
Methylation analysis.
DNA was extracted from frozen tissue using the QIAmp DNA mini kit according to the manufacturer's protocol (Qiagen). DNA was modified by sodium bisulfite to convert unmethylated cytosines to uracil using the EZ DNA Methylation kit (Zymo Research) according to the manufacturer's protocol. Target CpGs for MSLN were selected by consulting the Illumina Infinium Human Methylation panel and visually inspecting the promoter sequence; three targets were identified. Bisulfite pyrosequencing assays were designed for the three regions using Pyromark Assay Design software v2.0. Of these, one region was successfully pyrosequenced (corresponding to Illumina Infinium array CpG cg06100324, 214 base pairs upstream of the transcription start and two neighboring CpGs). A non-labeled forward primer (AGA GGT TTA AGT GTG GGT ATT GG) and biotin labeled reverse primer (CAC CAC AAC AAA AAA AAC TCA AAA ATT ATT) were used to amplify the target sequence using 50 ng of bisulfite converted DNA and 45 PCR cycles using an annealing temperature of 56°C. Bisulfite pyrosequencing was achieved using a sequencing primer (AAA GGG GTT GTT TTG) and the Pyromark MD (Qiagen) sequencer. The percentage of methylation was determined using the Pyromark MD software and was compared to methylated human DNA standards with known methylation percentages. Each sample was run in duplicate and the results averaged. Thirty-six tumors were available for study; they did not differ from the larger case group with respect to tumor histology.
Statistical analysis.
For SMRP positive definition, we used the manufacturer's recommended cutoff of 1.5 nM; for serum SMRP values of <0.30 nM (undetected), a value of 0.15 nM was assigned. Statistical analyses were performed using SAS version 9.2 (SAS Institute, Cary, NC) statistical software package. All p values are two sided. Statistical significance was determined at the 0.05 level.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures and Tables
Figure 2 CpG methylation at the MSLN gene is lower in tumor compared to normal pleura and lower in tumors from SMRP serum positive compared to serum negative cases.
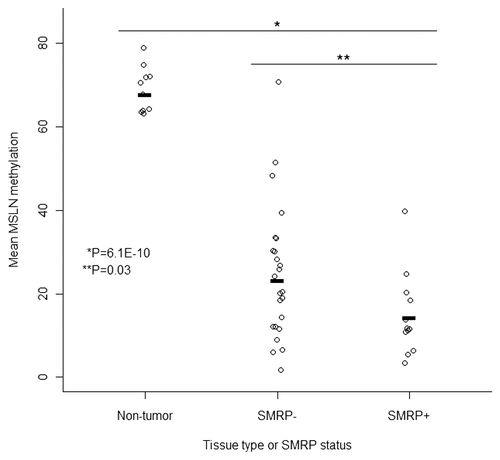
Figure 3 Methylation class, derived from a recursively partitioned mixed model of methylation at 1500 CpG sites, is associated with serum SMRP levels.
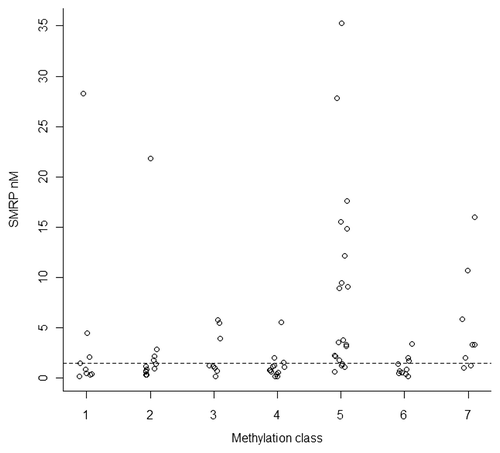
Table 1 Demographic and clinical traits of studied mesothelioma cases
Acknowledgments
This work was supported by the International Mesothelioma Program at Brigham and Women's Hospital and NIH grants R01CA126939, R01CA078609, P30 CA077598.
References
- Price B, Ware A. Mesothelioma trends in the United States: an update based on Surveillance, Epidemiology and End Results Program data for 1973 through 2003. Am J Epidemiol 2004; 159:107 - 112
- Peto J, Decarli A, La Vecchia C, Levi F, Negri E. The European mesothelioma epidemic. Br J Cancer 1999; 79:666 - 672
- Gaafar RM, Eldin NH. Epidemic of mesothelioma in Egypt. Lung Cancer 2005; 49:17 - 20
- Musk AW, de Klerk NH. Epidemiology of malignant mesothelioma in Australia. Lung Cancer 2004; 45:21 - 23
- Robinson BW, Lake RA. Advances in malignant mesothelioma. N Engl J Med 2005; 353:1591 - 1603
- Chang K, Pastan I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas and ovarian cancers. Proc Natl Acad Sci USA 1996; 93:136 - 140
- Beyer HL, Geschwindt RD, Glover CL, Tran L, Hellstrom I, Hellstrom KE, et al. MESOMARK: a potential test for malignant pleural mesothelioma. Clin Chem 2007; 53:666 - 672
- Di Serio F, Fontana A, Loizzi M, Capotorto G, Maggiolini P, Mera E, et al. Mesothelin family proteins and diagnosis of mesothelioma: analytical evaluation of an automated immunoassay and preliminary clinical results. Clin Chem Lab Med 2007; 45:634 - 638
- Pass HI, Wali A, Tang N, Ivanova A, Ivanov S, Harbut M, et al. Soluble mesothelin-related peptide level elevation in mesothelioma serum and pleural effusions. Ann Thorac Surg 2008; 85:265 - 272
- Scherpereel A, Grigoriu B, Conti M, Gey T, Gregoire M, Copin MC, et al. Soluble mesothelin-related peptides in the diagnosis of malignant pleural mesothelioma. Am J Respir Crit Care Med 2006; 173:1155 - 1160
- Grigoriu BD, Scherpereel A, Devos P, Chahine B, Letourneux M, Lebailly P, et al. Utility of osteopontin and serum mesothelin in malignant pleural mesothelioma diagnosis and prognosis assessment. Clin Cancer Res 2007; 13:2928 - 2935
- Creaney J, van Bruggen I, Hof M, Segal A, Musk AW, de Klerk N, et al. Combined CA125 and mesothelin levels for the diagnosis of malignant mesothelioma. Chest 2007; 132:1239 - 1246
- Cristaudo A, Foddis R, Vivaldi A, Guglielmi G, Dipalma N, Filiberti R, et al. Clinical significance of serum mesothelin in patients with mesothelioma and lung cancer. Clin Cancer Res 2007; 13:5076 - 5081
- Robinson BW, Creaney J, Lake R, Nowak A, Musk AW, de Klerk N, et al. Soluble mesothelin-related protein—a blood test for mesothelioma. Lung Cancer 2005; 49:109 - 111
- Christensen B, Houseman EA, Godleski JJ, Marsit CJ, Longacker JL, Roelofs CR, et al. Epigenetic profiles distinguish pleural mesothelioma from normal pleura and predict lung asbestos burden and clinical outcome. Cancer Res 2009; 69:227 - 234
- Destro A, Ceresoli GL, Baryshnikova E, Garassino I, Zucali PA, De Vincenzo F, et al. Gene methylation in pleural mesothelioma: correlations with clinico-pathological features and patient's follow-up. Lung Cancer 2008; 59:369 - 376
- Goto Y, Shinjo K, Kondo Y, Shen L, Toyota M, Suzuki H, et al. Epigenetic profiles distinguish malignant pleural mesothelioma from lung adenocarcinoma. Cancer Res 2009; 69:9073 - 9082
- Cristaudo A, Foddis R, Bonotti A, Simonini S, Vivaldi A, Guglielmi G, et al. Polymorphisms in the putative micro-RNA-binding sites of mesothelin gene are associated with serum levels of mesothelin-related protein. Occup Environ Med 2010; 67:233 - 236
- Applebaum KM, Furniss CS, Zeka A, Posner MR, Smith JF, Bryan J, et al. Lack of association of alcohol and tobacco with HPV16-associated head and neck cancer. J Natl Cancer Inst 2007; 99:1801 - 1810
- Schneider J, Hoffmann H, Dienemann H, Herth FJ, Meister M, Muley T. Diagnostic and prognostic value of soluble mesothelin-related proteins in patients with malignant pleural mesothelioma in comparison with benign asbestosis and lung cancer. J Thorac Oncol 2008; 3:1317 - 1324
- Tan K, Kajino K, Momose S, Masaoka A, Sasahara K, Shiomi K, et al. Mesothelin (MSLN) promoter is hypomethylated in malignant mesothelioma, but its expression is not associated with methylation status of the promoter. Hum Pathol 2010; 41:1330 - 1338
- Obulhasim G, Fujii H, Matsumoto T, Yasen M, Abe M, Matsuoka S, et al. Mesothelin gene expression and promoter methylation/hypomethylation in gynecological tumors. Eur J Gynaecol Oncol 2010; 31:63 - 71
- Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, et al. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res 2003; 63:4158 - 4166