Abstract
Osteoblasts are specialized cells that form new bone and also indirectly influence bone resorption by producing factors that modulate osteoclast differentiation. Although the methylation of CpG islands plays an important role in the regulation of gene expression, there is still scanty information about its role in human bone. The aim of this study was to investigate the influence of CpG methylation on the transcriptional levels of two osteoblast-derived critical factors in the regulation of osteoclastogenesis: the receptor activator of nuclear factor NF-κB ligand (RANKL) and its soluble decoy receptor osteoprotegerin (OPG). Quantitative methylation specific PCR (qMSP) and pyrosequencing analysis in various cell types showed that the methylation of regulatory regions of these genes, in the vicinity of the transcription start sites, repressed gene transcription, whereas an active transcription was associated with low levels of methylation. In addition, treatment with the DNA demethylating agent 5-azadeoxycitidine promoted a 170-fold induction of RANKL and a 20-fold induction of OPG mRNA expression in HEK-293 cells, which showed hypermethylation of the CpG islands and barely expressed RANKL and OPG transcripts at baseline. Transcriptional levels of both genes were also explored in bone tissue samples from patients with hip fractures and hip osteoarthritis. Although RANKL transcript abundance and the RANKL:OPG transcript ratio were significantly higher in patients with fractures than in those with osteoarthritis (RANKL: 0.76 ± 0.23 vs. 0.24 ± 0.08, p = 0.012; RANKL/OPG: 7.66 ± 2.49 vs. 0.92 ± 0.21, p = 0.002), there was no evidence for differential methylation across patient groups. In conclusion, the association between DNA methylation and the repression of RANKL and OPG expression strongly suggests that methylation-dependent mechanisms influence the transcription of these genes, which play a critical role in osteoclastogenesis. However, other mechanisms appear to be involved in the increased RANKL/OPG ratio of patients with osteoporotic fractures.
Introduction
Bone is a dynamic tissue that is under continuous remodeling. This process involves the synthesis of bone matrix by osteoblasts and its resorption by osteoclasts. Bone remodeling is tightly regulated by the RANKL-RANK-OPG system.Citation1 RANKL (Receptor activator of NF-kB ligand, encoded by the TNFSF11 gene) binds to its receptor, RANK, present in cells of the osteoclast lineage and stimulates osteoclast formation, activation and survival.Citation2,Citation3 On the other hand, OPG (Osteoprotegerin, the product of the TNFRSF11B gene) protects bone from excessive resorption by binding to RANKL and thus avoiding its interaction with RANKCitation4,Citation5. In addition, recent findings suggest that OPG may function as a traffic regulator of RANKL, modulating its ability to reach the cell membrane.Citation6 Thus, the relative concentrations of RANKL and OPG in bone are regarded as important determinants of bone mass and strength, and the inhibition of RANKL/RANK signaling has become a therapeutic target in osteoporosis and other disorders characterized by an increased bone resorption.Citation7,Citation8
Identifying the transcriptional mechanisms that drive RANKL and OPG expression is of great scientific interest and may help to identify new targets for bone therapies. Various cell types, including stromal cells and osteoblasts produce RANKL and OPG in the bone microenvironment.Citation9,Citation10 Recent studies also suggest that osteocytes, as well as hypertrophic chondrocytes, are other important sources of RANKL in the adult skeleton.Citation11,Citation12 PTH, 1,25(OH)2D3, glucocorticoids and IL 6-type cytokines stimulate RANKL expression.Citation13-Citation15 OPG expression is also regulated by different cytokines, hormones and growth factors, as well as by the Wnt/β-catenin pathway.Citation16,Citation17
Epigenetic mechanisms and particularly DNA methylation are known to contribute to gene transcription in many tissues. DNA methylation tends to block gene expression by incompletely known mechanisms, including the interference of the binding of transcription factors to the regulatory sites in DNA.Citation18 Although there are some indications for a role of DNA methylation of the RANKL/OPG genes in murine models and cancer cells,Citation19,Citation20 it is still unknown whether DNA methylation regulates the expression of these genes in human bone. Therefore, the aim of this study was to explore the influence of DNA methylation on RANKL and OPG expression in human osteoblastic cells and its possible involvement in osteoporotic fractures.
Results
Bioinformatics analysis of the RANKL and OPG gene sequence
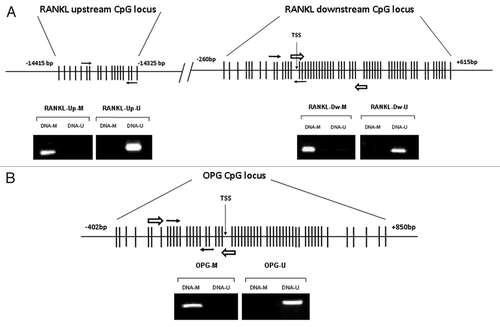
Our bioinformatics analysis revealed two CpG islands in the RANKL gene: the upstream one (18 CpG sites), located at -14415 bp from the transcription start site of isoform I (TSS I), the major RANKL transcript; and the downstream one (59 CpG sites), that spans from -260 bp to +615 bp of the TSS I (). One island was found in the OPG gene (56 CpG sites), spanning from -402 to +850 bp of the TSS ().
Figure 1. Locations of the RANKL and OPG CpG islands, qMSP and pyrosequencing amplicons. (A) Non-scaled representation of the RANKL gene. Two CpG-rich regions were identified, the upstream one located at -14415 bp from the TSS of the isoform I; and the downstream one, located -260 bp from isoform I TSS. One qMSP amplicon was designed for each region (white arrows). A pyrosequencing amplicon was designed for the downstream CpG region (black arrows). MSP was performed with control fully methylated DNA (DNA-M) or completely unmethylated (DNA-U) to verify the specificity of the MSP primers. (B) Non-scaled representation of the OPG gene. One CpG island was found spanning from -402 to +850 bp of the TSS. qMSP and pyrosequencing amplicons were designed within the CpG-rich area (white and black arrows respectively). MSP was performed using DNA-M or DNA-U to verify the specificity of the MSP primers.
RANKL gene expression and methylation
We studied RANKL and OPG gene expression in several cell lines and primary cells. RANKL mRNA was readily detected in human primary osteoblasts (hOBs) and the osteoblast-like cell line MG-63. However, minimum amounts of RANKL mRNA were detected in the osteoblastic cell line HOS-TE85 and in kidney-derived HEK-293 cells, less than 1% of the level of hOBs ().
Figure 2.RANKL expression and methylation analysis in cell lines and primary osteoblasts. (A) The abundance of RANKL transcripts was determined by RT-qPCR. Results are expressed as relative expression to the housekeeping gene TBP. Methylation of the downstream CpG region was studied by pyrosequencing. Bars represent average % methylation of the CpG dinucleotides studied. Mean and SD of three independent experiments for each cell line is represented. (B) Methylation was explored in the upstream (CpGs 5, 6 and 7 in the forward primer; CpGs 17 and 18 in the reverse primer) and downstream CpG islands (CpGs 9 and 10 in the forward primer; 15 and 16 in the reverse primer) by qMSP. Mean and SD of three independent experiments is showed. (C) All individual CpGs studied (CpGs 15 to 23) by pyrosequencing of the RANKL downstream island showed a similar methylation status. Black slices represent the percentage of methylation. Note that only CpG 1 to 40 are displayed in figure C.
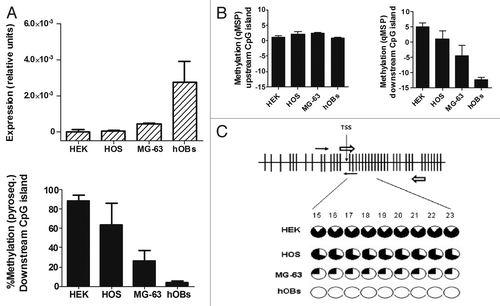
In parallel experiments we explored the methylation degree of the two CpG-rich regions of the RANKL gene by qMSP. MG-63 and hOBs, that expressed higher amounts of RANKL, showed lower methylation in the downstream CpG island than the two other cell types. However, no differences in the methylation of the upstream CpG island were found, which was strongly methylated in all cell types ().
To confirm the differences in DNA methylation we analyzed the downstream CpG-rich region by pyrosequencing (, bottom panel). Indeed, hOBs and MG-63 showed smaller degrees of methylation of the overall region than HOS-TE85 and HEK-293 cells (4.1 ± 1.4%; 26.6 ± 0.4%; 63.9 ± 2.2%; and 89.0 ± 5.6%, respectively). The single nucleotide analysis showed a very similar methylation pattern in the 9 cytosines included in the region, with methylation percentages between 1.2% and 5.4% in hOBs, and between 25.1% and 42.3% in MG-63. Much higher methylation percentages were found in the cells with low expression of RANKL: 68.7–91.2% in HOS-TE85 and 80.0–98.7% in HEK-293 ().
OPG gene expression and methylation
OPG mRNA was highly expressed in hOBs and MG-63 cells. HOS-TE85 cells expressed smaller amounts of OPG, whereas OPG mRNA was barely detected in the kidney-derived HEK-293 cell line (, upper panel).
Figure 3.OPG expression and methylation analysis in culture cell lines and primary osteoblasts. (A) OPG expression was detected by RT-qPCR. Results are expressed as relative expression in relation to the housekeeping gene TBP. Methylation was studied by qMSP (CpGs 10, 11 and 12 in the forward primer; CpGs 18, 19 and 20 in the reverse primer). For pyrosequencing bars represent the average % methylation of the CpG dinucleotides studied. Mean and SD of three independent experiments for each cell line is presented. (B) The CpG sites studied by pyrosequencing (CpGs 10–12) showed a similar degree of methylation. Each circle represents one CpG site. The black slices represent the percentage of methylation. Note that only CpGs 1 to 33 are displayed in figure B.
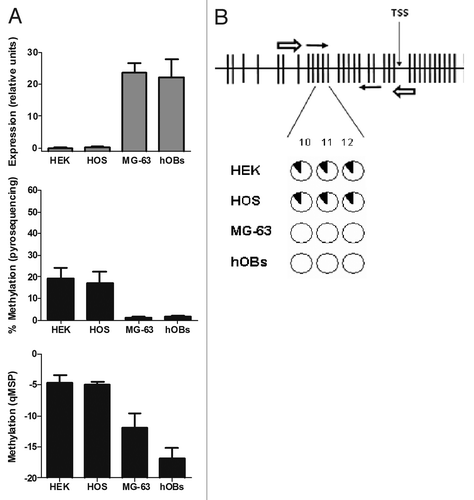
DNA methylation analysis by qMSP showed that HEK-293 and HOS-TE85 cells, that expressed smaller amounts of OPG, displayed higher methylation in the CpG-rich region of the OPG gene than hOBs or MG-63. The differences in methylation were also confirmed by pyrosequencing (, middle and bottom panels), which demonstrated that the OPG gene was more methylated in HEK-293 and HOS-TE85 cells (19.5 ± 4.5% and 17.3 ± 5.2%, respectively) than in MG-63 and hOBs (1.2 ± 0.3% and 1.6 ± 0.4%, respectively). The single-base analysis revealed a similar degree of methylation in the 3 cytosines studied (). In hOBs, the methylation of individual cytosines varied between 1.4% and 2.3%; in MG-63, between 0.8% and 1.6%. However, in HOS-TE85, it varied between 14.9% and 21.3%; and in HEK-293 between 12.3% and 21.5%.
5-Aza-2´-Deoxycytidine induces RANKL and OPG expression in HEK-293 cells
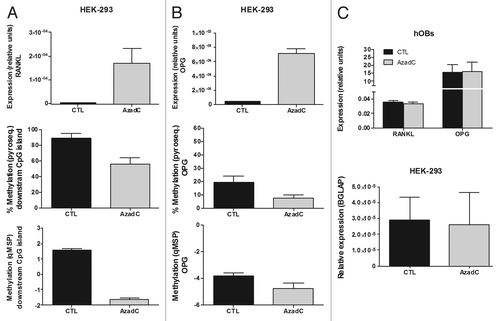
The inverse relationship observed between methylation and gene expression suggested that DNA methylation was indeed repressing RANKL and OPG expression. To confirm this hypothesis, we explored the ability of 5-Aza-2´-Deoxycytidine (AzadC) to induce gene expression in HEK-293 cells. AzadC is incorporated into DNA and blocks DNA methyltransferases. We found that AzadC treatment induced a decrease in the methylation of the CpG-rich regions of OPG and RANKL genes ( and , middle and bottom panel). It was associated with a marked increase in gene expression, with a 170-fold induction of RANKL and a 20-fold induction of OPG mRNA. However, interestingly enough, when hOBs, which had the OPG and RANKL regions already hypomethylated, were treated with AzadC, we did not detect any changes in OPG or RANKL gene expression. On the other hand, osteocalcin (BGLAP) expression was not affected by the demethylating treatment in the HEK-293 cells ().
Figure 4. Effect of AzadC on RANKL and OPG expression in HEK-293 cells and primary osteoblasts. (A) RANKL expression was explored by RT-qPCR in HEK-293 cells, either untreated or treated with AzadC 5μM for 4 d. Graph shows RANKL expression relative to the housekeeping gene TBP. AzadC effect on DNA methylation of the RANKL downstream CpG region was studied by pyrosequencing and qMSP. (B) OPG transcriptional levels were explored by RT-qPCR in HEK-293 cells, either untreated or treated with AzadC 5uM for 4 d. Graph shows OPG expression relative to the housekeeping TBP. DNA methylation was explored by pyrosequencing and qMSP. (C) RANKL and OPG expression was assayed in primary osteoblasts treated with 5 μM of AzadC for 4 d (upper plot). Osteocalcin expression (BGLAP) was studied in HEK-293, either untreated or treated with AzadC 5μM for 4 d (lower plot). Graphs show OPG, RANKL and BGLAP expression relative to TBP. Mean and SD of three independent experiments for each cell line is showed.
RANKL and OPG expression and methylation in osteoarthritic and osteoporotic bone samples
We explored if differences in RANKL-OPG expression were involved in the opposite changes in bone mass typical of osteoarthritis (OA) and osteoporotic (OP) fractures. RANKL expression was significantly higher in samples from patients with osteoporotic fractures (0.76 ± 0.23 vs. 0.24 ± 0.08, p = 0.012; ).No significant differences were observed in OPG expression (0.26 ± 0.06 in OP and 0.44 ± 0.11 in OA; ), but the RANKL:OPG ratio was much higher in OP bone tissue samples (7.66 ± 0.23 vs. 0.92 ± 0.21, p = 0.002; ).
Figure 5. Expression and methylation of RANKL and OPG genes in bone tissue from osteoporotic (OP) and osteoarthritic (OA) patients. (A) RANKL expression was studied in bone tissue samples. Bars represent RANKL expression relative to TBP. DNA methylation in the downstream CpG island was studied by pyrosequencing and qMSP, whereas methylation in the upstream region was studied by qMSP. (B) OPG studies were performed in the same samples. (C) RANKL:OPG transcript ratio. Each bar represents mean and SE values for each group.
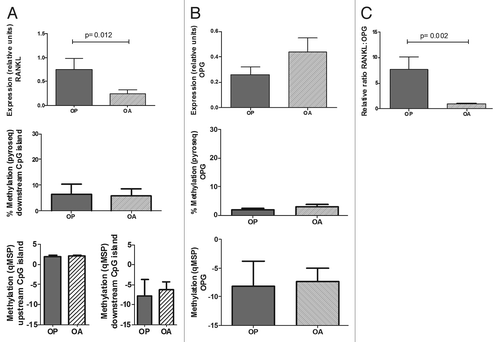
We next investigated if the differences in the RANKL/OPG expression pattern between the two groups of patients were related to differential methylation. DNA was extracted from femoral head samples from 9 patients with OA and 12 with fractures (age 77 ± 4 and 79 ± 3 y, respectively) and methylation was studied by qMSP. As shown above in cell cultures, the RANKL upstream region appeared highly methylated in all bone samples. However, the RANKL downstream and the OPG CpG islands appeared hypomethylated, without differences between OP and OA samples (, bottom panels). The results were confirmed by pyrosequencing (, middle panels). The average methylation of the RANKL downstream region was 6.6 ± 3.8% in hip fracture samples and 5.9 ± 2.8% in OA samples. The CpG-rich region of the OPG gene was also poorly methylated in both samples (1.9 ± 0.4% in OP and 3.0 ± 0.7% in OA).
Discussion
There is increasing experimental evidence showing that the methylation status of CpG islands plays an important role in the regulation of gene expression.Citation21 This has been extensively studied in neoplastic disorders, including bone tumors.Citation22,Citation23 However, little is known about the potential role of DNA methylation in normal bone and non-neoplastic bone disorders.
A bioinformatics search revealed two CpG-rich regions in the RANKL gene, the upstream CpG island and the downstream one. The human downstream CpG-rich sequence showed greater than 75% homology with the previously reported CpG island in the mouse RANKL gene.Citation19 In fact, there are not only sequence homologies, but also the location of the island is very similar in both species, around the transcription start site of the isoform I (TSS I). We did not find homologies for the upstream CpG-rich region between those species, which was located quite far from the TSS I (-14415 bp), but relatively close to the isoform II TSS (around -3000 bp). On the other hand, we found a CpG island in the vicinity of the OPG transcription start site, consistent with the location reported by Lu et al.Citation20 The presence of the CpG islands suggested that these genes might be regulated by cytosine methylation. Indeed, we found an inverse association between DNA methylation and gene expression. Thus, the CpG-rich regions of HEK-293 cells, which hardly expressed RANKL and OPG transcripts, were hypermethylated, whereas low percentage of methylation was observed in hOBs, which expressed large amounts of both genes. Pyrosequencing confirmed the semiquantitative results of qMSP and allowed us to establish that all individual CpG sites analyzed had a similar degree of methylation within a certain CpG island, a consistent result in all cell types studied. Interestingly enough, the methylation of the RANKL downstream CpG island was associated with the repression of transcription, but no differences in the methylation level of the upstream CpG island were found between the cells studied, despite the marked differences in RANKL expression. This observation, along the conservation, both in sequence and location, suggests that the methylation at the downstream CpG island, and not at the upstream one, actually modulates RANKL transcription. Nevertheless, in this study we analyzed the isoform I, which is the most widely expressed and active form of RANKL.Citation24 Thus, we cannot exclude an influence of the upstream CpG island on the transcription of alternative minor isoforms of RANKL. However, its consistent hypermethylation in all the samples studied raises doubts about the potential physiological role of methylation changes in this region, at least in the skeleton.
We have also studied the methylation profile of the RANKL downstream CpG-rich region in microdissected mature human osteocytes and lining osteoblasts and found that those cells show a low degree of methylation, similar to hOBs (unpublished observations). This strongly suggests that osteocytes are epigenetically ready to express RANKL. Indeed, recent studies suggest that osteocytes may be a major source of RANKL within the bone microenvironmentCitation11,Citation12. Furthermore, the expression of both genes in human mesenchymal stem cells (MSCs), which differentiate into various types of cells, including osteoblasts, is easily detectable. In line with the results here reported, the CpG islands of RANKL and OPG genes are also largely demethylated in MSCs (unpublished data). Therefore, the DNA methylation pattern of RANKL appears to be early established in osteoblast precursors and does not undergo significant changes during the process of differentiation toward osteocytes, which is consistent with reports demonstrating that RANKL is expressed throughout all stages of differentiation of the osteoblastic lineage.Citation25
To get further evidence for the role of DNA methylation in the expression of RANKL and OPG we studied the effect of AzadC on the heavily methylated HEK-293 cells. AzadC is a cytidine analog with a nitrogen atom replacing the carbon at the 5 position of the pyrimidine ring. When AzadC is incorporated into DNA, DNA-methyltransferases cannot methylate the modified cytidine, thus perturbing the methylation pattern in daughter cells.Citation26 AzadC was actually effective in decreasing gene methylation at the CpG sites studied, as shown by qMSP and pyrosequencing analysis. This hypomethylating effect was accompanied by a marked upregulation of OPG and RANKL (20‑fold and 170‑fold induction, respectively), a result consistent with a repressive role of CpG methylation in gene expression. This was not just the consequence of a non-specific increase in gene expression, because other genes typical of an osteblastic phenotype, such as osteocalcin, remained unaltered. Moreover, when primary osteoblasts, which show a low degree of methylation at the studied CpG sites, were treated with AzadC, no further upregulation of RANKL or OPG was found.
OP and OA are two common age-related, chronic disorders of the skeleton that tend to show opposite changes in bone mass.Citation27 Although OA has long been considered to be primarily a cartilage disorder, with secondary changes in the subchondral bone, several studies suggest a more active role of the bone tissue in the pathogenesis of the disease.Citation28,Citation29 As expected, RANKL and OPG transcripts were readily detected in bone tissue samples from both groups. No significant differences in OPG expression were observed. However we found that RANKL expression and the RANKL/OPG ratio were significantly higher in samples from patients with hip fractures than in those with osteoarthritis, which is consistent with enhanced osteoclastogenesis in the former group of patients. These results confirm previous reports by Logar et al.Citation30 and a recent report by D’AmelioCitation31 and are consistent with the hypothesis that an increased signaling of the RANK pathway, and the resultant stimulus of osteoclastogenesis, may contribute to osteoporotic fractures.
Emerging evidence suggests that epigenetic mechanisms may be involved in age related diseases and bone biologyCitation32,Citation33 and a few genes have been reported as epigenetically regulated in bone.Citation34,Citation35 Since we found an epigenetic modulation of RANKL and OPG gene expression levels, we explored if differential levels of DNA methylation could explain the differences in gene transcription between the OP and OA groups. In bone tissue samples, low DNA methylation was found in the downstream RANKL island and in the OPG island. This is in line with the results in cell cultures, showing an association between active expression and low methylation levels. However, despite the marked differences in gene expression, we found no differences in the methylation of RANKL between both groups of patients at the bone tissue level. These results would suggest that mechanisms other than DNA methylation would be responsible for the differences in RANKL expression between patients with OA and OP. We can speculate that DNA methylation represents an on/off regulator of RANKL-OPG gene expression, whereas other factors, such as hormones and cytokines in the bone microenvironment, are responsible for the fine-tuning of expression in cells with hypomethylated islands. In fact, PTH and estrogens, may be appealing candidates because they have been shown to modulate RANKL expression in several experimental system,Citation11,Citation15 in some cases by interacting with distant regulatory regions.Citation36 Besides these humoral factors, we cannot exclude the involvement of other epigenetic mechanisms, such as the post-translational modifications of histones or miRNA production, but their role, if any, remains speculative at the moment. Nevertheless, it is important to note that bone is formed by different cell types, many of which express RANKL. Since we analyzed whole bone tissue samples, we cannot rule out that DNA methylation differences in specific cell types, such as osteocytes or osteoblasts, could underlie the des-regulation of RANKL in osteoporotic patients.
In summary, we have shown that DNA methylation represses RANKL and OPG transcription. This was the case for the OPG island and the downstream RANKL CpG island, but not for the upstream RANKL island, suggesting that the latter does not play an important role in the regulation of gene expression. The silencing role of DNA methylation was further supported by the fact that treatment with a demethylating agent induced a marked upregulation of RANKL and OPG transcripts in cells that hardly expressed these genes at baseline. Together, these data suggest that changes in DNA methylation contribute to regulate the expression of these genes, which are critical for bone homeostasis. However, other mechanisms independent of DNA methylation appear to be involved in the increased RANKL/OPG ratio of patients with osteoporotic fractures.
Materials and Methods
Cell culture
The human osteoblastic cell line HOS-TE85Citation37 was maintained in culture with Eagle Minimum Essential Medium (MEM, Sigma-Aldrich, M0643) supplemented with 10% fetal bovine serum (FBS) and antibiotics. The human osteoblastic cell line MG-63Citation38 and the human embryonic kidney cell line HEK-293,Citation39 were routinely cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, 12800–116) supplemented as described above.
Bone tissue samples were obtained during arthroplasty, from patients with osteoporotic hip fractures (24 women, 2 men; age 80 ± 4 y) and with hip osteoarthritis (19 women, 5 men; age 75 ± 6 y), after obtaining informed consent. The study was approved by the Cantabria Clinical Research Ethics Committee and all participants gave informed written consent. Patients with secondary osteoporosis, fractures due to high-energy trauma or secondary osteoarthritis were excluded. Trabecular bone cylinders of the central part of the femoral head (thus avoiding the fractured and the subchondral regions) were obtained with a trephine, washed extensively in phosphate-buffered saline, snap-frozen in liquid nitrogen and stored at -80°C or used to generate primary human osteoblasts (hOBs) by the explant technique.Citation40 The osteoblastic phenotype was confirmed by the ability to express osteocalcin when stimulated with vitamin D and to form a mineralized matrix. hOBs were cultured in DMEM supplemented with 10% FBS and antibiotics. 5-Aza-2´-Deoxycytidine (AzadC) was purchased from Sigma-Aldrich (A2385). For demethylation experiments cells were plated at 5000 cells/cm2 and treated for 4 d in the presence/absence of AzadC, 5 μM, prior to the analysis of gene expression and DNA methylation.
Gene expression
RANKL and OPG expression was determined by reverse transcription followed by real time quantitative polymerase chain reaction (RT-qPCR). RNA was isolated from cell cultures or bone tissue samples with Trizol following the manufacturer’s protocol (Invitrogen, 15596–018). Then cDNA was synthesized from 1 μg total RNA, using random hexamers as primers, and the Superscript III First Strand kit (Invitrogen, 18080–051). Gene expression was measured using Taqman probes following the manufacturer’s protocol (Applied Biosystems). The following Taqman Gene Expression Assays were used: TNFSF11 (isoform I, RANKL): Hs00243519_m1; TNFRSF11B (OPG): Hs00900360_m1; and BGLAP (osteocalcin): Hs01587814_g1. Results are shown as the relative gene expression, using the expression of the TATA Box Binding Protein (TBP) housekeeping gene as a reference. The cycle thresholds of each gene were estimated and gene expression was computed as 2-ΔCt, where ΔCt is the difference between the gene of interest threshold cycle and the threshold cycle of TBP.
DNA bioinformatics search and methylation analysis by qMSP
We performed a bioinformatics analysis to identify potential CpG islands in the genomic sequences of RANKL and OPG genes. The USCS Human Genome Browser public database (http://genome.ucsc.edu), the Primer Express software (available from Applied Biosystems), and CpG Island Explorer software (cpgie.sourceforge.net) were used to explore the nucleotide sequences surrounding de transcription start site (TSS) of RANKL isoforms I and II (NG_008990.1 Ref Seq Gene), and OPG (NG_012202.1 Ref Seq Gene). A (C + G)/total bases ratio > 0.5 and CpG observed/CpG expected ratio > 0.6 were used as criteria. Quantitative methylation-specific PCR (qMSP) is based on the amplification of bisulfite converted DNA.Citation41 Sodium bisulfite converts unmethylated cytosines to uracils, whereas methylated cytosines are unaffected. Thus, it allows distinguishing between methylated and unmethylated DNA. Primers, which targeted CpG-rich regions within the RANKL and OPG regions, were designed with Methyl Primer Express software (available from Applied Biosystems). For each region, a pair of primers was selected; one specifically recognized the bisulfite-treated methylated DNA and other recognized only unmethylated bisulfite-treated DNA ().
Table 1. Primer sequences and amplicon sizes for the quantitative methylation-specific PCR (qMSP) of the RANKL and OPG gene. Abreviations: bp, base pairs; Fwd, forward; Rev, reverse
Genomic DNA from cells and bone tissue samples was isolated with phenol:chloroform: isoamyl alcohol (Invitrogen, 15593031). For optimized bisulfite conversion, we used 1000 ng of genomic DNA and the Zymo EZ DNA Methylation-Gold kit (Zymo Research, D5005). After bisulfite conversion, the DNA was bound to a Zymo spin column and desulfonated. The bisulfite converted DNA was eluted from the column in 10μl of elution buffer. In the qMSP reaction, duplicate aliquots of 1μl of the eluted bisulfite-treated genomic DNA were amplified with AmpliTaq Gold DNA Polymerase buffer (Applied Biosystems, N808–0240) with 2 mM MgCl2, 0.4 pmol primers, 400 μM dNTP mix, 8 μl of Sybr Green and 1U of AmpliTaq Gold DNA polymerase in a total volume of 25 μl. Amplification conditions were: 95°C for 5 min, followed by 40 cycles of 95°C for 30s, 57°C for 30s and 72°C for 30s, with a final extension of 72°C for 5 min. Epitec methylated and unmethylated human control DNA, bisulfite converted, as well as unmodified human DNA (Qiagen, 59695) were used as controls to check primer specificity (). The degree of DNA methylation was estimated as the unmethylated primer cycle threshold minus the methylated primer cycle threshold for each amplicon. Thus, smaller values are associated with smaller proportions of methylated DNA in the sample. The mean and SD was calculated from series of three experiments for each amplicon and cell line.
Pyrosequencing
The results obtained by qMSP were confirmed by pyrosequencing.Citation42,Citation43 Sodium bisulphite modification of 0.5 μg genomic DNA was performed as described above. Bisulphite-treated DNA was eluted in 15 μl, using 2 μl for each PCR. The set of primers for PCR amplification and sequencing were designed using the PyroMark assay design software, version 2.0.01.15. Primer sequences were designed to hybridize with CpG-free sites to ensure methylation-independent amplification. The primers were located flanking the qMSP amplicons (see Fig. S1). Primer sequences were:
Pyro-RANKL-F: 5′TTTTGGGAAGGTGGTTATTTAT3′;
Pyro-RANKL-R[Btn]: 5′CCAACAAAAACTACACCAAATAC 3′;
Pyro-RANKL-Seq: 5′GTTTTAGTTTTAGGAGGGTTA 3′;
Pyro-OPG-F: 5′GGGTTTTGTAATTTGAGGTTTTAGAA 3′;
Pyro-OPG-R[Btn]: 5′ACTTATATCTCCTCCACCCTAAA 3′;
Pyro-OPG-Seq: 5′GATAAAGGTTTGGGATATATT 3′.
PCR was performed with primers biotinylated to convert the PCR product to single-stranded DNA templates, using the Vacuum Prep Tool (Biotage), according to manufacturer’s instructions. Pyrosequencing reactions and methylation quantification were performed in a PyroMark Q24 System version 2.0.6 (Qiagen). Series of three independent experiments were performed for each cell line and condition.
Statistical analysis
The statistical significance of the differences between osteoporotic and osteoarthritic patients was tested by Mann-Whitney tests. All reported p values are two-tailed and 0.05 was the significance threshold.
Additional material
Download Zip (73.6 KB)Acknowledgments
This work was supported in part by grants from Instituto de Salud Carlos III-Fondo de Investigaciones Sanitarias (Spanish Ministry of Health) PI09/539, PI06/1267, PS09/02454, the Spanish National Research Council (CSIC 200820I172 to MFF), and the Community of Asturias (FICYT IB09–106). The IUOPA is supported by the Obra Social Cajastur, Spain. Jesús Delgado-Calle is recipient of a grant from IFIMAV. Agustín Fernández-Fernández is supported by the IUOPA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Boyce BF, Xing L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch Biochem Biophys 2008; 473:139 - 46; http://dx.doi.org/10.1016/j.abb.2008.03.018; PMID: 18395508
- Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci USA 1998; 95:3597 - 602; http://dx.doi.org/10.1073/pnas.95.7.3597; PMID: 9520411
- Lum L, Wong BR, Josien R, Becherer JD, Erdjument-Bromage H, Schlondorff J, et al. Evidence for a role of a tumor necrosis factor-alpha (TNF-alpha)-converting enzyme-like protease in shedding of TRANCE, a TNF family member involved in osteoclastogenesis and dendritic cell survival. J Biol Chem 1999; 274:13613 - 8; http://dx.doi.org/10.1074/jbc.274.19.13613; PMID: 10224132
- Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, Luthy R, et al. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell 1997; 89:309 - 19; http://dx.doi.org/10.1016/S0092-8674(00)80209-3; PMID: 9108485
- Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 1999; 397:315 - 23; http://dx.doi.org/10.1038/16852; PMID: 9950424
- Aoki S, Honma M, Kariya Y, Nakamichi Y, Ninomiya T, Takahashi N, et al. Function of OPG as a traffic regulator for RANKL is crucial for controlled osteoclastogenesis. J Bone Miner Res 2010; 25:1907 - 21; http://dx.doi.org/10.1002/jbmr.89; PMID: 20560139
- Bone HG, Bolognese MA, Yuen CK, Kendler DL, Wang H, Liu Y, et al. Effects of denosumab on bone mineral density and bone turnover in postmenopausal women. J Clin Endocrinol Metab 2008; 93:2149 - 57; http://dx.doi.org/10.1210/jc.2007-2814; PMID: 18381571
- Genant HK, Engelke K, Hanley DA, Brown JP, Omizo M, Bone HG, et al. Denosumab improves density and strength parameters as measured by QCT of the radius in postmenopausal women with low bone mineral density. Bone 2010; 47:131 - 9; http://dx.doi.org/10.1016/j.bone.2010.04.594; PMID: 20399288
- Silvestrini G, Ballanti P, Patacchioli F, Leopizzi M, Gualtieri N, Monnazzi P, et al. Detection of osteoprotegerin (OPG) and its ligand (RANKL) mRNA and protein in femur and tibia of the rat. J Mol Histol 2005; 36:59 - 67; http://dx.doi.org/10.1007/s10735-004-3839-1; PMID: 15704000
- Ikeda T, Utsuyama M, Hirokawa K. Expression profiles of receptor activator of nuclear factor kappaB ligand, receptor activator of nuclear factor kappaB, and osteoprotegerin messenger RNA in aged and ovariectomized rat bones. J Bone Miner Res 2001; 16:1416 - 25; http://dx.doi.org/10.1359/jbmr.2001.16.8.1416; PMID: 11499864
- Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, O'Brien CA. Matrix-embedded cells control osteoclast formation. Nat Med 2011; 17:1235 - 41; http://dx.doi.org/10.1038/nm.2448; PMID: 21909103
- Nakashima T, Hayashi M, Fukunaga T, Kurata K, Oh-Hora M, Feng JQ, et al. Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat Med 2011; 17:1231 - 4; http://dx.doi.org/10.1038/nm.2452; PMID: 21909105
- Fu Q, Jilka RL, Manolagas SC, O'Brien CA. Parathyroid hormone stimulates receptor activator of NFkappa B ligand and inhibits osteoprotegerin expression via protein kinase A activation of cAMP-response element-binding protein. J Biol Chem 2002; 277:48868 - 75; http://dx.doi.org/10.1074/jbc.M208494200; PMID: 12364326
- Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol 2006; 26:6469 - 86; http://dx.doi.org/10.1128/MCB.00353-06; PMID: 16914732
- O'Brien CA, Gubrij I, Lin SC, Saylors RL, Manolagas SC. STAT3 activation in stromal/osteoblastic cells is required for induction of the receptor activator of NF-kappaB ligand and stimulation of osteoclastogenesis by gp130-utilizing cytokines or interleukin-1 but not 1,25-dihydroxyvitamin D3 or parathyroid hormone. J Biol Chem 1999; 274:19301 - 8; http://dx.doi.org/10.1074/jbc.274.27.19301; PMID: 10383440
- Kieslinger M, Folberth S, Dobreva G, Dorn T, Croci L, Erben R, et al. EBF2 regulates osteoblast-dependent differentiation of osteoclasts. Dev Cell 2005; 9:757 - 67; http://dx.doi.org/10.1016/j.devcel.2005.10.009; PMID: 16326388
- Glass DA, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell 2005; 8:751 - 64; http://dx.doi.org/10.1016/j.devcel.2005.02.017; PMID: 15866165
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 2006; 31:89 - 97; http://dx.doi.org/10.1016/j.tibs.2005.12.008; PMID: 16403636
- Kitazawa R, Kitazawa S. Methylation status of a single CpG locus 3 bases upstream of TATA-box of receptor activator of nuclear factor-kappaB ligand (RANKL) gene promoter modulates cell- and tissue-specific RANKL expression and osteoclastogenesis. Mol Endocrinol 2007; 21:148 - 58; http://dx.doi.org/10.1210/me.2006-0205; PMID: 17008384
- Lu TY, Kao CF, Lin CT, Huang DY, Chiu CY, Huang YS, et al. DNA methylation and histone modification regulate silencing of OPG during tumor progression. J Cell Biochem 2009; 108:315 - 25; http://dx.doi.org/10.1002/jcb.22256; PMID: 19565568
- Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell 2010; 19:698 - 711; http://dx.doi.org/10.1016/j.devcel.2010.10.005; PMID: 21074720
- Esteller M. Epigenetics in cancer. N Engl J Med 2008; 358:1148 - 59; http://dx.doi.org/10.1056/NEJMra072067; PMID: 18337604
- Ulaner GA, Vu TH, Li T, Hu JF, Yao XM, Yang Y, et al. Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum Mol Genet 2003; 12:535 - 49; http://dx.doi.org/10.1093/hmg/ddg034; PMID: 12588801
- Suzuki J, Ikeda T, Kuroyama H, Seki S, Kasai M, Utsuyama M, et al. Regulation of osteoclastogenesis by three human RANKL isoforms expressed in NIH3T3 cells. Biochem Biophys Res Commun 2004; 314:1021 - 7; http://dx.doi.org/10.1016/j.bbrc.2003.12.191; PMID: 14751235
- Carda C, Silvestrini G, Gomez de Ferraris ME, Peydro A, Bonucci E. Osteoprotegerin (OPG) and RANKL expression and distribution in developing human craniomandibular joint. Tissue Cell 2005; 37:247 - 55; http://dx.doi.org/10.1016/j.tice.2005.03.002; PMID: 15899507
- Patra SK, Patra A, Rizzi F, Ghosh TC, Bettuzzi S. Demethylation of (Cytosine-5-C-methyl) DNA and regulation of transcription in the epigenetic pathways of cancer development. Cancer Metastasis Rev 2008; 27:315 - 34; http://dx.doi.org/10.1007/s10555-008-9118-y; PMID: 18246412
- Dequeker J, Aerssens J, Luyten FP. Osteoarthritis and osteoporosis: clinical and research evidence of inverse relationship. Aging Clin Exp Res 2003; 15:426 - 39; PMID: 14703009
- Neilson M, White A, Malik U, Morrison E, McGill PE, McDonald SW. Changes in bone architecture in the femoral head and neck in osteoarthritis. Clin Anat 2004; 17:378 - 91; http://dx.doi.org/10.1002/ca.10177; PMID: 15176035
- Jordan GR, Loveridge N, Bell KL, Power J, Dickson GR, Vedi S, et al. Increased femoral neck cancellous bone and connectivity in coxarthrosis (hip osteoarthritis). Bone 2003; 32:86 - 95; http://dx.doi.org/10.1016/S8756-3282(02)00920-1; PMID: 12584040
- Logar DB, Komadina R, Prezelj J, Ostanek B, Trost Z, Marc J. Expression of bone resorption genes in osteoarthritis and in osteoporosis. J Bone Miner Metab 2007; 25:219 - 25; http://dx.doi.org/10.1007/s00774-007-0753-0; PMID: 17593491
- D'Amelio P, Roato I, D'Amico L, Veneziano L, Suman E, Sassi F, et al. Bone and bone marrow pro-osteoclastogenic cytokines are up-regulated in osteoporosis fragility fractures. Osteoporos Int 2011; 22:2869 - 77; http://dx.doi.org/10.1007/s00198-010-1496-7; PMID: 21116815
- Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet 2007; 23:413 - 8; http://dx.doi.org/10.1016/j.tig.2007.05.008; PMID: 17559965
- Fraga MF. Genetic and epigenetic regulation of aging. Curr Opin Immunol 2009; 21:446 - 53; http://dx.doi.org/10.1016/j.coi.2009.04.003; PMID: 19500963
- Penolazzi L, Lambertini E, Giordano S, Sollazzo V, Traina G, del Senno L, et al. Methylation analysis of the promoter F of estrogen receptor alpha gene: effects on the level of transcription on human osteoblastic cells. J Steroid Biochem Mol Biol 2004; 91:1 - 9; http://dx.doi.org/10.1016/j.jsbmb.2004.02.005; PMID: 15261302
- Kang MI, Kim HS, Jung YC, Kim YH, Hong SJ, Kim MK, et al. Transitional CpG methylation between promoters and retroelements of tissue-specific genes during human mesenchymal cell differentiation. J Cell Biochem 2007; 102:224 - 39; http://dx.doi.org/10.1002/jcb.21291; PMID: 17352407
- Fu Q, Manolagas SC, O'Brien CA. Parathyroid hormone controls receptor activator of NF-kappaB ligand gene expression via a distant transcriptional enhancer. Mol Cell Biol 2006; 26:6453 - 68; http://dx.doi.org/10.1128/MCB.00356-06; PMID: 16914731
- McAllister RM, Gardner MB, Greene AE, Bradt C, Nichols WW, Landing BH. Cultivation in vitro of cells derived from a human osteosarcoma. Cancer 1971; 27:397 - 402; http://dx.doi.org/10.1002/1097-0142(197102)27:2<397::AID-CNCR2820270224>3.0.CO;2-X; PMID: 5100401
- Heremans H, Billiau A, Cassiman JJ, Mulier JC, de Somer P. In vitro cultivation of human tumor tissues. II. Morphological and virological characterization of three cell lines. Oncology 1978; 35:246 - 52; http://dx.doi.org/10.1159/000225298; PMID: 218153
- Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 1977; 36:59 - 74; http://dx.doi.org/10.1099/0022-1317-36-1-59; PMID: 886304
- Jonsson KB, Frost A, Nilsson O, Ljunghall S, Ljunggren O. Three isolation techniques for primary culture of human osteoblast-like cells: a comparison. Acta Orthop Scand 1999; 70:365 - 73; http://dx.doi.org/10.3109/17453679908997826; PMID: 10569267
- Sasaki M, Anast J, Bassett W, Kawakami T, Sakuragi N, Dahiya R. Bisulfite conversion-specific and methylation-specific PCR: a sensitive technique for accurate evaluation of CpG methylation. Biochem Biophys Res Commun 2003; 309:305 - 9; http://dx.doi.org/10.1016/j.bbrc.2003.08.005; PMID: 12951050
- Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc 2007; 2:2265 - 75; http://dx.doi.org/10.1038/nprot.2007.314; PMID: 17853883
- Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques 2003; 35:146 - 50; PMID: 12866414