Abstract
Maternal exposure to environmental agents throughout pregnancy and lactation may affect offspring’s mammary gland growth and alter the epigenome. This may predispose the offspring’s mammary glands to be more susceptible to carcinogenesis. The purpose of this study was to examine the effect of a maternal high-fat diet on the regulation of p16INK4a gene expression in the mammary gland of rat offspring. Timed-pregnant Sprague-Dawley rats were fed one of the two diets, a control (C, 16% of fat) or a high fat (HF, 45% of fat) diet, throughout gestation and lactation and sacrificed at 12 weeks of age. Compared with C, HF offspring showed a decrease of p16INK4a gene expression in the mammary gland at both mRNA and protein levels. Chromatin immunoprecipitation (ChIP) assay demonstrated that the downregulation of p16INK4a transcription in HF offspring was associated with reduced acetylation of histone H4 and increased recruitment of histone deacetylase 3 (HDAC3) within the p16INK4a promoter region, but was not associated with acetylation of histone H3 or HDAC1. Methylated DNA immunoprecipitation (MeDIP) did not detect differences in methylation at different regions of the p16INK4a gene between C and HF offspring. We conclude that maternal high fat exposure represses p16INK4a gene expression in the mammary gland of offspring through changes of histone modifications and HDAC3 binding activity within the regulatory regions of the p16INK4a gene.
Introduction
Gestation and lactation are critical periods for the development of offspring's mammary glands, since mammary buds and primitive mammary epithelial trees form during the fetal period.Citation1,Citation2 Exposure to certain diets during pregnancy and lactation can increase the risk of breast cancer in the next generation.Citation2–Citation7 Hilakivi-Clarke et al.Citation2 found that maternal exposure to a HF diet caused early puberty onset and it was associated with breast cancer incidence in offspring rats by raising 17β-estradiol (E2) levels in the dams. This finding was supported by another animal study,Citation6 which reported the deleterious effect of a diet with a high percentage of corn oil on the mammary gland in female offspring. However, the underlying mechanisms for these observations remain elusive. On this study, we aim to determine the effects of a maternal high fat diet on developmental transcriptional control.
Epigenetic programming of disease development by the maternal diet has been demonstrated by many studies.Citation8–Citation10 For example, the association between low birth weight and increased risk of developing diabetes, coronary heart disease and stroke is attributed to the epigenetic reprogramming of the genetic code during fetal development.Citation11–Citation13 Epigenetic alterations mainly include DNA methylation and histone modifications. The impact of DNA methylation on transcriptional regulation was verified by the finding that maternal methyl supplements in mice caused phenotypic change.Citation14 Dunn et al. reported that a maternal HF diet gave rise to increased body weight and reduced insulin sensitivity in offspring mice, and these changes were associated with decreased DNA methylation at the promoter of the GH secretagogue receptor (GHSR) gene. Histone modifications, especially acetylation of histones, play a critical role in the regulation of gene expression. Transcription factor CCAAT/enhancer-binding protein (C/EBPβ) was upregulated by increased acetylation of histone H3 and acetylated histone H4 in offspring female rats exposed to a maternal low protein diet.Citation15 Gene expression profiles under multiple conditions have revealed the importance of histone acetylation in altering the structure of the epigenome. However, the regulation of histone acetylation is complex, involving multiple histone acetyltransferases (HATs) and histone deacetylases (HDACs).
INK4a/ARF,Citation16 encoded by the tumor suppressor gene p16INK4a, inhibits cyclin-dependent kinases 4 and 6 by binding to both cyclin/cdk4 and cyclin/cdk6 kinase complexes, therefore blocking cell cycle at G1/S.Citation16 The expression of p16INK4a during mammary gland development was shown to be independent of DNA methylation.Citation17 Similarly, in our previous study in reference Citation3, we reported that histone modifications, rather than DNA methylation, downregulated the expression of p16INK4a in the mammary glands of offspring exposed to protein-restricted maternal diets. Histone deacetylase 1 (HDAC1) and HDAC3 have been found to function in the regulation of p16INK4a in vitro. Therefore, in the current study, we focused on the recruitment of these HDACs to the p16INK4a gene promoter.
Our study aimed to test the hypothesis that p16INK4a expression is regulated by epigenetic alterations in offspring rats exposed to a maternal HF diet. Compared with the gestational and lactational C diet, p16INK4a was significantly lower in offspring exposed to a maternal HF diet. Additionally, histone modifications, rather than promoter DNA methylation, corresponded with the altered p16INK4a gene expression, which further supports the role of HDACs in the regulation of p16INK4a gene transcription, especially in response to diet.
Results
Maternal and offspring observations.
Pregnant dams were monitored from day 2 of gestation. and B show mothers' body weight and food intake during the gestation period. There were no differences in the daily food intake or body weight gained between the C and HF dam groups (p > 0.05), indicating that experimental diet type did not influence dams' food intake or their body weight gained during gestation, thus only maternal diet should be considered as the contributing factor for the changes observed in the offspring. At birth, the female HF pups weighed less (p < 0.05) than C pups; however, the average weight of the HF offspring after weaning did not differ compared with C ().
Maternal HF diet repressed p16INK4a gene expression in offspring mammary glands.
The effect of a maternal high-fat diet on p16INK4a gene expression in offspring mammary gland was examined by real-time qPCR. shows the p16INK4a mRNA expression in C offspring rats vs. HF offspring. The mRNA expression of the p16INK4a gene was significantly decreased (p = 0.0004) by 78% in the mammary glands of HF pups when compared with C ().
Maternal HF diet decreased p16INK4a protein content in offspring mammary glands.
To examine whether the altered p16INK4a mRNA resulted in a change in protein content, p16INK4a protein content was measured in offspring mammary gland. p16INK4a protein was significantly decreased (p < 0.05) by 75% in the mammary glands of HF pups when compared with C (), which was consistent with mRNA expression.
Maternal HF diet increased the S-phase in offspring mammary glands.
To investigate whether decreased p16INK4a was associated with changes in the cell cycle regulation, flow cytometry assay was performed. Mammary cells arrested in S-phase were increased from 3.0% in the C group to 5.8% (p < 0.05) in the HF group, which corresponds to the downregulation of p16INK4a in the HF group when compared with C group (). G0/G1 or G2 phase were not significantly different between the two groups ().
Maternal HF diet affected histone acetylation, not methylation of p16INK4a in offspring mammary glands.
To determine whether the pattern of DNA methylation at the p16INK4a promoter corresponded to the gene's mRNA expression, its methylation status was assessed by MeDIP assay. Primer sets spanning −3,000 to +9,000 bp and targeting CpG islands 1 to 4 were designed to include all of potential regulatory regions associated with p16INK4a. represents the position of the p16INK4a gene promoter and its CpG islands on the intact genomic sequence. CpG methylation within the mammary glands was not significantly different between C and HF offspring in any of the regions tested (), indicating that the reduced p16INK4a mRNA expression in HF offspring was DNA methylation-independent.
Changes in chromatin structure caused by chemical modifications of histone proteins, such as methylation and acetylation, regulate gene transcription by affecting the action of eukaryotic RNA polymerase II.Citation18,Citation19 To determine whether the altered p16INK4a transcription was associated with changes of chromatin structure at the p16INK4a promoter or other regulatory regions, antibodies to either methylated or acetylated histones were utilized in the ChIP assay. A 38% decrease in acetylated histone H4 (H4Ac) (p < 0.05) at the p16INK4a promoter and an 84% decrease of H4Ac (p < 0.05) at the CpG-rich sites were observed in the mammary glands of HF offspring when compared with C ( and C), which correlates to p16INK4a transcriptional repression. No significant changes in histone modifications were detected within the p16INK4a gene body (+5 kb) between C and HF offspring (). These results suggest that the altered p16INK4a gene transcription is associated with histone modifications, specifically histone H4 acetylation.
Maternal HF diet increased the binding of the transcription factor HDAC3 within the p16INK4a promoter in offspring mammary glands.
To further investigate the mechanisms that contributed to the downregulated p16INK4a expression in terms of chromatin structure, the recruitment of histone deacetylase 1 and 3 (HDAC1 and HDAC3), which are known to act as transcriptional suppressors by affecting sp1 binding to the GC-rich binding sequence at the p16INK4a promoter,Citation20,Citation21 were tested by ChIP assay. Protein levels of HDAC3 () but not HDAC1 () were significantly higher (p < 0.05) in HF offspring when compared with C. Additionally, HDAC3 (), but not HDAC1 (), was recruited to the p16INK4a promoter in response to the maternal HF diet when HDAC levels were compared with IgG. The increased HDAC3 interaction within the p16INK4a promoter in HF offspring was 2.8 times higher than in C (p < 0.05). These data suggest that the decreased acetylation of histone H4 in mammary glands of HF offspring might be due to the recruitment of transcription factor HDCA3 within the p16INK4a promoter, which acts to downregulate gene transcription. Thus, HDAC3 may have an important role in modulating H4 acetylation to repress p16INK4a gene transcription.
Discussion
There is growing evidence that the maternal diet during gestation and lactation may program disease susceptibility in adulthood by altering the epigenetic state of the fetal genome. Prenatal and postnatal dietary modifications can induce persistent changes in offspring mammary gland that in turn impact breast cancer risk later in life.Citation22 These changes likely reflect epigenetic modulations, such as changes in DNA methylation and histone modifications that then affect gene transcription. Pre-pubertal exposure to a HF diet has been reported to upregulate genes that induce mammary cell proliferation and to downregulate genes that repair DNA damage and induce apoptosis, correlating to the elevated risk of breast cancer in adulthood.Citation23 In the current study, p16INK4a was significantly lower in offspring exposed to a maternal HF diet. Additionally, histone modifications, rather than promoter DNA methylation, corresponded with the altered p16INK4a gene expression. The downregulated p16INK4a gene in HF offspring also likely reflects the cell cycle state of the mammary cells.
In order to investigate the regulatory mechanism behind the decreased p16INK4a gene transcription, we examined the changes in DNA methylation and histone modifications in mammary glands of offspring in response to a maternal HF diet. Although the significance of hypermethylation within the p16INK4a gene promoter in tumorigenesis has been repeatedly shown in some studies in reference Citation24, little is known about its role in p16INK4a gene regulation in normal mammary tissues. Most mammalian methylation occurs at the cytosines of CpG islands, which are characterized by CpG dinucleotides, typically concentrated in the promoter regions and exons of target genes. We specifically tested all four CpG islands within the p16INK4a gene body, as well as its promoter and a distant up-stream region. However, there were no changes in DNA methylation at any of those regions, suggesting a DNA methylation independent regulation of p16INK4a expression in response to a maternal HF diet. This data was consistent with our previous report that the suppressed p16INK4a expression is DNA methylation independent in response to a maternal low protein diet.
Maternal hormone levels during pregnancy, such as estrogen and progesterone, have been proposed to play a role in increasing the risk of developing breast cancer both in mothers and offspring. However, in the present study the fat source is lard, which is not expected to induced maternal E2 levels during pregnancy.Citation25
We addressed the biological role of a maternal HF diet in inducing the recruitment of HDAC3 to the p16INK4a gene in rat mammary gland. Acetylation of the N-terminal tails of chromatin core histone proteins H3 and H4 loosens the compact conformation of the chromatic, possibly by disrupting the interactions between adjacent nucleosomes or by loosening contacts between histones and DNA. Some studies have investigated the various HDACs on the p16INK4a gene regulation in vivo, but only a few of them have been reported to repress the p16INK4a gene promoter activity.Citation20 In this study, we looked into HDAC1 and HDAC3, which have been shown to have significant effects on the p16INK4a gene promoter by other researchers. On the other hand, a few reports have shown that HDAC inhibitors can induce p16INK4a expression.Citation26,Citation27 HDAC inhibitors (HDACi) induce cell growth arrest, differentiation and apoptosis and play crucial roles in a wide range of biological processes, mainly through their repressive influence on transcription, and thus, become very influential and powerful targets for many diseases.Citation28 Unlike HDAC1, HDAC3 is required for cell growth and is involved in transcriptional regulation of genes important for cell cycle progression and development. HDAC3 has been implicated to play roles in governing cell proliferation via the inhibition of cell cycle genes.Citation29 In our rat model, we observed increased binding of HDAC3 to the p16INK4a promoter in the mammary gland of offspring exposed to a maternal HF diet. A similar phenomenon was also observed in a recent study in which Feng et al. verified that HDAC3 was recruited to the p16INK4a promoter by the transcription factor ZBP-89 and suppressed p16INK4a expression by histone acetylation modification.Citation20 Deletion of HDAC3 impaired DNA repair and greatly reduced chromatin compaction and heterochromatin contentCitation30 and it has been suggested that HDAC3 acts in the S phase of the cell cycle.Citation30 Murine embryonic fibroblasts (MEFs) required HDAC3 for cell viability, and the observed apoptosis upon deletion of HDAC3 was associated with an impaired S phase progression and DNA double-strand breaks.Citation31 It has been reported that HDAC1 could be recruited by lymphoid specific helicase in human fibroblasts and interact with the p16INK4a promoter to cause p16INK4a repression by the deacetylation of histone H3 at the p16 promoter.Citation21 However, we did not observe significant changes in HDAC1 binding within the p16INK4a gene in response to maternal diet. Thus, these recent reports and our data suggest that specificity of histone H3 and histone H4 acetylation status at the p16INK4a promoter region may depend on specific histone deacetylase activities resulting in the increased S phase arrest in this study.
Overall, the current study demonstrates for the first time that maternal exposure to a high-fat diet could decrease the expression of the cell cycle gene p16INK4a in offspring mammary gland through HDAC3-related histone acetylation. Because p16INK4a is closely related to cell cycle control, these changes are potentially critical for predisposing offspring toward disease development in adulthood.
Materials and Methods
Animals and treatment.
Timed-pregnant Sprague Dawley rats (Charles River Laboratories) were obtained on day 2 of gestation and randomly assigned into one of two pelleted diets (), C (16 kcal%) or HF (45 kcal%). Each group contained five dams and both groups had free access to food and drinking water. Animals were individually housed in standard polycarbonate cages in a temperature and humidity controlled room on a 12 h light-dark cycle. Twenty-four hours after birth, ten pups (five female and five male) were selected and used in the litters to minimize variation in pups' nutritional status during suckling. Pups body weights were measured once a week. Mothers were kept on the same diets, C or HF throughout lactation. On postnatal day 21, all pups were weaned from the mother to a C diet. Female pups were sacrificed when they were 12 weeks old and the fourth abdominal mammary gland was collected, snap-frozen in liquid nitrogen, and stored at −70°C. Mammary samples from five to six female pups in each treatment group were randomly chosen for all experiments.
Real-time quantitative RT-PCR (qPCR).
Frozen tissue samples from six offspring in each treatment group were individually ground with a mortar and pestle in liquid nitrogen (around 100 mg of mammary tissue per animal), and total RNA was isolated from each sample with TRI reagent (Sigma, St. Louis, MO) following the manufacturer's instructions. A high-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) was used for reverse transcription of 2 µg of total RNA. cDNA was then detected with SYBR Green fast master mix (2x, Quanta BioSciences) by 7300 real-time PCR system (Applied Biosystems). The reaction mixtures were activated at 95°C for 10 min, followed by 35 cycles of 95°C for 15 sec and 60°C for 1 min. mRNA was quantified by comparing to a serially diluted cDNA standard created from the same RNA isolation, and ribosomal protein L7a was used as the internal control. After PCR, a dissociation curve was generated through a stepwise increase of the temperature from 55°C to 95°C to ensure that a unique product was amplified. Primers for the qPCR were designed using Vector NTI software (InforMax Inc., Frederick, MD) to amplify the transcriptional region of the p16INK4a gene ().
Immunoblotting.
Protein samples were isolated from offspring mammary tissue according to a published protocol.Citation32 For immunoblotting, total 30 µg of proteins were size-fractionated on a Bio-Rad Precast Tris-HCl polyacrylamide gel and transferred at 14 V onto a PVDF membrane (Bio-Rad, Hercules, CA) at 4°C. Incubation with primary and secondary antibodies was done according to manufacturers' instructions. Briefly, PVDF membranes were incubated with blocking solution containing 10% (w/v) nonfat dry milk, 20 mmol/L Tris-HCl, pH 7.6, 137 mmol/L NaCl and 0.1% (v/v) Tween 20 for 1 h at room temperature. Primary antibodies were diluted in 10% fat-free milk blocking solution following manufacturers' instructions. After incubation with primary antibodies for 3 h at room temperature, the membranes were subsequently washed with blocking solution containing 5% (w/v) nonfat milk 5 times for 5 min each time. Goat anti-rabbit or goat anti-mouse HRP-conjugated secondary antibody was diluted 1:10,000 in blocking solution containing 5% (w/v) nonfat dry milk and incubated for 1 h at room temperature. Five 5 min washes in blocking solution containing 1% (w/v) nonfat dry milk were performed before the membranes were exposed to the enhanced chemiluminescence reagent SuperSignal West Dura (Pierce). Signals were detected and quantified using ChemiDoc XRS imaging system (Bio-Rad). Antibodies used in the present study were purchased from Santa Cruz: p16 (M-156) (sc-1207), HDAC1 (sc-7872), HDAC3 (sc-11417), actin (I-19) (sc-1616).
Isolation of genomic DNA from mammary gland.
Frozen mammary gland tissues from five offspring in each treatment group were ground in liquid nitrogen (around 50 mg of mammary tissue per animal). Homogenized mammary tissue was resuspended in 500 µL of TE. Lysis buffer (500 µL/sample; 20 mM Tris-HCl, pH = 8.0, 4 mM EDTA, 20 mM NaCl, 1% SDS) containing 10 µL proteinase K (20 mg/mL stock) was then added to the samples and incubated in a 55°C water bath for 5 h. Samples were then extracted with 600 µL of phenol and 600 µL of chloroform, respectively. The aqueous phase was collected to precipitate DNA by adding 1.2 mL ethanol containing 75 mM sodium acetate (pH = 5.2) at −20°C overnight. The next day, precipitated DNA was centrifuged at 16,000x g for 20 min at 4°C. After discarding the supernatant, the DNA pellet was washed with 70% ethanol. The pellet was air-dried, suspended with 100 µL of TE containing RNase A (20 ug/mL), incubated in a 37°C water bath for 30 min, and then stored at 4°C.
Cell cycle analysis by flow cytometry.
Frozen mammary gland tissues from three offspring from each treatment group were ground in liquid nitrogen (around 50 mg per animal). Ground powder was suspended in 1 mL suspension solution (PBS with of 5% fetal bovine serum) and further homogenized by a glass Teflon tissue grinder. Homogenized tissues were spun down at 100 rpm for 2 min. Supernatant were transferred to a fresh tube and spun down again at 100 rpm for 2 min. Then, 5 mL of suspension solution was mixed with supernatant and centrifuged at 800 rpm for 5 min. This step was repeated again to wash the cells and the pellet was collected and resuspended in 0.5 mL PBS. To fix the cells, 0.5 mL 100% cold ethanol was added to each sample and frozen at −20°C overnight. The next day, after centrifugation at 1,000 rpm for 5–7 min, ethanol was decanted and 0.5 mL of PBS containing Propidium Iodide (PI) and RNase A (final concentrations: 50 µg/mL PI + 100 µg/mL RNase A) was added to the pellet and mixed well. Samples were incubated at room temperature in the dark for a minimum of 20 min before analysis by flow cytometry. Cell cycle data was analyzed by FCS 3.0 software.
Methylated DNA immunoprecipitation (MeDIP).
MeDIP analysis was performed according to a modified protocol.Citation33 Genomic DNA (20 ug) in 300 µL of TE was sonicated 5 times for 10 sec each time using 20% power with the tube cooling in ice water. The sample was allowed to cool down after each pulse for 1 min. Sonicated product (5–10 µL) was loaded on an agarose gel to check the size of the DNA (mean size is within 300−1,000 bp). The sonicated DNA was immunoprecipitated with a monoclonal antibody against 5-Methy Cytidine (5mC; Abcam 10805, Cambridge, MA). A normal rabbit IgG antibody was used as the negative control to demonstrate non-specific binding. A portion of the sonicated DNA (30 µL) was left untreated to serve as input control. Briefly, the sonicated DNA (2 ug) was diluted in 450 µL TE, denatured for 10 min in boiling water and immediately cooled on ice for 10 min. The diluted sample was subsequently incubated overnight with 51 µL of 10x IP buffer (100 mM Na-Phosphate, pH = 7.0, 1.4 M NaCl, 0.5% Triton X-100) and 2 µg of 5mC or normal rabbit IgG antibody at 4°C with shaking. PureProteome Protein A Magnetic Beads (40 µL/sample; Millipore, Temecula, CA) were pre-washed twice with 800 µL of 0.1% BSA/PBS for 5 min at room temperature with shaking to avoid sedimentation of the beads. The beads were trapped on the wall of the tube using a magnetic rack for discarding supernatant. The beads were resuspended in 40 µL of 1x IP buffer and then added to the samples. The samples were incubated 2 h at 4°C with overhead shaking. The beads were washed 3 times with 700 µL 1x IP buffer for 10 min at RT with shaking. After trapping the beads on the magnetic rack, the beads were resuspended in 250 µL digestion buffer (50 mM Tris-HCl, pH = 8.0, 10 mM EDTA, 0.5% SDS) containing 7 µL proteinase K (10 mg/mL stock), and incubated in a 50°C water bath for 3 h. Samples were then extracted with 250 µL of phenol and 250 µL of chloroform. The aqueous phase was collected to precipitate DNA overnight by adding 500 µL ethanol containing 400 mM NaCl and 1 µL of glycogen (2 mg/mL stock) at −20°C. The next day, precipitated DNA was centrifuged at 16,000 g for 20 min at 4°C. After discarding the supernatant, the DNA pellet was washed with 70% ethanol, airdried, resuspended in 50 µL TE and stored at 4°C. The amount of 40 ug of input DNA and 2 µL of MeDIP DNA were used for real time PCR reaction. The methylation levels were expressed as ratio to the input.
Chromatin immunoprecipitation (ChIP).
ChIP analysis was performed according to a modified protocol.Citation15 Briefly, 200 mg of frozen tissue was ground in liquid nitrogen, re-suspended in PBS and cross-linked in 1% formaldehyde for 10 min at room temperature. The tissue pellet was re-suspended in nuclei swelling buffer containing protease inhibitor. The separated nuclei were lysed in SDS lysis buffer containing protease inhibitors. The resulting chromatin was sonicated (Fisher Scientific model 100 Sonic Dismembrator, Pittsburgh, PA) on ice with 8 bursts for 40 sec at power setting 5, with a 2 min cooling interval between each burst. The average length of sonicated chromatin was determined by resolving it on a 1.6% agarose gel, and was found to be around 200–500 bp. The sample was then centrifuged at 13,000 rpm for 10 min at 4°C to remove cell debris from the crude chromatin lysate. One milliliter of sheared chromatin was diluted in 10 mL of ChIP dilution buffer. Ten percent of the diluted lysate was subsequently incubated overnight on a hematology mixer (model 346, Fisher Scientific) with 2 µg of primary antibodies at 4°C. Normal rabbit IgG antibody was used as the negative control to demonstrate non-specific binding. Antibodies were considered negative for binding if the resulting value was equal to or less than the IgG value (ratio to input). Pre-blocked salmon sperm DNA/protein G agarose beads (60 µL, 50% slurry; Upstate Biotechnology, Lake Placid, NY) were then added to the chromatin for 2 h, followed by centrifugation at 2000 rpm for 1 min at 4°C. The supernatant of normal rabbit IgG was saved as the input control for PCR after clean-up. The pellets containing immunoprecipitated complexes were washed sequentially with salt solutions and twice with TE (pH 8.0). Antibody/protein/DNA complexes were eluted from Protein A agarose beads by adding 250 µL of the elution buffer (50 mmol/L NaHCO3 and 1% SDS) two times, followed by 15 min of shaking at 37°C and finally by flash spinning the samples down at room temperature. The combined supernatants were incubated at 65°C for 4–5 h, after the addition of 20 µL of 5 mol/L NaCl and 1 µg of RNase A to reverse the formaldehyde cross-linking and release the DNA fragments. Samples were then treated with proteinase K at 37°C for 1 h to remove any protein, and DNA was purified with a QiaPrep miniprep kit (Qiagen). Each real time PCR reaction used 5% of immunoprecipitated DNA. The modified histones binding were expressed as ratios to the input. Standards and the samples were simultaneously amplified in a 20 µL reaction volume, and primers were designed to amplify genomic sequences at the promoter region of p16INK4a (see ).
Statistical analysis.
Results are expressed as the mean ± SEM. Growth curve and food intake were analyzed using a repeated-measures ANOVA using the Proc Mixed procedure (SAS 9.1; SAS Institute Inc.). Comparison of mRNA expression and protein levels between OR and OP groups were performed by one-way ANOVA (SAS v. 9.1.2). A p < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Abbreviations
| HDAC | = | histone deacetylase |
| H3Ac | = | acetylated histone H3 |
| H4Ac | = | acetylated histone H4 |
| ChIP | = | chromatin immunoprecipitation |
| H3K4me2 | = | di-methylated histone H3 at lysine 4 residues |
| HF | = | high fat |
| H3K9me3 | = | tri-methylated histone H3 at lysine 9 residues |
| MeDIP | = | methylated DNA immunoprecipitation |
Figures and Tables
Figure 1 Maternal food intake and offspring body weight. (A) Gestational growth curve for (C) and high-fat (HF) fed dams. (B) Food intake for (C) and high-fat (HF) fed dams. (C) Growth curve after weaning for offspring of (C) and high-fat (HF) fed dams. The values are presented as the mean ± SEM.
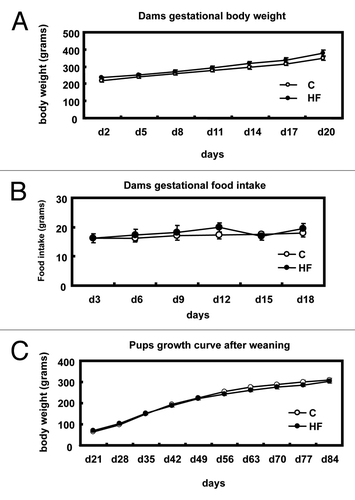
Figure 2 p16INK4a mRNA and protein levels in offspring mammary glands. (A) Expression of p16INK4a mRNA in mammary glands of offspring of control (C) and high-fat (HF) fed dams (n = 6) presented as the ratio to L7a housekeeping gene. The values are presented as the mean ± SEM; *p < 0.05 when compared with C group. (B) Expression of protein in mammary glands of offspring of control (C) and high-fat (HF) fed dams (n = 4). The right part is a representative image of p16INK4a protein as measured by immunoblotting, and values in the left part are presented as the mean ± SEM *p < 0.05 when compared with C group.
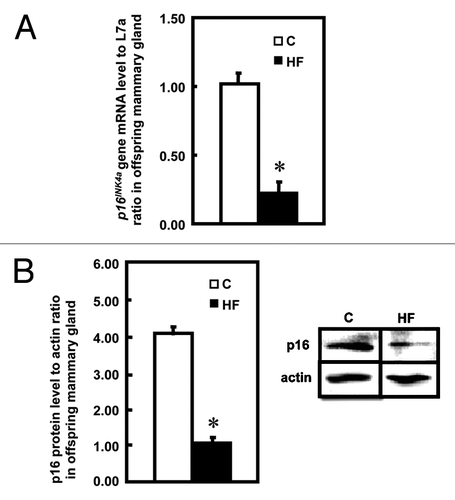
Figure 3 Cell cycle analysis in offspring mammary glands. Cell cycle analysis was performed in cells from mammary glands of offspring of control (C) and high-fat (HF) fed dams using flow cytometry (n = 3). Data are shown as the percentages of total cells corresponding to G0/G1, S and G2/M phases. The values are presented as the relative mean ± SEM, *p < 0.05 when compared with C group.
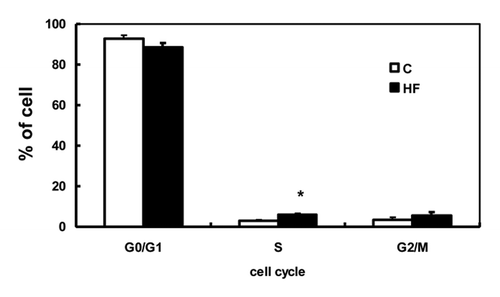
Figure 4 DNA methylation at the p16INK4a gene in offspring mammary glands. (A) CpG islands at the rat p16INK4a gene. (B) Relative methylation levels relative to input corresponding to the negative 3 kb region, promoter region and CpG islands 1 to 4 of the p16INK4a gene in mammary glands of offspring of control (C) and high-fat (HF) fed dams (n = 5). (C) Non-specific IgG levels relative to input corresponding to the negative 3 kb region, promoter region and CpG islands 1 to 4 of the p16INK4a gene in mammary glands of offspring of control (C) and high-fat (HF) fed dams (n = 5). The values presented as the mean ± SEM.
Figure 5 Histone modifications within the p16INK4a gene in offspring mammary glands. (A) Histone modifications at the p16INK4a promoter region in mammary glands of offspring of control (C) and high-fat (HF) fed dams (n = 5). (B) Histone modifications at the p16INK4a gene body (around + 5 kb region) in mammary glands of offspring of control (C) and high-fat (HF) fed dams (n = 5). (C) Histone modifications at CpG-rich regions on the p16INK4a gene in mammary glands of offspring of control (C) and high-fat (HF) fed dams (n = 5). Data are shown as a ratio to the input DNA. H4Ac: acetylated histone 4; H3Ac: acetylated histone 3; H3K4me2: di-methylated histone 3 at lysine 4 residues; H3K9me3: tri-methylated histone 3 at lysine 9 residues. The values are presented as the relative mean ± SEM, *p < 0.05 when compared with C group for each modification.
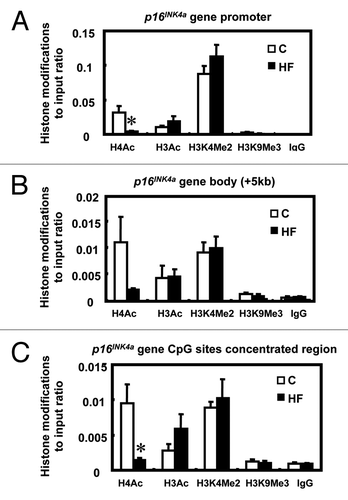
Figure 6 The binding of HDCA3 and HDAC1 within the p16INK4a promoter. (A and B) Expression of HDAC3 and HDAC1 protein levels in mammary glands of offspring of control (C) and high-fat (HF) fed dams (n = 4). The lower parts are a representative image of HDAC3 (left) and HDAC1 (right) protein as measured by immunoblotting. (C) Recruitment of HDAC3 at the p16INK4a promoter in mammary glands of offspring of control (C) and high-fat (HF) fed dams (n = 5). (D) Recruitment of HDAC1 at the p16INK4a promoter in mammary glands of offspring of control (C) and high-fat (HF) fed dams (n = 5). The values present the mean ± SEM, *p < 0.05.
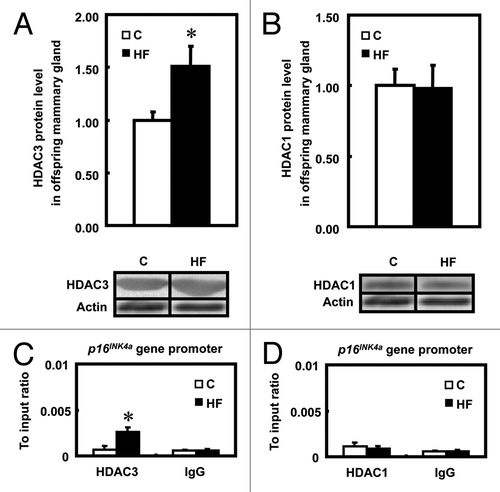
Table 1 Maternal diet composition
Table 2 p16 primers (ENSRNOT00000066011)
Funding
This project was supported, in whole or in part, by the USDA Cooperative State Research, Education and Extension Service, Hatch project number # ILLU-698-374 and by National Institutes of Health Grants CA-139557 (to Y.X.P.).
References
- Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol 2005; 6:715 - 725; PMID: 16231422; http://dx.doi.org/10.1038/nrm1714
- Hilakivi-Clarke L, Clarke R, Onojafe I, Raygada M, Cho E, Lippman M. A maternal diet high in n - 6 polyunsaturated fats alters mammary gland development, puberty onset and breast cancer risk among female rat offspring. Proc Natl Acad Sci USA 1997; 94:9372 - 9377; PMID: 9256489; http://dx.doi.org/10.1073/pnas.94.17.9372
- Zheng S, Pan YX. Histone modifications, not DNA methylation, cause transcriptional repression of p16 (CDKN2A) in the mammary glands of offspring of protein-restricted rats. J Nutr Biochem 2010; 22:567 - 573; PMID: 20934317
- Hilakivi-Clarke L. Nutritional modulation of terminal end buds: its relevance to breast cancer prevention. Curr Cancer Drug Targets 2007; 7:465 - 474; PMID: 17691906; http://dx.doi.org/10.2174/156800907781386641
- Walker BE. Tumors in female offspring of control and diethylstilbestrol-exposed mice fed high-fat diets. J Natl Cancer Inst 1990; 82:50 - 54; PMID: 2293656; http://dx.doi.org/10.1093/jnci/82.1.50
- Stark AH, Kossoy G, Zusman I, Yarden G, Madar Z. Olive oil consumption during pregnancy and lactation in rats influences mammary cancer development in female offspring. Nutr Cancer 2003; 46:59 - 65; PMID: 12925305; http://dx.doi.org/10.1207/S15327914NC4601_08
- Fernandez-Twinn DS, Ekizoglou S, Gusterson BA, Luan J, Ozanne SE. Compensatory mammary growth following protein restriction during pregnancy and lactation increases early-onset mammary tumor incidence in rats. Carcinogenesis 2007; 28:545 - 552; PMID: 16952910; http://dx.doi.org/10.1093/carcin/bgl166
- Dunn GA, Bale TL. Maternal high-fat diet promotes body length increases and insulin insensitivity in second-generation mice. Endocrinology 2009; 150:4999 - 5009; PMID: 19819967; http://dx.doi.org/10.1210/en.2009-0500
- Zeisel SH. Epigenetic mechanisms for nutrition determinants of later health outcomes. Am J Clin Nutr 2009; 89:1488 - 1493; PMID: 19261726; http://dx.doi.org/10.3945/ajcn.2009.27113B
- Fernandez-Twinn DS, Ozanne SE. Early life nutrition and metabolic programming. Ann NY Acad Sci 2010; 1212:78 - 96; PMID: 21070247; http://dx.doi.org/10.1111/j.1749-6632.2010.05798.x
- Gluckman PD, Hanson MA. Living with the past: evolution, development and patterns of disease. Science 2004; 305:1733 - 1736; PMID: 15375258; http://dx.doi.org/10.1126/science.1095292
- Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction 2004; 127:643 - 651; PMID: 15175501; http://dx.doi.org/10.1530/rep.1.00221
- Morley R. Fetal origins of adult disease. Semin Fetal Neonatal Med 2006; 11:73 - 78; PMID: 16368278; http://dx.doi.org/10.1016/j.siny.2005.11.001
- Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr 2002; 132:2393 - 2400; PMID: 12163699
- Zheng S, Rollet M, Pan YX. Maternal protein restriction during pregnancy induces CCAAT/enhancer-binding protein (C/EBPβ) expression through the regulation of histone modification at its promoter region in female offspring rat skeletal muscle. Epigenetics 2011; 6:161 - 170; PMID: 20930553; http://dx.doi.org/10.4161/epi.6.2.13472
- Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 1998; 395:237 - 243; PMID: 9751050; http://dx.doi.org/10.1038/26155
- Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res 2001; 264:42 - 55; PMID: 11237522; http://dx.doi.org/10.1006/excr.2000.5149
- Conaway RC, Conaway JW. General transcription factors for RNA polymerase II. Prog Nucleic Acid Res Mol Biol 1997; 56:327 - 346; PMID: 9187058; http://dx.doi.org/10.1016/S0079-6603(08)61009-0
- Roeder RG, Rutter WJ. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature 1969; 224:234 - 237; PMID: 5344598; http://dx.doi.org/10.1038/224234a0
- Feng Y, Wang X, Xu L, Pan H, Zhu S, Liang Q, et al. The transcription factor ZBP-89 suppresses p16 expression through a histone modification mechanism to affect cell senescence. FEBS J 2009; 276:4197 - 4206; PMID: 19583777; http://dx.doi.org/10.1111/j.1742-4658.2009.07128.x
- Zhou R, Han L, Li G, Tong T. Senescence delay and repression of p16INK4a by Lsh via recruitment of histone deacetylases in human diploid fibroblasts. Nucleic Acids Res 2009; 37:5183 - 5196; PMID: 19561196; http://dx.doi.org/10.1093/nar/gkp533
- de Assis S, Warri A, Cruz MI, Hilakivi-Clarke L. Changes in mammary gland morphology and breast cancer risk in rats. J Vis Exp 2010; 16:2260; PMID: 20972418
- Olivo-Marston SE, Zhu Y, Lee RY, Cabanes A, Khan G, Zwart A, et al. Gene signaling pathways mediating the opposite effects of prepubertal low-fat and high-fat n-3 polyunsaturated fatty acid diets on mammary cancer risk. Cancer Prev Res 2008; 1:532 - 545; PMID: 19139003; http://dx.doi.org/10.1158/1940-6207.CAPR-08-0030
- Auerkari EI. Methylation of tumor suppressor genes p16(INK4a), p27(Kip1) and E-cadherin in carcinogenesis. Oral Oncol 2006; 42:5 - 13; PMID: 15978859; http://dx.doi.org/10.1016/j.oraloncology.2005.03.016
- de Assis S, Khan G, Hilakivi-Clarke L. High birth weight increases mammary tumorigenesis in rats. Int J Cancer 2006; 119:1537 - 1546; PMID: 16646052; http://dx.doi.org/10.1002/ijc.21936
- Munro J, Barr NI, Ireland H, Morrison V, Parkinson EK. Histone deacetylase inhibitors induce a senescence-like state in human cells by a p16-dependent mechanism that is independent of a mitotic clock. Exp Cell Res 2004; 295:525 - 538; PMID: 15093749; http://dx.doi.org/10.1016/j.yexcr.2004.01.017
- Wu LP, Wang X, Li L, Zhao Y, Lu S, Yu Y, et al. Histone deacetylase inhibitor depsipeptide activates silenced genes through decreasing both CpG and H3K9 methylation on the promoter. Mol Cell Biol 2008; 28:3219 - 3235; PMID: 18332107; http://dx.doi.org/10.1128/MCB.01516-07
- Feng J, Fang H, Wang X, Jia Y, Zhang L, Jiao J, et al. Discovery of N-hydroxy-4-(3-phenylpropanamido) benzamide derivative 5j, a novel histone deacetylase inhibitor, as a potential therapeutic agent for human breast cancer. Cancer Biol Ther 2011; 11:477 - 489; PMID: 21263218; http://dx.doi.org/10.4161/cbt.11.5.14529
- Huang W, Tan D, Wang X, Han S, Tan J, Zhao Y, et al. Histone deacetylase 3 represses p15(INK4b) and p21(WAF1/cip1) transcription by interacting with Sp1. Biochem Biophys Res Commun 2006; 339:165 - 171; PMID: 16298343; http://dx.doi.org/10.1016/j.bbrc.2005.11.010
- Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 2010; 18:436 - 447; PMID: 21075309; http://dx.doi.org/10.1016/j.ccr.2010.10.022
- Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell 2008; 30:61 - 72; PMID: 18406327; http://dx.doi.org/10.1016/j.molcel.2008.02.030
- Mukhopadhyay S, Ballard BR, Mukherjee S, Kabir SM, Das SK. Beneficial effects of soy protein in the initiation and progression against dimethylbenz [a] anthracene-induced breast tumors in female rats. Mol Cell Biochem 2006; 290:169 - 176; PMID: 16941229; http://dx.doi.org/10.1007/s11010-006-9184-9
- Mohn F, Weber M, Schübeler D, Roloff TC. Methylated DNA immunoprecipitation (MeDIP). Methods Mol Biol 2009; 507:55 - 64; PMID: 18987806; http://dx.doi.org/10.1007/978-1-59745-522-0_5
- Voorhoeve PM, Agami R. Unraveling human tumor suppressor pathways: a tale of the INK4A locus. Cell Cycle 2004; 3:616 - 620; PMID: 15044859; http://dx.doi.org/10.4161/cc.3.5.859