Abstract
The Drosophila lethal(2)denticleless (l(2)dtl) gene was originally reported as essential for embryogenesis and formation of the rows of tiny hairs on the larval ventral cuticle known as denticle belts. It is now well-established that l(2)dtl (also called cdt2) encodes a subunit of a Cullin 4-based E3 ubiquitin ligase complex that targets a number of key cell cycle regulatory proteins, including p21, Cdt1, E2F1 and Set8, to prevent replication defects and maintain cell cycle control. To investigate the role of l(2)dtl/cdt2 during development, we characterized existing l(2)dtl/cdt2 mutants and generated new deletion alleles, using P-element excision mutagenesis. Surprisingly, homozygous l(2)dtl/cdt2 mutant embryos developed beyond embryogenesis, had intact denticle belts, and lacked an observable embryonic replication defect. These mutants died during larval stages, affirming that loss of l(2)dtl/cdt2 function is lethal. Our data show that L(2)dtl/Cdt2 is maternally deposited, remains nuclear throughout the cell cycle, and has a previously unreported, elevated expression in the developing gonads. We also find that E2f1 regulates l(2)dtl/cdt2 expression during embryogenesis, possibly via several highly conserved putative E2f1 binding sites near the l(2)dtl/cdt2 promoter. Finally, hypomorphic allele combinations of the l(2)dtl/cdt2 gene result in a novel phenotype: viable, low-fertility males. We conclude that “denticleless” is a misnomer, but that l(2)dtl/cdt2 is an essential gene for Drosophila development.
Introduction
Lethal(2)denticleless (l(2)dtl) was first identified as a heat shock gene located on chromosome 2R in Drosophila.Citation1 The authors of that initial study reported that mutation of l(2)dtl resulted in both embryonic lethality and the absence of the rows of tiny hair-like projections known as denticle belts that are characteristic of late embryonic cuticles in 7 reported alleles: l(2)dtlAo-2, l(2)dtlAj-3, l(2)dtlAl-3, l(2)dtlAd-3, l(2)dtlAb-2, l(2)dtlAg-4, and l(2)dtlAt. Subsequently, the yeast homolog of l(2)dtl, cdt2, was shown to be required for both mitotic and pre-meiotic DNA replication.Citation2 More recently, the L(2)dtl /Cdt2 gene product has been characterized as a member of the Cullin4-based CRL4Cdt2 complex, a PCNA-dependent E3 ubiquitin ligase that targets a growing number of cell cycle proteins to the proteasome for destruction.Citation3,Citation4 These proteins include the replication licensing factor Cdt1, the transcription factor E2f1, the CDK inhibitor p21, and the histone methyltransferase Set8. Deletion of l(2)dtl/cdt2 in either zebrafish or mice results in replication defects and embryonic lethality.Citation5,Citation6 Indeed, L(2)dtl/Cdt2 has been dubbed a “master regulator” of the cell cycle based on its role in promoting the timely destruction of key cell cycle regulatory proteins.
Here we characterize strains in which the Drosophila l(2)dtl/cdt2 gene has been disrupted by transposable elements and describe newly generated null alleles as tools for future investigations into the role of the l(2)dtl/cdt2 gene product. In contrast to earlier reports of embryonic lethality, we find that l(2)dtl/cdt2 null mutants survive beyond embryogenesis with intact denticle belts and die in early larval stages. Furthermore, zygotic null mutant embryos do not display replication defects. Instead, maternally deposited L(2)dtl/Cdt2 is sufficient for normal embryogenesis in zygotic mutants. In wild-type embryos, we are first to report two novel findings: (1) l(2)dtl/cdt2 expression appears to be regulated by E2f1 in vivo, and (2) l(2)dtl/cdt2 has elevated expression in the gonads. Interestingly, hypomorphic male flies are viable, but predominantly sterile: a previously unreported phenotype for the l(2)dtl/cdt2 gene. Taken together, we demonstrate that although the lethal(2)denticleless name appears to be a misnomer, l(2)dtl/cdt2 is an essential gene in Drosophila that is required for normal development and adult male fertility.
Results
Generation of new l(2)dtl/cdt2 deletion strains
To begin investigating the function of l(2)dtl/cdt2 in Drosophila development, we obtained three strains with a transposable element insertion in l(2)dtl/cdt2 and we used these to create and characterize two new deletion alleles. The three original strains included one P-element and two piggyBac insertion strains: l(2)dtl/cdt2KG02590, l(2)dtl/cdt2c02261 and l(2)dtl/cdt2f04209 (Bloomington and Harvard Stock Centers, respectively). The positions of these three insertion elements were confirmed by PCR (flanking primers , and data not shown). The P-element, l(2)dtl/cdt2KG02590 is inserted within the 5′UTR, while the two piggyBac elements are positioned within the solitary intron (l(2)dtl/cdt2f04209) and the second exon (l(2)dtl/cdt2c02261). We measured the effect of each insertion on l(2)dtl/cdt2 mRNA expression in homozygous mutant embryos 8–12 h after egg deposition (A.E.D.) by quantitative RT-PCR (). In homozygous l(2)dtl/cdt2f04209, l(2)dtl/cdt2KG02590, and Df(2R)BSC136 embryos, l(2)dtl/cdt2 expression is undetectable in qualitative PCR (). The level of KG02590 expression detected with quantitative PCR is not significantly different compared with the deficiency strain (p value = 0.04, ). Expression of l(2)dtl/cdt2 mRNA is substantially elevated in l(2)dtl/cdt2c02261 homozygous embryos relative to control embryos. These data indicate that transcription of l(2)dtl/cdt2 is initiated in the l(2)dtl/cdt2c02261 line and the level of mRNA accumulation is significantly misregulated via an unknown mechanism. Thus, molecular null alleles would be beneficial for definitive investigations into the role of L(2)dtl/Cdt2 during Drosophila development.
Figure 1. Generation of l(2)dtl/cdt2 deletion strains. (A) Illustration of the position for the three transposable elements with inverted triangles indicating positions and colored arrows denoting the primers used to determine position: KG02590 (purple), f04209 (green), and c02261 (red). The l(2)dtl/cdt2 gene has one intron and is flanked by two genes, CG4091 and l(2)not. The Del-166 and Del-172 strains were generated from KG02590. Breakpoints are at 2,300 bp into the 5′ region of the 11 kb element for both strains and at 379 bp or 637 bp within the l(2)dtl/cdt2 coding region, respectively. A scale bar indicating 400 bp is shown. (B) Qualitative RT-PCR showing that l(2)dtl/cdt2 is zygotically transcribed in 8–12 h old homozygous w1118 (control) and c02261 embryos, but no expression was detected in lines KG02590, f04209, Df(2R)BSC136 (Bloomington stock #9424), Del-166, or Del-172. Expression of the rp49 gene serves as a control to demonstrate equivalent cDNA amounts were used for each strain. Genomic DNA yields larger bands as described in Experimental Procedures. Twenty-six cycles were completed for the rp49 primers; 30 cycles were required to visualize all l(2)dtl/cdt2 expression products. The gel image on the left is a composite of two separate gels, with the upper gel having a longer exposure time. The gel image on the right was created by cropping photos from the same double-comb gel. (C) Quantitative PCR on 8–12 h old homozygous mutant embryos. Results are shown with bars representing the average Relative Quantitation (RQ) observed for l(2)dtl/cdt2 expression relative to the rp49 expression pattern. The w1118 RQ was arbitrarily set at 1.0. The RQ of the other genotypes (c02261, f04209, KG02590, Df(2R)BSC136, Del-166, and Del-172) was compared with w1118. A graph with all seven genotypes is shown on the left. A graph without the highly expressed c02261 genotype is shown on the right. KG02590 expression is significantly less than control w1118 expression (RQ = 0.21 ± 0.04 vs. RQ = 1, p value = 0.0005) and not significantly different from the deficiency strain, Df(2R)BSC136 (RQ = 0.18 ± 0.14, p value = 0.32). Four independent assays were run in triplicate. Error bars indicate standard deviation as calculated by Excel (Microsoft 2007).
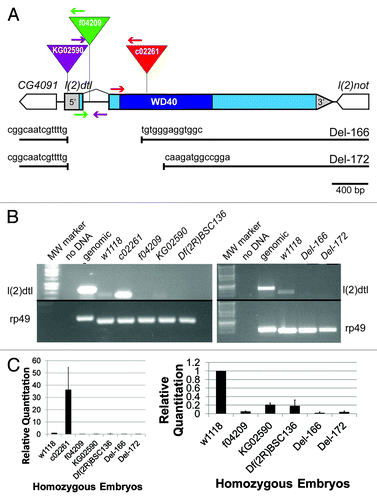
Having confirmed the location of the KG02590 transposable element, we mobilized this element to generate new mutant alleles of l(2)dtl/cdt2. Of the 300 strains created, two were selected for characterization based on PCR analysis: Deletion-166 (Del-166) and Deletion-172 (Del-172). PCR of genomic DNA from Del-166 and Del-172 homozygous embryos indicated that neighboring genes, CG4091 and l(2)not, remained intact and that part of the KG02590 P-element was present (, and data not shown). However, PCR analysis also indicated that exon 1 (including the first 14 amino acids of the L(2)dtl/Cdt2 protein) and a portion of exon 2 had been removed from both the l(2)dtl/cdt2Del-166 and l(2)dtl/cdt2Del-172 strains. Subsequent sequence and BLAST analyses confirmed that only 2,300 bp of the 11 kB P-element had been retained in each strain. While in Del-166, the first 379 bp of coding sequence had been deleted, slightly more coding sequence had been deleted from Del-172 (637 bp); both deletions have lost the translational start site and the 5′ portion of exon 2 (). RT-PCR analyses verified that expression of l(2)dtl/cdt2 mRNA is disrupted by the deletion: l(2)dtl/cdt2 mRNA was undetectable in both homozygous Del-166 and Del-172 embryos while rp49 control gene expression was observed in all strains (). Therefore, we characterized the expression of l(2)dtl/cdt2 in three previously reported disruption strains and introduced two new l(2)dtl/cdt2 deletion strains for further investigation into the role of l(2)dtl/cdt2 in Drosophila.
Neither disruption nor deletion of l(2)dtl/cdt2 causes a replication defect in Drosophila embryos
Since deletion of l(2)dtl/cdt2 in other metazoans results in replication defects,Citation7,Citation8 we investigated whether zygotic loss of l(2)dtl/cdt2 would affect replication in Drosophila embryos. During the final embryonic epidermal cell cycles, cycles 15 and 16 (approximately 5 h after egg deposition (A.E.D.)), homozygous l(2)dtl/cdt2 mutant embryos incorporate BrdU in similar patterns and quantities when compared with w1118 control embryos (). Since L2dtl/Cdt2 targets the replication licensing factor, Cdt1/Dup, to the proteosome and Cdt1/Dup disruption is known to cause observable replication defects, we also examined double-parked (dup) mutant embryos for comparison. In contrast to the l(2)dtl/cdt2 mutants, dupa1 mutant embryos exhibit a clear replication defect in cycle 16, as previously reported ().Citation9 These data suggest that l(2)dtl/cdt2 mutant embryos are able to progress normally through S phase without replication defects. We also examined BrdU incorporation in mitotic CNS cells and endocycling gut cells in later embryos, but again overt replication defects were absent (data not shown). We conclude that zygotic expression of l(2)dtl/cdt2 is not required for normal replication during Drosophila embryogenesis.
Figure 2.l(2)dtl/cdt2 mutant embryos undergo normal replication. (A-H) BrdU labeling of embryonic epidermis in S phase of cycles 15 and 16 (Stage 10, or approximately 5 h A.E.D.) is shown. (A) depicts control pattern and levels of BrdU incorporation in a w1118 (control) embryo. (B) depicts BrdU incorporation in dupa1 mutant embryos, which have a previously characterized replication defect; these mutants incorporate BrdU during S phase of cycle 15, but BrdU incorporation is absent from cycle 16 nuclei.Citation9 (C-H) similarly staged l(2)dtl/cdt2 mutant embryos do not differ significantly from control in either the pattern or levels of BrdU incorporation in these cell cycles. The deficiency strain in this figure was Df(2R)BSC779 (Bloomington stock #27351).
Zygotic lethal(2)denticleless/cdt2 mutants exhibit a larval lethal phenotype marked by the presence of intact denticle belts
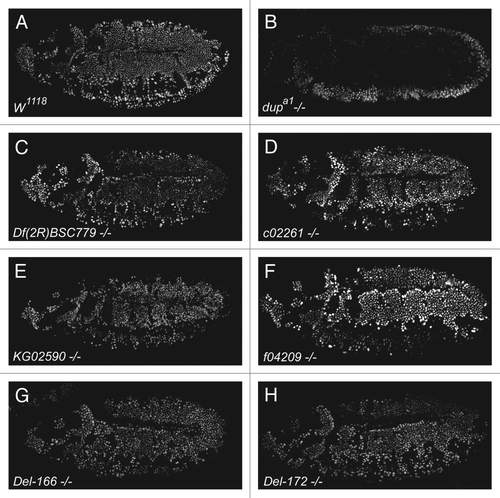
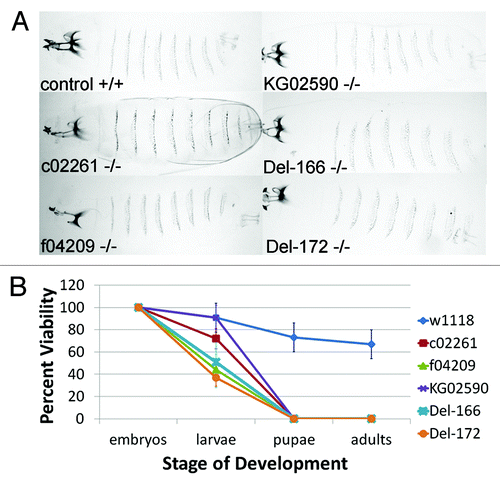
Previous work characterizing seven l(2)dtl/cdt2 alleles reported that mutation of the Drosophila lethal(2)denticleless gene results in embryonic lethality and loss of the ventral denticle belts from the cuticle.Citation1 However, since we did not detect a replication defect during embryogenesis, we questioned whether l(2)dtl/cdt2 was required for completion of embryogenesis. Therefore, we determined the age of l(2)dtl/cdt2 lethality. Consistent with our failure to detect replication defects in l(2)dtl/cdt2 mutant embryos, we found that embryos homozygous for each l(2)dtl/cdt2 mutant allele progressed beyond embryogenesis with intact denticle belts (). In comparison to the normal progression of w1118 embryos to adulthood, most l(2)dtl/cdt2 mutant strains developed only to the 1st instar larval stage, dying within 4 d after egg deposition (A.E.D.) and measuring between 0.5–1 mm in length with small mouth parts. Furthermore, we were able to obtain 4 of the original 7 lethal(2)denticleless strains l(2)dtlAd-3, l(2)dtlAb-2, l(2)dtlAg-4, and l(2)dtlAt from Dr. Kurzik-Dumke and determine their age of lethality.Citation1 In contrast to the previous report, these homozygous mutant embryos survived to an early larval stage with intact denticle belts (data not shown). Notably, a few embryos homozygous for l(2)dtl/cdt2KG02590 (the P-element insertion in the 5′UTR) survived beyond 4 d A.E.D., and grew larger (~1.5 mm in length), suggesting this allele may be hypomorphic. The KG02590 larvae were smaller than control and never progressed to adulthood (). Thus, in contrast to previously reported data, disruption or deletion of the l(2)dtl/cdt2 gene appears to result in lethality at the larval stage of Drosophila development and does not impair denticle belt formation.
Figure 3.lethal(2)denticleless/cdt2 zygotic mutants have intact denticle belts and are larval lethal. (A) Cuticle preparations of control (w1118) and homozygous l(2)dtl/cdt2 mutant larvae (c02261, f04209, KG02590, Del-166 and Del-172) imaged under BF lighting using 10x objectives and ImagePro software (Olympus BX-51 Microscope system). (B) Graph indicating average percent viability of larvae, pupae, and adults developing from w1118 (control) or l(2)dtl/cdt2 mutant embryos: mutant embryos survive beyond embryogenesis and die as larvae. Percent viability is calculated: # hatched larvae/# eggs x 100 = percent viability of larvae. Error bars indicate standard deviation in average for 100 embryos per genotype (10 sets of 10 embryos).
Maternal L(2)dtl/Cdt2 contribution supports embryogenesis
The lack of an obvious phenotype in the l(2)dtl/cdt2 mutant embryos might suggest that this gene is not essential for Drosophila embryogenesis. However, many essential genes are not zygotically required in Drosophila embryos due to an abundance of maternally supplied RNA or protein. We found that the L(2)dtl/Cdt2 protein is maternally supplied in syncytial w1118 embryos (). L(2)dtl/Cdt2 protein co-localized with DNA throughout the cell cycle in these early syncytial cycles (). L(2)dtl/Cdt2 protein is broadly expressed throughout the epidermis in cycles 15 and 16, although nuclear localization of the protein is not always apparent at this stage (). However, in older embryos L(2)dtl/Cdt2 protein levels decline, except in the gonads; there, L(2)dtl/Cdt2 protein staining remained robust in all embryos observed, suggesting this expression is not gender-specific (; arrow in ’). Immunoblots indicate that L(2)dtl/Cdt2 protein persists but is greatly depleted in all 14–20 h mutant embryos relative to control (). Of interest, the L(2)dtl/Cdt2 protein runs as a doublet, suggesting that it may be post-translationally modified in the Drosophila embryo. Previous reports suggest that the mammalian Cdt2 protein can be phosphorylated, and this could also be the case in Drosophila.Citation10 Because our data indicated a lack of significant zygotic l(2)dtl/cdt2 transcription in KG02590, f04209, Del-166, and Del-172 homozygotes, we conclude that L(2)dtl/Cdt2 in these mutant embryos must reflect maternal l(2)dtl/cdt2 gene function. Interestingly, despite elevated l(2)dtl/cdt2 expression in c02261 mutant embryos (), L(2)dtl/Cdt2 protein is severely depleted in these embryos, suggesting that the transcript generated from this allele does not yield normal protein. In addition, we noted that more protein accumulated in KG02590 embryos relative to other mutant strains, in agreement with our previous observation that this allele is hypomorphic (), and can produce small amounts of L(2)dtl/Cdt2 protein zygotically. We conclude that maternally supplied L(2)dtl/Cdt2 supports Drosophila embryogenesis in l(2)dtl/cdt2 mutants.
Figure 4. L(2)dtl/Cdt2 protein is maternally loaded, is localized in the nucleus throughout the cell cycle, and is expressed in the gonads. (A-D) Immunofluorescently labeled L(2)dtl/Cdt2 protein and phopho-histone H3 (pH3, a marker of mitosis). (E) shows a zoom of the embryos in (B) and (C); white boxes in (B) and (C) represent the area shown in the zoom in (E). Note that the brightness of the DAPI stain was decreased in (E) to allow better visualization of Cdt2 and pH3 staining. In syncytial, control embryos (which lack zygotically transcribed gene products), there is an abundant supply of maternally loaded L(2)dtl/Cdt2 protein (A’ and B’). In the early embryo, the cell cycle lacks gap phases, and therefore embryos that are not in mitosis (pH3 negative) are presumably undergoing S phase. L(2)dtl/Cdt2 protein co-localizes with DNA (A and B) in both S phase (A, A”) and in mitosis (B, B”). L(2)dtl/Cdt2 is broadly expressed in the proliferating epidermis, although nuclear localization is not always apparent at this stage (C, E). L(2)dtl/Cdt2 protein expression appears to be somewhat diminished in older embryos (D), with the notable exception being the gonads, where robust L(2)dtl/Cdt2 expression is localized to the gonadal cell nuclei in all embryos observed (D’, arrow). (F) Immunoblot of embryonic extracts from 14–20 h embryos shows abundant L(2)dtl/Cdt2 protein in w1118 control embryos at this stage, but depleted protein in all l(2)dtl/cdt2 mutants; note that the L(2)dtl/Cdt2 protein runs as a doublet: the protein may be post-translationally modified. In those strains which entirely lack zygotic transcription of l(2)dtl/cdt2 [KG02590, Df(2R)BSC779, f04209, Del-166, Del-172, ()], remaining protein likely represents maternal protein. Anti-β-tubulin: loading control. L(2)dtl/Cdt2 and β-tubulin blots in were taken from the same blot at the same exposure.
![Figure 4. L(2)dtl/Cdt2 protein is maternally loaded, is localized in the nucleus throughout the cell cycle, and is expressed in the gonads. (A-D) Immunofluorescently labeled L(2)dtl/Cdt2 protein and phopho-histone H3 (pH3, a marker of mitosis). (E) shows a zoom of the embryos in (B) and (C); white boxes in (B) and (C) represent the area shown in the zoom in (E). Note that the brightness of the DAPI stain was decreased in (E) to allow better visualization of Cdt2 and pH3 staining. In syncytial, control embryos (which lack zygotically transcribed gene products), there is an abundant supply of maternally loaded L(2)dtl/Cdt2 protein (A’ and B’). In the early embryo, the cell cycle lacks gap phases, and therefore embryos that are not in mitosis (pH3 negative) are presumably undergoing S phase. L(2)dtl/Cdt2 protein co-localizes with DNA (A and B) in both S phase (A, A”) and in mitosis (B, B”). L(2)dtl/Cdt2 is broadly expressed in the proliferating epidermis, although nuclear localization is not always apparent at this stage (C, E). L(2)dtl/Cdt2 protein expression appears to be somewhat diminished in older embryos (D), with the notable exception being the gonads, where robust L(2)dtl/Cdt2 expression is localized to the gonadal cell nuclei in all embryos observed (D’, arrow). (F) Immunoblot of embryonic extracts from 14–20 h embryos shows abundant L(2)dtl/Cdt2 protein in w1118 control embryos at this stage, but depleted protein in all l(2)dtl/cdt2 mutants; note that the L(2)dtl/Cdt2 protein runs as a doublet: the protein may be post-translationally modified. In those strains which entirely lack zygotic transcription of l(2)dtl/cdt2 [KG02590, Df(2R)BSC779, f04209, Del-166, Del-172, (Fig. 1B)], remaining protein likely represents maternal protein. Anti-β-tubulin: loading control. L(2)dtl/Cdt2 and β-tubulin blots in Figure 4F were taken from the same blot at the same exposure.](/cms/asset/444ebd7c-7336-4bc6-85f8-9db689658ec6/kfly_a_10920247_f0004.gif)
The l(2)dtl/cdt2 gene is regulated by E2f1 during Drosophila embryogenesis
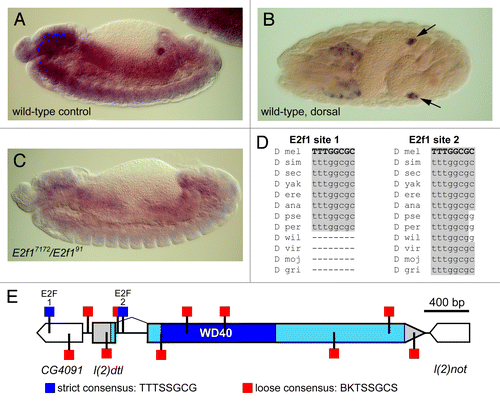
We also examined transcriptional regulation of l(2)dtl/cdt2 in the Drosophila embryo. We found that l(2)dtl/cdt2 mRNA is expressed throughout the gut and CNS in a pattern that is a hallmark of genes involved in S phase, and we also observed a marked expression in embryonic gonads (Fig. Five A-B). Many S phase genes expressed in this pattern are directly regulated by the transcription factor E2f1. Indeed, we identified several E2f1 consensus binding sites in close proximity to the l(2)dtl/cdt2 promoter (Fig. Five D,E). Two of these sites, sites 1 and 2 (), perfectly match the strict E2f1 consensus binding site (TTTSSGCG)Citation11 located at -381 and +232 relative to the l(2)dtl/cdt2 transcription start site, respectively. In addition, sites that match the loose consensus for E2f1 binding are located as close as -39 relative to the l(2)dtl/cdt2 transcription start site (). Furthermore, l(2)dtl/cdt2 expression is greatly diminished in E2f1 mutant embryos (E2f191/E2f17172) (). Interestingly, l(2)dtl/cdt2 mRNA expression was detected in all embryos of the appropriate age, suggesting that the l(2)dtl/cdt2-expressing gonads are not gender-specific. Thus, the Drosophila l(2)dtl/cdt2 gene appears to be expressed in the epidermis, gut, and presumptive gonads and is regulated by E2f1.
Figure 5.l(2)dtl/cdt2 is a probable E2f target gene. (A-C) In situ hybridization of embryos using anti-DIG-labeled l(2)dtl/cdt2 mRNA which was detected using anti-DIG antibody conjugated to alkaline phosphatase (AP) and exposed to AP substrate. The expression pattern appears to primarily include the epidermis, gut, CNS, and gonads (arrows in B) as shown in control embryos at 8–10 h A.E.D. (A) and 12–14 h A.E.D. (B). In E2f17172/E2f191 mutant embryos, l(2)dtl/cdt2 expression is significantly diminished (C). Note: A and C are sibling embryos. (D) Alignment of putative E2F sites that match the strict consensus sequence reveals high conservation of these sequences within the Drosophila genus (adapted from UCSC Genome BrowserCitation34). (E) Map of the l(2)dtl/cdt2 promoter region showing the locations of putative E2F binding sites, including sites that match either the strict (blue) or loose (red) consensus sequences (binding site definitions previously describedCitation11 and adapted from GenePalette).Citation12 A scale bar indicating 400 bp is shown.
Hypomorphic allele combinations of the l(2)dtl/cdt2 gene results in reduced male fertility
L(2)dtl/Cdt2 is a known member of the CRL4 complex and it has been previously reported that a CRL4 complex component, Cul4A, is required for male fertility and spermatogenesis in mice.Citation13,Citation14 In addition, we observed high levels of l(2)dtl/cdt2 mRNA and protein expression in the embryonic gonads. We wondered, therefore, whether disruption of l(2)dtl/cdt2 might affect adult fertility. To address this question, we examined the fertility of viable, hypomorphic l(2)dtl/cdt2 mutant males. w1118 virgin females were crossed to 30 individual males of either the l(2)dtl/cdt2KG02590/l(2)dtl/cdt2c02261 or the l(2)dtl/cdt2KG02590/l(2)dtl/cdt2f04209 paternal genotype and we measured fertility by tracking the percentage of progeny that progressed to adulthood. Of the ~100 eggs laid by the w1118 control females fertilized by each of 30 l(2)dtl/cdt2 hypomorphic males, few progressed to adulthood: 0% for KG02590/f04209 and 4.8 ± 11% for KG02590/c02261 (). Precise excision of the l(2)dt/cdt2lKG02590 P-element restored male fertility (compare w1118 to pHOP#1/c02261 and pHOP#1/f04209 in ). PCR analysis of pHOP#1 genomic DNA indicated that the P-element was completely removed, restoring the l(2)dtl/cdt2 5′UTR (data not shown). Since the L(2)dtl/Cdt2 protein and l(2)dtl/cdt2 mRNA staining patterns suggested that the gene may be expressed in both male and female gonads, we wanted to determine whether the low-fertility phenotype was male-specific or a general fertility issue. To accomplish this, l(2)dtl/cdt2 hypomorphic virgin females were tested for fertility issues, but appeared to be fertile with progeny progressing to adulthood at percentages that were not significantly different from control w1118 females: 79 ± 13% KG02590/f04209 and 74 ± 19% w1118 (p value = 0.11, data not shown). Thus, disruption of the l(2)dtl/cdt2 gene causes reduced fertility in Drosophila males.
Figure 6. Hypomorphic l(2)dtl/cdt2 adult males have reduced fertility or complete sterility. Graphic representation of the fertility of l(2)dtl/cdt2 hypomorphic adult males as measured by the percent of embryos progressing to adulthood (# adult/ # embryos x 100 = Percent Fertility). At least 100 eggs per cross were collected from w1118 virgin females that were crossed with 30 individual male flies of the following paternal genotypes (~100 egg x 30 individual males = 3000 eggs per genotype were scored): w1118 (control), KG02590/c02261, KG02590/f04209, pHOP#1/c02261 or pHOP#1/f04209. Zero percent (KG02590/f04209) or 4.8% ± 11% (KG02590/c02261) of the embryos progressed to adulthood as compared with w1118 (66% ± 11%) [p value = 2.14x10-23]. Fertility was rescued by P-element mobilization repair of KG02590 (pHOP#1). Error bars indicate standard deviation.
![Figure 6. Hypomorphic l(2)dtl/cdt2 adult males have reduced fertility or complete sterility. Graphic representation of the fertility of l(2)dtl/cdt2 hypomorphic adult males as measured by the percent of embryos progressing to adulthood (# adult/ # embryos x 100 = Percent Fertility). At least 100 eggs per cross were collected from w1118 virgin females that were crossed with 30 individual male flies of the following paternal genotypes (~100 egg x 30 individual males = 3000 eggs per genotype were scored): w1118 (control), KG02590/c02261, KG02590/f04209, pHOP#1/c02261 or pHOP#1/f04209. Zero percent (KG02590/f04209) or 4.8% ± 11% (KG02590/c02261) of the embryos progressed to adulthood as compared with w1118 (66% ± 11%) [p value = 2.14x10-23]. Fertility was rescued by P-element mobilization repair of KG02590 (pHOP#1). Error bars indicate standard deviation.](/cms/asset/cca37d70-2008-4f7c-b9e0-d866a3e44532/kfly_a_10920247_f0006.gif)
Discussion
The function of the lethal(2)denticleless/cdt2 gene product is conserved from yeast to man, playing an essential role in marking factors involved in cell cycle regulation (p21, E2f1, Set8, and Cdt1) for timely proteasomal destruction.Citation3,Citation4 In the absence of l(2)dtl/cdt2 gene function, DNA replication defects can occur. We are the first to report the following four conclusions regarding lethal(2)denticleless/cdt2: (1) the ‘denticleless’ phenotype is a misnomer, as l(2)dtl/cdt2 mutant embryos progress to larval stages with intact denticle belts; (2) l(2)dtl/cdt2 expression appears to be regulated by one of the L(2)dtl/Cdt2 target proteins, E2f1, during embryogenesis; (3) expression of the L(2)dtl/Cdt2 mRNA and protein is elevated in embryonic gonads; and (4) l(2)dtl/cdt2 appears to be essential for male fertility which is a novel phenotype for the l(2)dtl/cdt2 gene.
“Denticleless” is a misnomer for zygotic loss of lethal(2)denticleless
In this report, we characterized existing Drosophila l(2)dtl/cdt2 transposon insertion alleles and generated two new mutant alleles in which the l(2)dtl/cdt2 gene is deleted. Of the strains characterized, one appears to be hypomorphic (KG02590), two act as genetic nulls (f04209 and c02261), and two are new deletion alleles (Del-166 and Del-172) that are both genetic and molecular null alleles. We found that, contrary to a previous report,Citation1 zygotic l(2)dtl/cdt2 mutants survive beyond embryogenesis and retain intact denticle belts. In metazoans, including fish, mice and human cells, depletion or deletion of the l(2)dtl/cdt2 gene results in embryonic lethality and DNA replication defects.Citation5,Citation6,Citation8 Our data indicate that Drosophila L(2)dtl/Cdt2 protein is maternally deposited and that maternal l(2)dtl/cdt2 gene function is sufficient for normal embryonic DNA replication and the completion of embryogenesis. Interestingly, we observe that the L(2)dtl/Cdt2 protein appears to be post-translationally modified in the embryo, and in addition, the higher molecular weight form of the protein is slightly more stable. Nevertheless, in the absence of zygotic L(2)dtl/Cdt2 expression, larval development was halted in the 1st instar. Therefore, our data confirm that l(2)dtl/cdt2 is an essential gene in Drosophila as previously reported; however, we find that lethality occurs beyond embryogenesis and the name “denticleless” appears to be a misnomer.
In the original lethal(2)denticleless report, Kurzik-Dumke and colleagues observed that the l(2)dtl-containing genomic region is tightly packed with multiple genes that may be represented by several lethal complementation groups mapping to the region under study: l(2)dtl, l(2)not, l(2)tid, and l(2)rot. Citation1 While current models of the l(2)dtl/cdt2 genomic region support l(2)not and l(2)tid as the 3′ proximal neighbors of l(2)dtl, the chromosomal location of l(2)rot is unknown and the CG4091 gene is indicated as the 5′ proximal neighbor of l(2)dtl (flybase.org). Kurzik-Dumke et al. characterized 7 lethal mutations (l(2)dtlAo-2, l(2)dtlAj-3, l(2)dtlAl-3, l(2)dtlAd-3, l(2)dtlAb-2, l(2)dtlAg-4, and l(2)dtlAt) mapping to this region that they assigned to one gene that they named l(2)dtl based on an embryonic denticleless phenotype they observed in at least 3 of the mutants (At, Al-3, and Ag-4). However, the complementation data for these mutants was complex, and the authors noted that this data suggested intra-genic complementation and that mutation in more than one gene may contribute to the l(2)dtl lethality phenotype.Citation1 Furthermore, this study did not determine whether the EMS-generated mutations in the seven l(2)dtl alleles contained molecular lesions in the gene encoding L(2)dtl/Cdt2. Finally, the original report stated that germ-line rescue of lethality occurred with constructs to all four genes reported in the l(2)dtl region, but did not mention whether the lethal(2)denticleless construct specifically rescued the denticleless phenotype. Indeed, we found that complementation occurred between a subset of the original l(2)dtl mutants (Ab-2, Ag-4, and At) when crossed to the c02261 allele, suggesting that lethality in these l(2)dtl strains is not due to loss of l(2)dtl/cdt2 function (data not shown). Therefore, it is possible that the denticleless phenotype previously observed was a result of mutation in a gene other than that encoding L(2)dtl/Cdt2. Significantly, the ‘L2DTL’ nomenclature is used in the mammalian literature in association with human hepatocellular carcinoma.Citation15 We propose that the “denticleless” nomenclature be discontinued and that the field simply refer to the gene as cdt2. At present, the direct cause of larval lethality is unclear; it is worth noting, however, that Cul4 and Ddb1 mutants also die as larvae upon depletion of maternal protein,Citation16,Citation17 indicating that the CRL4Cdt2 complex is required for Drosophila development.
Significance of E2f1 regulation of l(2)dtl/cdt2 expression
E2f1 regulates transcription of multiple genes required for DNA replication, such as DNA polymerase and RnrS, which are expressed in the gut and CNS when G1 regulation appears during embryogenesis around 8–10 h A.E.D.Citation18,Citation19 Our data show that l(2)dtl/cdt2 expression requires E2f1, similar to other factors involved in S phase. Moreover, we identified several highly conserved, consensus E2f1 binding sites located in close proximity to the l(2)dtl/cdt2 promoter, suggesting that E2f1 might directly regulate l(2)dtl/cdt2 expression. E2f1 binding sites for characterized E2f1 target genes in Drosophila, including CycE, DNA pol α, and PCNA, have been identified as far as 497 bases from the target gene’s transcription start site.Citation20,Citation21 We, have identified 12 putative E2f1 binding sites within 400 bases of the l(2)dtl/cdt2 transcription start site, two of which match the E2f1 strict consensus binding site and are highly conserved. Thus, the l(2)dtl/cdt2 locus contains multiple putative E2f1 sites that are well within the reported range for direct transcriptional regulation by E2f1. Further, it was recently shown that the human Cdt2 gene is directly regulated by E2F1–4 in vitro via four E2F binding sites that are evolutionarily conserved in mammals.Citation22 We are the first to show that l(2)dtl/cdt2 appears to be regulated in vivo during embryogenesis. Thus, transcriptional regulation of l(2)dtl/cdt2 by E2F-family members appears to be conserved among metazoans.
L(2)dtl/Cdt2 role in Drosophila male fertility
The L(2)dtl/Cdt2 Cullin E3 ubiquitin RING Ligase (CRL) complex, Cul4-DDB1-Cdt2 (CRL4Cdt2), is involved in multiple cellular processes including meiosis. Our data indicate that l(2)dtl/cdt2 is highly expressed in the embryonic gonad, a novel expression pattern for this gene. Furthermore, we report that hypomorphic allele combinations of the l(2)dtl/cdt2 gene were associated with reduced male fertility (KG02590/c02261) or complete sterility (KG02590/f04209). Several other members of CRL E3 ubiquitin ligase complexes have been implicated in male fertility.Citation14,Citation23-Citation27 Interestingly, another component of the CRL4Cdt2 E3 ubiquitin ligase complex, Cul4A, is reportedly an important factor in male fertility. Inactivation of Cul4 in either C.elegans or mice results in either premature spermatogenesis or male-specific infertility due to reduced spermatogenesis, respectively.Citation13,Citation14,Citation26 A recent publication has uncovered that there is a testes-specific isoform of Drosophila Cullin3, Cul3testes. During the late stages of sperm differentiation, the Cul3testes-Roc1b-Klhl10 E3 ubiquitin complex triggers localized caspase activity necessary for sperm individualization.Citation23 Further, loss of any individual component of the Cul3-Roc1b-Klhl10 complex results in male sterility. Once protein substrates are polyubiquinated, they are destroyed in the proteasome. The 26S proteasome itself appears to have unique components used only in Drosophila testes. Of the 33 proteasomal subunits, almost half (12 subunits) have alternative forms that are specifically expressed in the Drosophila testes; deletion of a subset of these factors has resulted in male sterility.Citation27 Significantly, a number of the components of Drosophila ubiquitin-mediated proteolysis are conserved in humans and perturbation of at least one of these components (Klhl10) is associated with infertility in men.Citation28 Thus, it may be that in the testes there is a unique requirement for ubiquitin-mediated proteolysis to ensure rapid protein turnover. It remains to be determined whether L(2)dtl/Cdt2 could participate in another E3 ubiquitin ligase complex other than CRL4Cdt2 to potentially affect testes-specific processes.
Materials and Methods
Drosophila stocks
The l(2)dtl (FBgn0013548) alleles in this paper include: (1) Transposable element strains used in this study include: y1w67c23P{SUPor-P}l(2)dtlKG02590 (FBal0130128, Bloomington (BL) Stock Center #13317), PBac{PB}l(2)dtlc02261(FBal0179150), and PBac{WH}l(2)dtlf04209 (FBal0179149) (both PBacs from Exelixis Collection at Harvard Medical School) and (2) EMS-generated strain: l(2)dtlAd-3 (FBal0061315), l(2)dtlAg-4 (FBal0061314), l(2)dtlAt (FBal0061309) and l(2)dtlAl-3 (FBal0061312). The control and deficiency strains used were: w1118 (BL#3605), w1118; Df(2R)BSC136/SM6a (BL#9424), and w1118;Df(2R)BSC779/SM6a (BL#27351). The E2f1 (FBgn0011766) mutant alleles E2f17172 and E2f191 are previously described, as are Dupa1 mutant alleles.Citation9,Citation19 All homozygous embryos from the transposable insertion strains l(2)dtlKG02590, l(2)dtlc02261, and l(2)dtlf04209 or new deletion strains l(2)dtlDel-166 and l(2)dtlDel-172 were negatively selected using the CyO; twist-GFP balancer.
Generation and analysis of l(2)dtl/cdt2 deletion strains
The new l(2)dtl/cdt2Del-166, l(2)dtl/cdt2Del-172, and pHOP#1 strains were created using a P-element excision screen as previously describedCitation29. The y1w*;CyO, H{w+mC = PDelta2–3}HoP2.1/Bc1 strain (Bloomington Stock #2078) served as the transposase source. Deletion or repair events (pHOP#1) were identified by PCR using primers directed to the l(2)dtl/cdt2 gene, l(2)dtl/cdt2 promoter region, and P-element. PCR products were cloned into pCR4-TOPO vector and purified using the PureLink system according to the manufacturer’s instructions (Invitrogen Cat# K457502) and then subjected to DNA sequence analysis (Eurofins MWG Operon, Huntsville, AL). NCBI BLAST analysis of sequence data was accomplished by alignment of the two sequences (l(2)dtl FASTA from flybase.org or KG element (flybase.org/cgi-bin/get_construct_seq.cgi?id = FBtp0001587) to the FASTA of sequence data) in blastn searches (blast.ncbi.nlm.nih.gov/Blast.cgi).
RNA isolation, embryonic cDNA synthesis and standard PCR analysis
Total RNA was extracted from Drosophila samples (30 embryos per genotype) using TriZol (Cat# 15596–026, Invitrogen) extraction according to manufacturer’s protocol. To extract all traces of Trizol an additional chloroform extraction was used before RNA precipitation. This was followed by RQ1 DNase (Promega Corp., Cat# M6101) treatment followed by RNA cleanup using TriZol. RNA was checked for integrity by observing two bands related to 28S and 18S, respectively, by gel electrophoresis. PCR analysis using rp49 primers and RNA sample (250 ng) as template was performed to check for chromosomal contamination. The total RNA was quantified using a Nano Drop ND-1000 Spectrophotometer. All RNA samples were kept at -80C until use.
RNA samples were reversed transcribed using two micrograms of total RNA per sample. The reverse transcription was performed using 2.5 uM oligo(dT) to prime cDNA synthesis, 500 nM dNTP mix, 40 units RNasin Ribonuclease Inhibitor (Promega Cat# N2511) and 200 Units of M-MLV reverse Transcriptase (Promega Cat# M170)1 in a final 25 ul volume. The preparation of cDNA was performed as previously described,Citation30 with the addition of one step at the end of synthesis: 1 ul RNase H (Promega Cat# M4281) was added and each sample was incubated at 37°C for 30 min. The quality of cDNA libraries was checked by amplifying the riboprotein49 (rp49/RpL32) housekeeping gene using the following primers: rp49 FP: 5′ATGACCATCCGCCCAGCATAC 3′ and rp49 RP: 5′TAAACGCGGTTCTGCATGAGC 3′, which amplify a 290-bp product from a gDNA template and 230-bp product from a cDNA template. Standard PCR was performed with 250 ng cDNA in Eppendorf Personal Mastercycler settings: 94°C for 2 min followed by 26 cycles of 94°C for 15 sec, 60°C for 22 sec, 72°C for 1 min, and the PCR reaction ended with a final incubation at 72°C for 5 min. Thirty cycle were required for l(2)dtl/cdt2 amplification. The sense and antisense primers for l(2)dtl/cdt2 mRNA were CSKdtl F2: 5′CCAAGAGGAGTGCACAATGA3′, CSKdtl R2: 5′ACAGTCTGGCAGTGTGATCG 3′, which yielded, a 396-bp PCR product (cDNA) or 672-bp fragment (gDNA).
Real time quantitative PCR analysis
Quantitative PCR analysis of l(2)dtl/cdt2 mRNA expression was performed on a 7500 Real Time PCR System (Applied Biosystems). PCR products were generated from cDNA samples using AbsoluteTM Blue QPCR SYBR ®low ROX Mix, Cat.#: AB-4323/A from Thermo Fisher Scientific Inc. Assays were run on 96-well plates so that each sample was run in triplicate for the target gene and for the endogenous control. Primers were used to a final concentration of 240 nM per reaction. Besides rp49, two other housekeeping genes, actin and tubulin, were used for normalization of most reactions: Actin FP: 5′GCGGCATCCACGAGACCAC 3′, Actin RP: 5′GGACAGCGAAGCCAGGATG 3′, Tubulin FP: 5′ CACTGGTAAGGAGGATGCGGC 3′ and Tubulin RP: 5′CAGCAGCGAGGTGAAGCCG 3′. Actin and Tubulin primers yielded 236-bp and 181-bp PCR fragments respectively.
A concentration of 480 ng per well of cDNA template was used for each assay. All results were normalized to endogenous gene levels. Relative quantification of gene expression was the method of choice for assessing results. Relative Quantification method is also known as ∆∆Ct method, and it is based in the comparison of the cDNA content of two samples following the ∆∆ median threshold cycle number (Ct).Citation31 Cycle conditions were as follows: After an initial stage 1 at 50°C for 2 min and stage 2 at 95°C for 15 min, samples were cycled 40 times at 95°C for 15 sec and at 60°C for 60 sec.
Cuticle preparations
To verify the phenotypic characterization of lack of ventricle denticle belt, cuticle preparations were performed on GFP-negative homozygous mutant embryos using 1:1 lactic acid: Hoyer’s Mountant as previously described.Citation32 Cuticle preparations were observed and digitally photographed by standard bright-field microscopy under 10x objective lense on an Olympus BX-51 Microscope. The w1118, c02261, and f04209 images were converted from color to grayscale. Automatic contrast was applied to all whole cuticle prep images and then images were resized, organized, and saved as 8-bit TIFF with LZW compression using Adobe Photoshop.
Age of lethality and sterility assays
Embryos were collected, aged 8–13 h, dechorionated using 50% bleach, and sorted in 1x Embryo Wash BufferCitation30 using a fluorescent stereomicroscope (Zeiss Discovery.V12 with X-CITE series 120 UV source) to select CyO; twi-GFP negative homozygous mutant Drosophila melanogaster embryos. Selected embryos were placed onto grape juice agar plates (ten embryos per plate: 10 plates for a total of 100 eggs) with 1 g of yeast paste, placed at 25°C, and monitored for development.
Thirty individual heterozygous males (1–2 d post-eclosion) of each genotype (KG02590/c02261, pHOP#1/c02261, KG02590/f04209, and pHOP#1/f04209) were mated with three virgin w1118 females in cages. Eggs were collected for 12-h intervals until at least 100 embryos were collected. Thus, ~3000 eggs per genotype were scored.
For each collection, the number of eggs, larvae, pupae and adult flies were counted and the percent of viability for each stage of development calculated. Calculations for average, standard deviation, and student TTEST were performed using Excel formulas (Microsoft Office 2007).
BrdU labeling
Four to seven hour embryos were bleached for two minutes to dechorionate, air-dried for 3–5 min, permeabilized for 3 min in octane, and then incubated in 1 mg/ml BrdU in Schneider’s for 15 min. Embryos were then fixed in 7% formaldehyde/heptane for 30 min. The formaldehyde layer was removed and replaced with methanol before shaking to devittelinize. Devittelinized embryos were washed with methanol, and rehydrated for 2 min in 3:1 methanol to PBS-Tx, 5 min in 1:3 methanol:PBS-Tx, 5 brief washes in PBS-Tx, and a 30 min incubation in PBS-Tx. At this point embryos were incubated overnight at 4°C in 1:100 chick anti-GFP (Millipore, # 06–896) to amplify the twi-GFP marker. Embryos were washed and fixed again for 20 min in 7% formaldehyde. Embryos were washed again and incubated in freshly made 2M HCl with 0.1% Tween-20, then washed 2x2 min in 0.1 M Sodium Borate and 3x5 min in PBS-Tx. They were then incubated in 1:100 mouse anti-BrdU (BD, Cat # 347580) for 1–2 h at room temperature. After washing with PBS-Tx, embryos were incubated in 1:200 donkey anti-chick Cy2 and 1:500 goat anti-mouse Cy3 (Jackson Immunoresearch, Cat # 703–225–155 and Cat # 115–165–003) for 1–2 h at room temperature. Embryos were then incubated for 5 min in 1:1000 DAPI, washed 2x5 min in PBS-Tx, and mounted in FluorSave (Calbiochem, Cat # 345789). Homozygous mutant embryos were identified by the absence of the twi-GFP marker and imaged on a Zeiss 710 confocal microscope.
Immunohistochemistry
An antibody against the full-length Cdt2 protein was raised in guinea pig by Rockland Immunochemicals. Wild-type 0–12 h embryos were bleached, fixed in 7% formaldehyde/heptane, devittelinized with methanol, and rehydrated as described above. Embryos were then incubated for 48 h at 4°C in 1:200 affinity purified guinea pig anti-Cdt2 and 1:500 rabbit anti-phospho-histone H3 (Millipore, Cat # 07–679). Embryos were washed and incubated for 1–2 h in 1:500 goat anti-guinea pig Cy3 and 1:1000 goat anti-rabbit Cy2 (Jackson Immunoresearch, Cat # 106–165–003 and Cat # 111–225–047). Embryos were then incubated in 1:1000 DAPI for 5 min, washed 2x5 min in PBS-Tx, and mounted in FluorSave (Calbiochem, Cat # 345789). Embryos were imaged on a Zeiss 710 confocal microscope.
In order to make embryonic protein extracts, 14–20 h homozygous mutant embryos were hand-sorted based on absence of twi-GFP expression under a fluorescent dissecting scope. Embryos were homogenized in Lysis Buffer (1% Triton X-100, 50 mM TRIS-HCl (pH 8.0), 150 mM NaCl,1X Fermentas ProteoBlock protease inhibitor) in a 1 ml Dounce homogenizer. Homogenate was centrifuged at 14,000g for 10 min at 4°C and the supernatant was transferred to a clean tube. We loaded 30 ug of protein into each lane. After transfer, the nitrocellulose blot was washed for 5 min in PBST, blocked for 15 min in PBST + 5% powdered milk, and then incubated in 1:500 affinity purified guinea pig anti-Cdt2 and 1:1000 rabbit anti β-tubulin (Abcam) overnight at 4°C. The blot was washed 3x5 min in PBST, incubated for 2 h in 1:1000 HRP anti-guinea pig (Jackson Immunoresearch, Cat # 106–035–003) and 1:1000 HRP anti-rabbit (GE, Cat # NA934V), and washed 3x5 min. HRP was detected by 5 min incubation with the SuperSignal Chemiluminescent Substrate (Pierce, Cat # 34080) followed by exposure of film.
In situ hybridization
Whole Drosophila embryo in situ hybridization was performed using DIG-labeled l(2)dtl/cdt2 RNA transcribed from the position UG-14-E-2 (LD21861) in the cDNA library version 1 (Drosophila Genomics Resource Center, FBcl0172441) as previously described.Citation33 The l(2)dtl/cdt2 cDNA content of UG-14-E-2 clone was verified by sequence analysis. DIC illuminated embryos were photographed using the 20x objective (Olympus BX-51 Microscope) and digital camera (FinePix F20SE).
Identification and alignment of E2f1 binding sites
We used the GenePalette program to locate putative E2F1 binding sites as previously definedCitation11 near the l(2)dtl/cdt2 promoter.Citation12 Binding site alignments were adapted from the UCSC Genome Browser.Citation34
| Abbreviations: | ||
| (l(2)dtl) | = | lethal(2)denticleless |
| cdt2 | = | Cdc10-dependent transcript 2 |
Acknowledgments
The authors would like to thank the reviewers for valuable suggestions that strengthened the manuscript, Ursula Kurzik-Dumke for sending the l(2)dtl Ad-3 l(2)dtl Ag-4, l(2)dtl At, and l(2)dtl Ab-2 stocks, the Indiana University Bloomington Stock Center, Harvard Stock Center and Drosophila Genome Resource Center for Drosophila transposable element stocks and the l(2)dtl/cdt2 cDNA clone. We thank http://flybase.org for valuable database information; L.G. was supported by MBRS grant S06 GM 008049 (A.M. Abu-Shakra, PI). K.N.S. is supported by the S-STEM grant NSF-DUE 0807055 (PI: A. Tokuta). C.S. is supported by the Seeding Postdoctoral Innovators in Research and Education (SPIRE) program NIH-NIGMS grant #K12GM000678. This work funded by NIH: R01 GM57859 to R.J.D. and NIH-NIGMS SC2 GM083764 to S.C.S.K.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Kurzik-Dumke U, Neubauer M, Debes A. Identification of a novel Drosophila melanogaster heat-shock gene, lethal(2)denticleless [l(2)dtl], coding for an 83-kDa protein. Gene 1996; 171:163 - 70; http://dx.doi.org/10.1016/0378-1119(95)00885-3; PMID: 8666267
- Yoshida SH, Al-Amodi H, Nakamura T, McInerny CJ, Shimoda C. The Schizosaccharomyces pombe cdt2(+) gene, a target of G1-S phase-specific transcription factor complex DSC1, is required for mitotic and premeiotic DNA replication. Genetics 2003; 164:881 - 93; PMID: 12871901
- Havens CG, Walter JC. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev 2011; 25:1568 - 82; http://dx.doi.org/10.1101/gad.2068611; PMID: 21828267
- Abbas T, Dutta A. CRL4Cdt2: master coordinator of cell cycle progression and genome stability. Cell Cycle 2011; 10:241 - 9; http://dx.doi.org/10.4161/cc.10.2.14530; PMID: 21212733
- Sansam CL, Shepard JL, Lai K, Ianari A, Danielian PS, Amsterdam A, et al. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev 2006; 20:3117 - 29; http://dx.doi.org/10.1101/gad.1482106; PMID: 17085480
- Liu CL, Yu IS, Pan HW, Lin SW, Hsu HC. L2dtl is essential for cell survival and nuclear division in early mouse embryonic development. J Biol Chem 2007; 282:1109 - 18; http://dx.doi.org/10.1074/jbc.M606535200; PMID: 17107960
- Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev 2008; 22:2507 - 19; http://dx.doi.org/10.1101/gad.1703708; PMID: 18794348
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 2006; 23:709 - 21; http://dx.doi.org/10.1016/j.molcel.2006.08.010; PMID: 16949367
- Whittaker AJ, Royzman I, Orr-Weaver TL. Drosophila double parked: a conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes Dev 2000; 14:1765 - 76; PMID: 10898791
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316:1160 - 6; http://dx.doi.org/10.1126/science.1140321; PMID: 17525332
- Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res 2008; 18:1763 - 77; http://dx.doi.org/10.1101/gr.080622.108; PMID: 18836037
- Rebeiz M, Posakony JW. GenePalette: a universal software tool for genome sequence visualization and analysis. Dev Biol 2004; 271:431 - 8; http://dx.doi.org/10.1016/j.ydbio.2004.04.011; PMID: 15223345
- Yin Y, Lin C, Kim ST, Roig I, Chen H, Liu L, et al. The E3 ubiquitin ligase Cullin 4A regulates meiotic progression in mouse spermatogenesis. Dev Biol 2011; 356:51 - 62; http://dx.doi.org/10.1016/j.ydbio.2011.05.661; PMID: 21624359
- Kopanja D, Roy N, Stoyanova T, Hess RA, Bagchi S, Raychaudhuri P. Cul4A is essential for spermatogenesis and male fertility. Dev Biol 2011; 352:278 - 87; http://dx.doi.org/10.1016/j.ydbio.2011.01.028; PMID: 21291880
- Pan HW, Chou HY, Liu SH, Peng SY, Liu CL, Hsu HC. Role of L2DTL, cell cycle-regulated nuclear and centrosome protein, in aggressive hepatocellular carcinoma. Cell Cycle 2006; 5:2676 - 87; http://dx.doi.org/10.4161/cc.5.22.3500; PMID: 17106265
- Lin HC, Wu JT, Tan BC, Chien CT. Cul4 and DDB1 regulate Orc2 localization, BrdU incorporation and Dup stability during gene amplification in Drosophila follicle cells. J Cell Sci 2009; 122:2393 - 401; http://dx.doi.org/10.1242/jcs.042861; PMID: 19531585
- Lee HO, Zacharek SJ, Xiong Y, Duronio RJ. Cell type-dependent requirement for PIP box-regulated Cdt1 destruction during S phase. Mol Biol Cell 2010; 21:3639 - 53; http://dx.doi.org/10.1091/mbc.E10-02-0130; PMID: 20826610
- Duronio RJ, O’Farrell PH. Developmental control of the G1 to S transition in Drosophila: cyclin Eis a limiting downstream target of E2F. Genes Dev 1995; 9:1456 - 68; http://dx.doi.org/10.1101/gad.9.12.1456; PMID: 7601350
- Duronio RJ, O’Farrell PH, Xie JE, Brook A, Dyson N. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev 1995; 9:1445 - 55; http://dx.doi.org/10.1101/gad.9.12.1445; PMID: 7601349
- Ohtani K, DeGregori J, Nevins JR. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci U S A 1995; 92:12146 - 50; http://dx.doi.org/10.1073/pnas.92.26.12146; PMID: 8618861
- Yamaguchi M, Hayashi Y, Matsukage A. Essential role of E2F recognition sites in regulation of the proliferating cell nuclear antigen gene promoter during Drosophila development. J Biol Chem 1995; 270:25159 - 65; http://dx.doi.org/10.1074/jbc.270.42.25159; PMID: 7559650
- Nakagawa H, Tategu M, Yamauchi R, Sasaki K, Sekimachi S, Yoshida K. Transcriptional regulation of an evolutionary conserved intergenic region of CDT2-INTS7. PLoS One 2008; 3:e1484; http://dx.doi.org/10.1371/journal.pone.0001484; PMID: 18213392
- Arama E, Bader M, Rieckhof GE, Steller H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol 2007; 5:e251; http://dx.doi.org/10.1371/journal.pbio.0050251; PMID: 17880263
- Bader M, Benjamin S, Wapinski OL, Smith DM, Goldberg AL, Steller H. A conserved F box regulatory complex controls proteasome activity in Drosophila. Cell 2011; 145:371 - 82; http://dx.doi.org/10.1016/j.cell.2011.03.021; PMID: 21529711
- Merlet J, Burger J, Tavernier N, Richaudeau B, Gomes JE, Pintard L. The CRL2LRR-1 ubiquitin ligase regulates cell cycle progression during C. elegans development. Development 2010; 137:3857 - 66; http://dx.doi.org/10.1242/dev.054866; PMID: 20978077
- Zhong W, Feng H, Santiago FE, Kipreos ET. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 2003; 423:885 - 9; http://dx.doi.org/10.1038/nature01747; PMID: 12815436
- Belote JM, Zhong L. Duplicated proteasome subunit genes in Drosophila and their roles in spermatogenesis. Heredity (Edinb) 2009; 103:23 - 31; http://dx.doi.org/10.1038/hdy.2009.23; PMID: 19277057
- Yatsenko AN, Roy A, Chen R, Ma L, Murthy LJ, Yan W, et al. Non-invasive genetic diagnosis of male infertility using spermatozoal RNA: KLHL10 mutations in oligozoospermic patients impair homodimerization. Hum Mol Genet 2006; 15:3411 - 9; http://dx.doi.org/10.1093/hmg/ddl417; PMID: 17047026
- Hummel TaCK. P-Element Mutagenesis. In: Dahmann C, ed. Drosophila Methods and Protocols. Totowa, NJ: Humana Press, a part of Springer Science+Business Media, LLC, 2008:97-117.
- Donaldson TD, Noureddine MA, Reynolds PJ, Bradford W, Duronio RJ. Targeted disruption of Drosophila Roc1b reveals functional differences in the Roc subunit of Cullin-dependent E3 ubiquitin ligases. Mol Biol Cell 2004; 15:4892 - 903; http://dx.doi.org/10.1091/mbc.E04-03-0180; PMID: 15331761
- Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s CT difference” formula. J Mol Med (Berl) 2006; 84:901 - 10; http://dx.doi.org/10.1007/s00109-006-0097-6; PMID: 16972087
- Stern DLaES. Preparation of Larval and Cuticles for Light Microscopy. In: Ashburner M, Hawley, R.S., and W. Sullivan, ed. Drosophila Protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2000:601-16.
- Cayirlioglu P, Ward WO, Silver Key SC, Duronio RJ. Transcriptional repressor functions of Drosophila E2F1 and E2F2 cooperate to inhibit genomic DNA synthesis in ovarian follicle cells. Mol Cell Biol 2003; 23:2123 - 34; http://dx.doi.org/10.1128/MCB.23.6.2123-2134.2003; PMID: 12612083
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res 2002; 12:996 - 1006; PMID: 12045153