Abstract
The monoallelic expression of imprinted genes is controlled by epigenetic factors including DNA methylation and histone modifications. In mouse, the imprinted gene Gtl2 is associated with two differentially methylated regions: the IG-DMR, which serves as a gametic imprinting mark at which paternal allele-specific DNA methylation is inherited from sperm, and the Gtl2-DMR, which acquires DNA methylation on the paternal allele after fertilization. The timeframe during which DNA methylation is acquired at secondary DMRs during post-fertilization development and the relationship between secondary DMRs and imprinted expression have not been well established. In order to better understand the role of secondary DMRs in imprinting, we examined the methylation status of the Gtl2-DMR in pre- and post-implantation embryos. Paternal allele-specific DNA methylation of this region correlates with imprinted expression of Gtl2 during post-implantation development but is not required to implement imprinted expression during pre-implantation development, suggesting that this secondary DMR may play a role in maintaining imprinted expression. Furthermore, our developmental profile of DNA methylation patterns at the Cdkn1c- and Gtl2-DMRs illustrates that the temporal acquisition of DNA methylation at imprinted genes during post-fertilization development is not universally controlled.
Introduction
Genomic imprinting is controlled at the level of transcription and results in the parent of origin-specific monoallelic expression of a subset of mammalian genes.Citation1 To ensure appropriate expression of imprinted genes, the transcriptional machinery must be able to distinguish between the maternal and paternal alleles and regulate their expression accordingly. The elements responsible for controlling imprinted gene regulation must be both heritable and reversible in order to maintain the appropriate regulatory state following somatic cell division while still allowing the mark to be reset during gametogenesis such that the appropriate parent of origin-specific imprinting mark will be transmitted via the gamete to the next generation.Citation2 Comprehensive studies of the various epigenetic elements responsible for controlling imprinted gene expression are necessary in order to fully understand the regulation of imprinted genes.
To date, approximately 140 imprinted genes have been identified in mammals (www.mgu.har.mrc.ac.uk/research/genomic_imprinting; www.otago.ac.nz/IGC) and the vast majority of these imprinted genes are located in clusters spread throughout the genome. Recent efforts to identify imprinting marks and elucidate the mechanisms by which they regulate the expression of imprinted genes have focused on the imprinting control regions (ICRs) associated with imprinted gene clusters. Each ICR serves as a shared regulatory element that functions both to specify parental origin and control expression (reviewed in refs. Citation3 and Citation4). ICRs are comprised of CpG-rich regions at which the cytosines in CpG dinucleotides are methylated on one parental allele but not the other, thereby conferring parental identity on the maternal and paternal alleles. Parent of origin-specific DNA methylation of the ICRs is established during gametogenesis; paternal allele-specific DNA methylation inherited via the sperm has been observed at the imprinting clusters including H19/Igf2, Dlk1/Gtl2, Rasgrf1 and Zdbf2, while maternal allele-specific DNA methylation originates in the oocyte for clusters including the imprinted Snurf/Snrpn, Peg3, Mest/Peg1 and Air/Igf2r genes.Citation5–Citation10 Establishment of germline DNA methylation at imprinted loci requires the activity of both the de novo DNA methyltransferase Dnmt3A and the related Dnmt3L.Citation11–Citation13 These gametic DNA methylation differences are maintained on the parental alleles throughout development, thereby serving as an imprinting mark that differentiates the parental alleles.
In addition to gametic ICRs, many imprinted genes are associated with other epigenetic modifications that are differentially distributed on the parental alleles. Non-gametic secondary differentially methylated regions (DMRs) have been identified at several imprinted genes.Citation6,Citation14,Citation15 These secondary DMRs acquire DNA methylation on one parental allele after fertilization and may function to maintain imprinted expression throughout development. For example, monoallelic expression of the maternal Cdkn1c allele in mouse requires paternal allele-specific DNA methylation at the promoter, despite the fact that DNA methylation is not acquired on the paternal allele until post-implantation development.Citation14,Citation16 Furthermore, recent reports have implicated a role for differential allelic distribution of modified histones in the regulation of imprinted genes.Citation17–Citation20 Thus, parent of originspecific expression of imprinted genes is likely achieved via the coordinated activity of ICRs, DMRs and additional differential epigenetic modifications, such as histone acetylation and histone methylation.
The Dlk1-Dio3 locus is one example of an imprinting cluster containing multiple epigenetic regulatory elements. This imprinting cluster spans 1 Mb on mouse chromosome 12, and contains three paternally expressed protein-coding genes (Dlk1, Rtl1 and Dio3) as well as multiple maternally expressed untranslated RNAs.Citation21–Citation25 Three distinct DMRs have been identified at this locus.Citation6 The IG-DMR, centered 12 kb upstream of Gtl2, functions as the ICR for this cluster (); maternal transmission of an IG-DMR deletion confers a paternal expression pattern on the maternally inherited genes within this imprinted locus, while paternal transmission of the deletion has no effect.Citation26 Paternal allele-specific DNA methylation of the IG-DMR is inherited from sperm, implicating differential DNA methylation of this region as the primary imprinting mark.Citation6 Preferential DNA methylation of the paternal allele has also been observed at two other DMRs within this imprinting cluster: the Dlk1-DMR, located at the 3′ end of the Dlk1 gene, and the Gtl2-DMR, which flanks the 5′ end of Gtl2.Citation6 Unlike DNA methylation at the IG-DMR, paternal allele-specific DNA methylation at the Dlk1- and Gtl2-DMRs must be acquired post-fertilization, as sperm DNA is unmethylated in these regions.Citation6
The developmental dynamics of DNA methylation at secondary DMRs have not been well established, and it is not known whether the acquisition of methylation at post-fertilization DMRs is coordinately regulated across the genome. To address these questions, we have conducted a temporal analysis of DNA methylation patterns at the IG- and Gtl2-DMRs in pre- and post-implantation mouse embryos. This study has allowed us to determine the developmental dynamics of DNA methylation and imprinted expression at the mouse Gtl2 locus on chromosome 12 and compare these results with data we previously obtained detailing the post-fertilization acquisition of DNA methylation at the Cdkn1c locus on chromosome 7.Citation14 Our findings indicate that differential methylation at secondary DMRs is not required to implement imprinted expression and that the temporal acquisition of DNA methylation at secondary DMRs is variable at distinct imprinted loci.
Results
Paternal allele-specific methylation of the IG-DMR serves as a primary imprinting mark.
The IG-DMR, centered approximately 12 kb upstream of the Gtl2 transcriptional start site, has been shown to harbor paternal allele-specific methylation in uniparental disomic embryos.Citation6 To confirm this pattern of methylation in mice bearing both paternally and maternally derived alleles, bisulfite analysis was performed on F1 hybrid somatic tissue derived from reciprocal crosses between C57BL/6J (B6) and Mus musculus castaneus (CAST) mice. We identified five polymorphisms between B6 and CAST mice in a 1 kb region of the IG-DMR that contains 40 CpG residues (). Thirty-two of 40 CpG dinucleotides are located within a 458 bp region that includes two polymorphisms, allowing the parental alleles to be distinguished. Multiple independent PCR reactions were subcloned and sequenced to assess the methylation status at all 32 cytosines in this region. We observed methylation on the paternal strands in somatic tissue, while the maternally inherited strands generally lacked methylation ().
To determine if paternal allele-specific methylation of the IG-DMR can serve as a primary imprinting mark, we assessed the methylation status of this region in F1 hybrid spermatozoa and blastocysts. The IG-DMR was methylated on both paternal and maternal strands in spermatozoa (), consistent with previous observations that DNA methylation is acquired during spermatogenesis and that sperm contribute a methylated paternal allele at fertilization.Citation27,Citation28 Analysis of DNA from 3.5 d.p.c. blastocysts yielded methylated paternal strands and unmethylated maternal strands (). As the blastocyst genome is generally hypomethylated,Citation29,Citation30 this result supports the hypothesis that methylation of the paternal IG-DMR serves as the primary imprinting mark for this imprinting cluster.
We further investigated IG-DMR methylation in 6.5–9.5 d.p.c. F1 hybrid post-implantation embryos. Consistent with the hypothesis that differential methylation is maintained at the IG-DMR throughout development, we observed methylated paternal strands and unmethylated maternal strands at all stages analyzed ().
Paternal allele-specific methylation is present in a CpG-rich region flanking the Gtl2 transcriptional start site.
The transcriptional start site of the Gtl2 gene is flanked by a 2.35 kb CpG-rich region, located from −1,400 bp to +950 bp (NC_000078.5, with +1 corresponding to bp 110,779,206). We identified three polymorphisms between B6 and CAST, clustered at the 3′ end of the CpG-rich region ().
Using bisulfite mutagenesis followed by DNA sequencing, we analyzed a 722 bp region, containing 29 CpG residues, spanning from +126 to +847 bp (). This region is exclusively methylated on the paternal allele in somatic tissue derived from reciprocal crosses between B6 and CAST (). To determine if paternal allele-specific methylation spans the CpG-rich region, we also performed bisulfite analysis on a 589 bp region containing 15 CpGs located between −828 and −240 bp (). We were unable to definitively assign parental identity to the subclones obtained from this region as there were no polymorphisms; however, analysis of both B6 × CAST and CAST × B6 F1 hybrid somatic DNA yielded two distinct classes of subclones: those that were highly methylated and those that were unmethylated (data not shown). Therefore, we conclude that paternal allele-specific methylation is distributed across the 2.35 kb CpG-rich region.
Takada et al.Citation6 reported that the Gtl2-DMR is not methylated in sperm. To verify this result in our strain background, we examined the 722 bp region in sperm derived from B6 × CAST F1 hybrid mice. We observed a striking absence of methylation on both the paternal and maternal alleles in sperm in this region (), indicating that the paternal allele-specific methylation of this region observed in somatic tissue must be acquired during post-fertilization development.
Paternal allele-specific methylation at the Gtl2-DMR is acquired before 6.5 d.p.c.
To determine when paternal allele-specific methylation is acquired at the Gtl2-DMR, we examined the methylation status of this region in pre- and post-implantation embryos. We observed an absence of methylation on both the paternal and maternal alleles in 3.5 d.p.c. blastocysts, indicating that the paternal allele does not acquire DNA methylation during pre-implantation development (). This result is consistent with the observation that the paternal allele is unmethylated at fertilization () and with the low levels of global DNA methylation observed in pre-implantation embryos.Citation29,Citation31
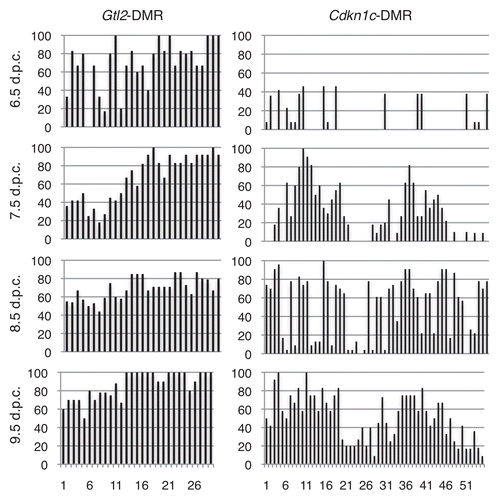
We next analyzed the methylation status at the Gtl2-DMR in 6.5–9.5 d.p.c. post-implantation embryos. At all four stages, we consistently observed methylation on the paternal allele and an absence of methylation on the maternal allele (). The extent of methylation on the paternal allele did not change appreciably between 6.5 and 9.5 d.p.c., indicating that the acquisition of methylation on the paternal allele is primarily accomplished by 6.5 d.p.c.
Gtl2 is imprinted in pre- and post-implantation embryos.
Repression of the paternal Gtl2 allele correlates directly with its paternal allele-specific methylation: the silent paternal allele is methylated at both the IG-DMR and the Gtl2-DMR, while the expressed maternal allele lacks methylation in both of these regions ( and ). When a deletion of the IG-DMR is maternally inherited, the Gtl2-DMR located in cis becomes methylated, concurrent with repression of the maternal Gtl2 allele.Citation26 Therefore, the methylation of the Gtl2-DMR may directly influence Gtl2 expression.
We performed an RT-PCR assay to assess Gtl2 expression in preand post-implantation embryos. Expression of Gtl2 was consistently observed in 6.5–9.5 d.p.c. embryos ( and C). In order to distinguish between maternally- and paternally-derived RT-PCR products, we performed allele-specific restriction digestion, taking advantage of B6-specific restriction endonuclease recognition sites contained within the RT-PCR products. For the AB primer pair, the paternal CAST and maternal B6 products were expected to be 306 bp and 216 + 90 bp, respectively,Citation22 while the FR primer pair was expected to yield 424 bp and 216 + 208 bp products following allele-specific restriction digestion. Our analysis of products derived from both sets of primers indicated that Gtl2 was expressed solely from the maternal allele in 6.5–9.5 d.p.c. F1 hybrid embryos (). This result is consistent with the hypothesis that parental allele-specific methylation plays a role in regulating imprinted expression at Gtl2.
Since the paternally-inherited Gtl2-DMR is unmethylated in 3.5 d.p.c. blastocysts, we investigated the expression of Gtl2 at this stage of development. Although the assay was not quantitative, we regularly obtained much less RT-PCR product from 3.5 d.p.c. RNA than the RNA derived from post-implantation embryos; low levels of expression were detected in 86% of our replicates. To increase the amount of product derived from blastocysts, we developed a semi-nested RT-PCR assay. As controls, we performed both unnested and semi-nested assays using B6 and CAST brain RNA, and showed that both the original and new assay yielded the same products ( and F). We found that Gtl2 was expressed solely from the maternal allele in 3.5 d.p.c. blastocysts, despite the absence of paternal allele-specific methylation at this stage of development ( and F and data not shown).
When developing the allele-specific RT-PCR assay, we observed that the FR primer pair consistently amplified two distinct products ( and E). We hypothesized that this RT-PCR reaction was amplifying splice variants, as alternative splicing of Gtl2 has been described previously ().Citation32,Citation33 To test this hypothesis, we subcloned and sequenced the two distinct PCR products. The sequence data we obtained indicated that the longer RT-PCR product included exon 6, while exon 6 was absent from the shorter product (data not shown). B6-specific restriction digestion of the shorter product yields 216 + 127 bp products. The 127 bp digestion product is less apparent in due to its low relative concentration as compared to the 216 and 208 bp products.
CpG-rich regions internal to the Gtl2 transcription unit are biallelically methylated.
In order to assess the methylation status of other sequences within the Gtl2 transcription unit, we examined the genomic sequence for the presence of additional CpG-rich regions. We identified two CpG-rich regions located 3′ relative to the Gtl2-DMR (). A 350 bp CpG-rich region (E1) is located in exon 5, beginning approximately 600 bp downstream from a putative alternate promoter. An additional 360 bp CpG-rich region (E3) is located 4 kb further downstream, spanning exons 7 and 8. To determine if methylation of either of these two regions correlates with imprinted expression, we conducted bisulfite analysis on somatic DNA isolated from reciprocal crosses between B6 and CAST mice.
The E1 region lacks B6 vs. CAST polymorphisms; therefore, we were unable to definitely assign parental identity to the subclones we analyzed. However, bisulfite analysis of two independent PCRs for each of the reciprocal crosses yielded only methylated subclones, suggesting that both the maternal and the paternal alleles are methylated in this region (data not shown). The E3 region contains one polymorphism, which allowed us to distinguish between the maternal and paternal alleles. Although our amplifications of the E3 region were biased towards the B6 allele, we consistently observed methylation of both parental alleles, suggesting that this region is not differentially methylated (data not shown). Based on our results, it is unlikely that the methylation status of these CpG-rich regions plays a role in regulating imprinted expression of Gtl2.
Discussion
The functional inequivalence of the parental genomes as a consequence of genomic imprinting means that genetic contribution from both the maternal and paternal genomes is required for normal mammalian development.Citation34,Citation35 Loss of imprinting can result in developmental disorders and disease, such as Beckwith-Wiedemann Syndrome, transient neonatal diabetes mellitus and cancer.Citation36 The Dlk1-Dio3 imprinting cluster has long been implicated as playing a role in mammalian growth and development and, along with Igf2-H19, serves as a block to parthenogenesis.Citation37–Citation40 Gene knockout studies have begun to pinpoint the relative contributions of individual genes in the Dlk1-Dio3 cluster,Citation41,Citation42 and recent work has highlighted the importance of Gtl2 in maintaining the imprinting status of the genes contained within this locus. Deletion of the paternally methylated, maternally expressed Gtl2 results in the misregulation of other genes in the Dlk1-Dio3 cluster, ultimately causing lethality.Citation21,Citation22,Citation43,Citation44 Therefore, comprehensive analysis of the allele-specific acquisition of epigenetic modifications responsible for controlling the expression of Gtl2 provides an important step towards understanding the regulation of imprinting across this cluster.
Paternal allele-specific methylation of the Gtl2-DMR may be directly responsible for silencing Gtl2, based on the observation that a paternally inherited insertional mutation causes loss of imprinting concordant with a loss of DNA methylation.Citation45,Citation46 To fully understand the relationship between DNA methylation and expression at Gtl2, we investigated the dynamics of DNA methylation acquisition and maintenance at this locus. The IG-DMR serves as the imprinting control region, and it is well established that paternal allele-specific methylation of this region is inherited from sperm and is maintained throughout development (data herein).Citation6,Citation26 In contrast, although both Steshina et al.Citation45 and Sekita et al.Citation46 provide evidence that DNA methylation at the Gtl2-DMR is directly responsible for repression, our current study demonstrates that methylation of this region is acquired during post-fertilization development, after the onset of imprinted expression.
The lack of a direct correlation between the onset of allele-specific DNA methylation and imprinted expression at Gtl2 highlights the complex relationship between secondary DMRs and the expression of imprinted genes. Previous studies have established a link between DNA methylation and transcriptional silencing of imprinted genes.Citation13,Citation16,Citation47 Despite this correlation, there are several examples of imprinted genes that exhibit monoallelic expression prior to the acquisition of paternal allele-specific methylation at their repressed promoters. For example, the paternal alleles of both Cdkn1c and Igf2r are silenced before paternal allele-specific methylation is achieved.Citation14,Citation48 Similarly, in this study we showed that Gtl2 is imprinted in blastocysts that lack paternal allele-specific methylation at the Gtl2-DMR. If imprinting can be achieved in the absence of differential methylation at secondary DMRs, what role do secondary DMRs play? We propose that the role of secondary DMRs is to reinforce imprinting that has been established by other epigenetic elements, such as the ICR. Several lines of evidence support this hypothesis. For example, although paternal allele-specific DNA methylation is not required to implement imprinted expression of Cdkn1c, Dnmt1 mutant mice lacking DNA methylation at this locus exhibit biallelic expression of Cdkn1c, suggesting that DNA methylation at this secondary DMR plays a role in maintaining an imprinted expression pattern.Citation14
Evidence for a functional role for the human homolog of the Gtl2-DMR (MEG3-DMR, human chromosome 14) has recently been reported. Kagami et al.Citation49 analyzed DNA from patients with paternal uniparental disomy 14 [upd(14)pat] phenotypes: one patient with both body and placental phenotypes was heterozygous for a deletion of the IG-DMR, while a second patient had only body phenotypes and was heterozygous for a deletion of the MEG3-DMR. Deletion of the maternally inherited IG-DMR in patient 1 resulted in the paternalization of the maternally-inherited allele in body tissues, including hypermethylation of the MEG3-DMR and a concordant absence of MEG3 expression. These data illustrate that the methylation status of the MEG3-DMR is dependent on the methylation status of the IG-DMR and directly influences expression in cis, consistent with data from mouseCitation45,Citation46 and supporting the hypothesis that the methylation status of the Gtl2/MEG3-DMR is responsible for maintaining imprinted expression of Gtl2/MEG3 during development. Based on the absence of a placental phenotype in patient 2, Kagami et al.Citation49 conclude that the MEG3-DMR is not required to regulate imprinting in the placenta, raising questions about tissue-specific differences in the regulation of imprinted genes. Further studies are necessary to address this issue, as well as to clarify the precise functional role of the Gtl2-DMR in mice.
To fully understand the role of secondary DMRs in maintaining imprinted expression, it is important to define when differential methylation is acquired at these loci. A comparison of the temporal acquisition of methylation at Cdkn1c and at the Gtl2-DMR illustrates that the establishment of allele-specific methylation during post-fertilization development is not universally controlled. Methylation of the paternal Cdkn1c allele is acquired between 7.5–9.5 d.p.c. and is not fully established until 9.5 d.p.c. ().Citation14 In contrast, methylation at the paternal Gtl2-DMR is acquired prior to 6.5 d.p.c. and does not change appreciably after that stage of development (). Furthermore, paternal allele-specific methylation at Igf2r is acquired during late embryogenesis.Citation48 Combined, these data indicate that different imprinted loci acquire DNA methylation at secondary DMRs at various stages of development, and that this process is not globally coordinated across the genome. It is possible that differences in the temporal acquisition of methylation at secondary DMRs may correlate with the maintenance of imprinted expression during early embryonic development. Perhaps the earlier establishment of allele-specific DNA methylation helps to ensure monoallelic expression, while the absence of a secondary region of differential methylation permits a more labile expression pattern.
Materials and Methods
Mice.
C57BL/6J (B6) and Mus musculus castaneus (CAST) mice were purchased from the Jackson Laboratory. To facilitate the isolation of F1 hybrid mice, a strain of mice that served as the source of the M. m. castaneus allele [B6(CAST12)] was constructed. M. m. castaneus males were mated to B6 females. The resulting B6/C F1 hybrid females were backcrossed to B6 males. The backcross progeny were screened for the CAST12 allele at microsatellite markers D12Mit101 (50 cM), D12Mit80 (53 cM), D12Mit133 (56 cM) and D12Mit20 (58 cM), as well as at an NcoI RFLP located within the IG-DMR; Gtl2 is located at 54 cM. CAST12/B6 females were backcrossed to B6 males and CAST12/B6 male and female offspring were identified among the progeny from the second backcross. CAST12/B6 males and females were mated, and their offspring were screened for male and female progeny homozygous for the CAST12 allele; these animals were interbred to produce the CAST12 strain. Natural matings between B6 and B6(CAST12) or CAST were used to generate F1 hybrid tissues for bisulfite and RT-PCR analyses. Procedures involving animals were approved by the Bryn Mawr College Institutional Animal Care and Use Committee.
DNA purification and bisulfite analysis.
DNA was isolated and subjected to bisulfite modification, PCR amplification, subcloning and sequencing either as previously described in reference Citation50, or using an EZ DNA methylation-direct kit (Zymo Research, cat# D5020). All mutagenized DNAs were subjected to multiple independent PCR amplifications to ensure analysis of different strands of DNA. The following primer pairs were used for nested or semi-nested amplification of the mutagenized DNA, with all base pair numbers from NC_000078.5. Gtl2 IG-DMR, with the first nucleotide of IG-BS-F1 corresponding to position 110,766,235: 30 cycles of 94°C for 30 sec, 52°C for 1 min, 72°C for 1 min, using primers IG-BS-F1/IG-BS-R, followed by 35 cycles using IG-BS-F2/IG-BS-R and the same cycling conditions as above. Identical reaction conditions were used to amplify the Gtl2-DMR, with the first nucleotide of Gtl2BI4F1 corresponding to position 110,779,293: Gtl2BI4F1/Gtl2BI4R1 followed by Gtl2BI4F2/Gtl2BI4R2. Primer sequences follow. IG-BS-F1, 5′-GTA TGT GTA TAG AGA TAT GTT TAT ATG GTA-3′; IG-BS-F2, 5′-GTG TTA AGG TAT ATT ATG TTA GTG TTA GGA-3′; IG-BS-R, 5′-GCT CCA TTA ACA AAA TAA TAC AAC CCT TCC-3′; Gtl2BI4F1, 5′-GAA GAA TTT TTT ATT TGG TGA GTG G-3′; Gtl2BI4F2, 5′-GTT TGA AAG GAT GTG TAA AAA TG-3′; Gtl2BI4R1, 5′-CAA CAC TCA AAT CAC CCC CC-3′; Gtl2BI4R2, 5′-GCC CCC CAC ATC TAT TCT ACC-3′. Primer sequences and reaction conditions for amplifying CpG-rich regions E1 and E3 are available upon request.
RT-PCR.
Ten milligrams B6 liver, B6 brain, CAST brain or single 6.5–9.5 d.p.c. embryos were disrupted using a pestle and total RNA was isolated using a High Pure RNA Tissue Kit (Roche Applied Science, cat# 12033674001). Reverse transcription was performed according to manufacturer's instructions using 10% of the isolated RNA in a 20 µl reaction containing SuperScript II and oligo dT (Invitrogen, cat# 18064022, 18418012). One microliter of the RT product was subjected to PCR in the presence of 1× PCR Master Mix (Promega, cat# M7502) and 0.5 µM of each primer. For all tissues, 30 cycles of PCR were performed under the following conditions: 94°C, 30 sec; 55°C, 1 min; 72°C, 1 min. Primer pairs: Gtl2-RTA, 5′-GCC AAA GCC ATC ATC TGG ATT C-3′/Gtl2-RTB, 5′-CAC AGA TGT AGA CTC AAC AGT GAA G-3′ and Gtl2RT-F, 5′-CGC TCT TCT CCA TCG AAC GG-3′/Gtl2RT-R, 5′-TTA CAG TTG GAG GGT CCT GG-3′. Gtl2-RTA/B and F/R PCR products were digested with SfcI and NgoMIV (New England Biolabs, cat# R0561, R0564), respectively, and resolved on a 10% acrylamide gel or a 2% agarose gel to distinguish between B6 and CAST products. To obtain sufficient RT-PCR product for clear visualization from 3.5 d.p.c. blastocysts, a modified semi-nested RT-PCR protocol was developed. Briefly, pools of 6–8 3.5 d.p.c. blastocysts were used to prepare RNA and cDNA as described above, and 0.5 µl brain RT or 2 µl of blastocyst RT product was subjected to 30 cycles of PCR as described above, using primers Gtl2-RTF and Gtl2-RTB to yield a 650 bp product. F/B PCR products were diluted in ddH2O (1:1,000 for brain, 1:10 for blastocysts) and subjected to an additional 30 cycles of PCR using either primer pair Gtl2-RTA/B or Gtl2-RTF/R, as described above.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Abbreviations
| ICR | = | imprinting control region |
| DMR | = | differentially methylated region |
| B6 | = | C57BL/6 |
| CAST | = | Mus musculus castaneus |
| d.p.c. | = | days post coitum |
Figures and Tables
Figure 1 (A) Dlk1-Gtl2 imprinting cluster, including transcriptional start sites (arrows) and transcription units (hatched boxes). (B) Portion of IG-DMR analyzed in this study. The 458 bp region analyzed by bisulfite mutagenesis and DNA sequencing corresponds to positions 110,766,298-110,766,755, NC_000078.5 (black box). Polymorphisms (*) between C57BL/6J and Mus musculus castaneus are as follows: (B6/CAST): 110,766,439 (A/G), 110,776,579 (G/A), 110,766,774 (G/A), 110,766,902-110,766,904 (TTT/TT), 110,767,052 (A/G). (C) Schematic of the Gtl2-DMR, including the Gtl2 transcriptional start site (arrow) and exon 1 (hatched box); +1 corresponds to position 110,779,206. Regions analyzed correspond to positions 110,778,378-110,778,966 and 110,779,331-110,780,052. Polymorphisms are as follows: 110,779,741 (G/A), 110,779,881 (A/G), 110,780,030-110,780,031 (AA/GC).
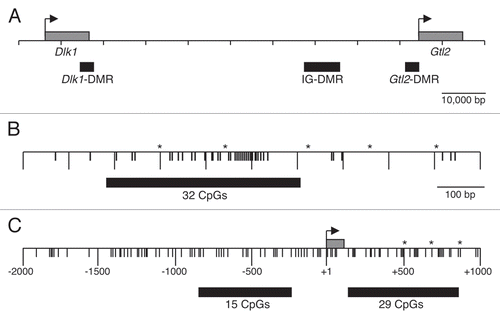
Figure 2 Paternal allele-specific methylation of the IG-DMR is inherited from sperm. Bisulfite mutagenesis and sequencing of DNA from B6 × CAST and CAST × B6 F1 hybrid liver and B6 × CAST F1 hybrid spermatozoa. Each circle represents one of 32 potentially methylated CpG dinucleotides, the first one located at position 110,766,345 (NC_000078.5). Each row of circles represents an individual strand sequenced. Filled circles represent methylated cytosines, open circles represent unmethylated cytosines, absent circles represent positions at which methylation data was not obtained.
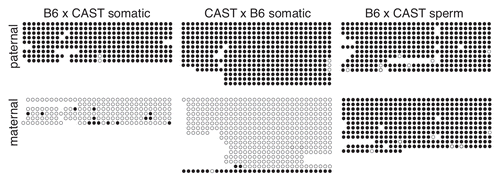
Figure 3 Methylation of the paternal IG-DMR is maintained during pre- and post-implantation development. Bisulfite mutagenesis and sequencing of DNA from B6 × CAST F1 hybrid embryos. Details as described in .
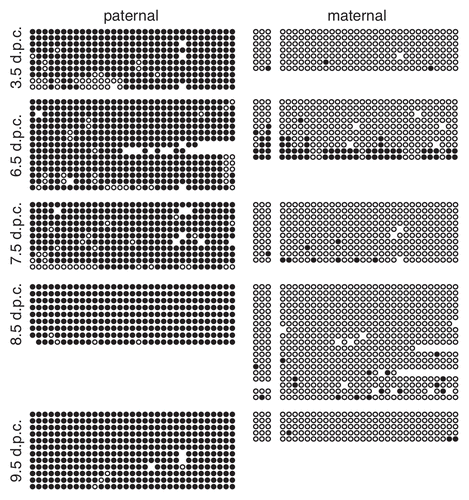
Figure 4 Paternal allele-specific methylation of the Gtl2-DMR is not inherited from sperm. Each circle represents one of 29 potentially methylated CpG dinucleotides, the first one located at position 110,779,349 (NC_000078.5). Details as described in .
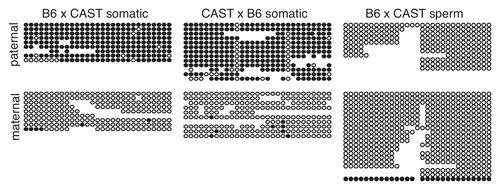
Figure 5 Methylation of the paternal Gtl2-DMR is acquired between 3.5 and 6.5 d.p.c. Bisulfite analysis and sequencing of DNA from B6 × CAST F1 hybrid embryos. Details as described in and .
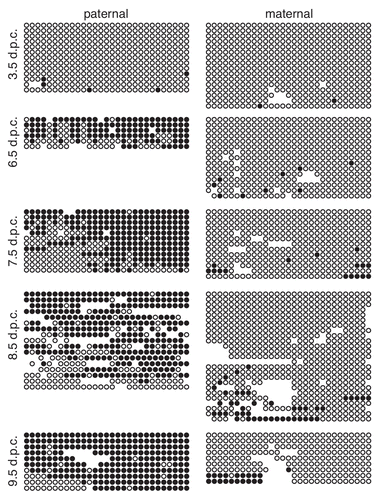
Figure 6 Expression analysis of Gtl2 in embryonic tissues. (A) Genomic structure of Gtl2, including exons (black boxes), B6 vs. CAST polymorphisms (*), major transcripts, primers used for RT-PCR and CpG-rich regions analyzed in this study (grey boxes). (B and C) RT-PCR of RNA from neonatal B6 liver and 6.5–9.5 d.p.c. B6 × CAST embryos using PCR primers A/B (B) and F/R (C). MW, molecular weight marker; 0, no template; + and − represent samples with and without reverse transcriptase (RT). (D) RT-PCR products from (B and C) were digested with SfcI and NgoMIV, respectively, to distinguish between products derived from maternal B6 vs. paternal CAST alleles. (E) RT-PCR of RNA from B6 brain, CAST brain and 3.5 d.p.c. B6 × CAST blastocysts using PCR primers F/R; B-N, C-N and 3.5 d.p.c. products were generated using a nested PCR approach described in the Materials and Methods. Alternative splice forms were detected in B6 and CAST samples; only the larger splice variant was detected in the +RT 3.5 d.p.c. sample. (F) RT-PCR products from E were digested with NgoMIV to distinguish between products derived from digested B6 vs. undigested CAST alleles.
Figure 7 Methylation is acquired at the Gtl2 and Cdkn1c DMRs during different developmental stages. Graphs summarizing the percentage of methylation on paternal strands at each CpG dinucleotide at the Gtl2- and Cdkn1c-DMRs in 6.5–9.5 d.p.c. embryos. Cdkn1c data derived from Bhogal et al.Citation14
Acknowledgments
We thank Mahvish Qureshi and Nelly Khaselev for technical support and Marisa Bartolomei for discussion and critical reading of the manuscript. This work was supported by U.S. Public Health Service Grant HD041444 to T.L.D.
References
- Verona RI, Mann MRW, Bartolomei MS. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu Rev Cell Dev Biol 2003; 19:237 - 259; PMID: 14570570
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science 2001; 293:1089 - 1093; PMID: 11498579
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol 2007; 19:281 - 289; PMID: 17467259
- Wan LB, Bartolomei MS. Regulation of imprinting in clusters: noncoding RNAs versus imsulators. Adv Genet 2008; 61:207 - 223; PMID: 18282507
- Tremblay KD, Duran KL, Bartolomei MS. A 5′ 2-kilo-base-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol 1997; 17:4322 - 4329; PMID: 9234689
- Takada S, Paulsen M, Tevendale M, Tsai CE, Kelsey G, Cattanach BM, et al. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2-H19. Hum Mol Genet 2002; 11:77 - 86; PMID: 11773001
- Shibata H, Yoda Y, Kato R, Ueda T, Kamiya M, Hiraiwa N, et al. A methylation imprint mark in the mouse imprinted gene Grf1/Cdc25Mm locus shares a common feature with the U2afbp-rs gene: an association with a short tandem repeat and a hypermethylated region. Genomics 1998; 49:30 - 37; PMID: 9570946
- Kobayashi H, Yamada K, Morita S, Hiura H, Fukuda A, Kagami M, et al. Identification of the mouse paternally expressed imprinted gene Zdbf2 on chromosome 1 and its imprinted human homolog ZDBF2 on chromosome 2. Genomics 2009; 93:461 - 472; PMID: 19200453
- Shemer R, Birger Y, Riggs AD, Razin A. Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci USA 1997; 94:10267 - 10272; PMID: 9294199
- Lucifero D, Mertineit C, Clarke H, Bestor T, Trasler J. Methylation dynamics of imprinted genes in mouse germ cells. Genomics 2002; 79:530 - 538; PMID: 11944985
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science 2001; 294:2536 - 2539; PMID: 11719692
- Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 2002; 129:1983 - 1993; PMID: 11934864
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 2004; 429:900 - 903; PMID: 15215868
- Bhogal B, Arnaudo A, Dymkowski A, Best A, Davis TL. Methylation at mouse Cdkn1c is acquired during postimplantation development and functions to maintain imprinted expression. Genomics 2004; 84:961 - 970; PMID: 15533713
- Hanel M, Wevrick R. Establishment and maintenance of DNA methylation patterns in mouse Ndn: implications for maintenance of imprinting in target genes of the imprinting center. Mol Cell Biol 2001; 21:2384 - 2392; PMID: 11259587
- Caspary T, Cleary MA, Baker CC, Guan X, Tilghman SM. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol Cell Biol 1998; 18:3466 - 3474; PMID: 9584186
- Carr MS, Yevtodiyenko A, Schmidt CL, Schmidt JV. Allele-specific histone modification regulate expression of the Dlk1-Gtl2 imprinted domain. Genomics 2007; 89:280 - 290; PMID: 17126526
- Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J 2007; 26:720 - 729; PMID: 17255950
- Fournier C, Goto Y, Ballestar E, Delaval K, Hever AM, Esteller M, et al. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J 2002; 21:6560 - 6570; PMID: 12456662
- Verona RI, Thorvaldsen JL, Reese KJ, Bartolomei MS. The transcriptional status but not the imprinting control region determines allele-specific histone modifications at the imprinted H19 locus. Mol Cell Biol 2008; 28:71 - 82; PMID: 17967893
- Takada S, Tevendale M, Baker J, Georgiades P, Campbell E, Freeman T, et al. Delta-like and Gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol 2000; 10:1135 - 1138; PMID: 10996796
- Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev 2000; 14:1997 - 2002; PMID: 10950864
- Yevtodiyenko A, Carr MS, Patel N, Schmidt JV. Analysis of candidate imprinted genes linked to Dlk1-Gtl2 using a congenic mouse line. Mamm Genome 2002; 13:633 - 638; PMID: 12461649
- Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J. A large imprinted microRNA cluster at the mouse Dlk1-Gtl2 domain. Genome Res 2004; 14:1741 - 1748; PMID: 15310658
- Hagan JP, O'Neill BL, Stewart CL, Kozlov SV, Croce CM. At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS One 2009; 4:4352; PMID: 19194500
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet 2003; 35:97 - 102; PMID: 12937418
- Li JY, Lees-Murdock DJ, Xu GL, Walsh CP. Timing of establishment of paternal methylation imprints in the mouse. Genomics 2004; 84:952 - 960; PMID: 15533712
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, et al. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 2007; 16:2272 - 2280; PMID: 17616512
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 1987; 99:371 - 382; PMID: 3653008
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet 2005; 14:47 - 58; PMID: 15809273
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 2002; 241:172 - 182; PMID: 11784103
- Schuster-Gossler K, Bilinski P, Sado T, Ferguson-Smith A, Gossler A. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev Dyn 1998; 212:214 - 228; PMID: 9626496
- Croteau S, Charron MC, Latham KE, Naumova AK. Alternative splicing and imprinting control of the Meg3/Gtl2-Dlk1 locus in mouse embryos. Mamm Genome 2003; 14:231 - 241; PMID: 12682775
- Surani MAH, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 1984; 308:548 - 550; PMID: 6709062
- McGrath J, Solter D. Completion of mouse embryo-genesis requires both the maternal and paternal genomes. Cell 1984; 37:179 - 183; PMID: 6722870
- Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005; 6:597 - 610; PMID: 16136652
- Georgiades P, Watkins M, Surani MA, Ferguson-Smith AC. Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development 2000; 127:4719 - 4728; PMID: 11023874
- Berghmans S, Segers K, Shay T, Georges M, Cockett N, Charlier C. Breakpoint mapping positions the callipyge gene within a 450-kilobase chromosome segment containing the DLK1 and GTL2 genes. Mamm Genome 2001; 12:183 - 185; PMID: 11210193
- Kagami M, Sekita Y, Nishimura G, Irie M, Kato F, Okada M, et al. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet 2008; 40:237 - 242; PMID: 18176563
- Wu Q, Kumagai T, Kawahara M, Ogawa H, Hiura H, Obata Y, et al. Regulated expression of two sets of paternally imprinted genes is necessary for mouse parthenogenetic development to term. Reprod 2006; 131:481 - 488; PMID: 16514191
- Moon YS, Smas CM, Lee K, Villena JA, Kim KH, Yun EJ, et al. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol 2008; 22:5585 - 5592; PMID: 12101250
- Sekita Y, Wagatsuma H, Nakamura K, Ono R, Kagami M, Wakisaka N, et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat Genet 2008; 40:243 - 248; PMID: 18176565
- Takahashi N, Okamoto A, Kobayashi R, Shirai M, Obata Y, Ogawa H, et al. Deletion of Gtl2, imprinted non-coding RNA, with its differentially methylated region induces lethal parent-origin-dependent defects in mice. Hum Mol Genet 2009; 18:1879 - 1888; PMID: 19264764
- Zhou Y, Cheunsuchon P, Nakayama Y, Lawlor MW, Zhong Y, Rice KA, et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development 2010; 137:2643 - 2652; PMID: 20610486
- Steshina EY, Carr MS, Glick EA, Yevtodiyenko A, Appelbe OK, Schmidt JV. Loss of imprinting at the Dlk1-Gtl2 locus caused by insertional mutagenesis in the Gtl2 5′ region. BMC Genet 2006; 7:44; PMID: 17014736
- Sekita Y, Wagatsuma H, Irie M, Kobayashi S, Kohda T, Matsuda J, et al. Aberrant regulation of imprinted gene expression in Gtl2lacZ mice. Cytogenet Genome Res 2006; 113:223 - 229; PMID: 16575184
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature 1993; 366:362 - 365; PMID: 8247133
- Stöger R, Kubicka P, Liu CG, Kafri T, Razin A, Cedar H, et al. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell 1993; 73:61 - 71; PMID: 8462104
- Kagami M, O'Sullivan MJ, Green AJ, Watabe Y, Arisaka O, Masawa N, et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet 2010; 6:1 - 13; PMID: 20585555
- Dockery L, Gerfen J, Harview C, Rahn-Lee C, Horton R, Park Y, et al. Differential methylation persists at the mouse Rasgrf1 DMR in tissues displaying monoallelic and biallelic expression. Epigenetics 2009; 4:241 - 247; PMID: 19502804