Abstract
Comparative genomics has identified regions of chromosomes susceptible to participate in rearrangements that modify gene order and genome architecture. Additionally, despite the high levels of genome rearrangement, unusually large regions that remain unaffected have also been uncovered. Functional constraints, such as long-range enhancers or local coregulation of neighboring genes, are thought to explain the maintenance of gene order (i.e., collinearity conservation) among distantly related species since the disruption of these protected regions would cause detrimental misregulation of gene expression. Local enrichment of certain genetic elements in regions of conserved collinearity has been used to support the existence of regulatory-based constraints, although the evidence is largely circumstantial. Indeed, a mechanism of chromosome evolution based only on the existence of fragile regions (i.e., those more susceptible to breaks) can also give rise to extended collinearity conservation, making it difficult to determine whether conserved gene organization is actually caused by functional constraints. Chromosome engineering coupled with genome wide expression profiling and phenotypic assays can provide unambiguous evidence for the presence of functional constraints acting on particular genomic regions. We have recently used this integrated approach to evaluate the presence and nature of putative constraints acting on one of the largest chromosomal regions conserved across nine species of Drosophila. We propose that regulatory-based constraints might not suffice to explain the maintenance of gene organization of some chromosome domains over evolutionary time.
The malleability of Diptera genomes, which can largely be explained by their short generation time and a type of meiosis especially suited to accommodate chromosomal inversions (Citation1 and references therein), makes them ideal systems for studying genome architecture and evolution. Several mechanisms have been proposed for how genomes evolve, including the random breakage model,Citation2 whereby chromosome breaks occur at random sites throughout the genome, and a non-random model where breakpoints are re-used due to the presence of constraintsCitation3-Citation5 and/or fragile regions.Citation6 Comparative studies based on nine Drosophila species that shared a common ancestor ~63 myr ago revealed that many genomic regions participated recurrently at the edge of chromosomal inversions.Citation7 Subsequent simulation studies unambiguously showed that the mode of evolution that better recapitulated the observed patterns of gene rearrangement across species was one dominated by the presence of fragile regions in the Drosophila genome. Nevertheless, this most parsimonious evolutionary scenario also pointed to ~15% of the intergenic regions being under some kind of constraints.Citation7 Determining how these protected intergenic regions contribute to collinearity conservation is complicated by the fact that regions of extended collinearity can be caused both by chromosome fragility and by constraints ().
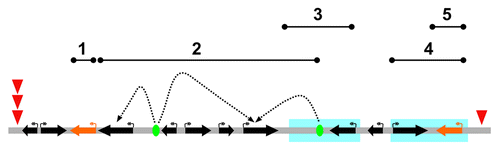
Figure 1. Outline of the mechanisms acting on the gene organization of a region of extended collinearity conservation across Diptera species. The region shown is a composite of constraints of several types (top solid lines, 1–5) acting on independent or on overlapping domains. Domain 2 encompasses two genes whose expression is mediated by regulatory sequences (green ovals) acting at long distance or coordinating the expression of several genes (dotted arrowhead lines). Between those particular regulatory sequences and their target genes other structurally and functionally unrelated genes might exist (bystander genes). Domain 1 and 5 correspond to chromosome stretches that encompass yet-to-be annotated genes or functional features (orange arrows), which are assumed to be relevant for organismal fitness. Domains 3 and 4 (blue boxes) correspond to chromosomal stretches that are exposed to molecular environments associated with reduced probabilities of chromosomal breakage in the germline. Red arrowheads denote chromosomal breakpoints that demarcate the boundaries of the region conserved across species. The left boundary corresponds to a fragile region susceptible to chromosomal breakage due to particular sequence features. The right boundary corresponds to the physical limit of two overlapping constraints. Black arrows, protein- and non-coding genes.
Complex regulatory inputs, such as gene interdigitation, long-range enhancers, or enhancer sharing, have been proposed to act as ‘regulatory’ constraints, thereby inhibiting chromosomal breakpoints. However, the data are mostly indirect.Citation3-Citation5 In a recent paper,Citation8 we evaluated the existence of regulatory-based constraints in one of the largest collinear regions across nine Drosophila species.Citation7 A similar approach has only previously been used in the case of the mouse Hoxd cluster,Citation9 which is conserved in vertebrates, and in several expression neighborhoods of D. melanogaster, which are conserved only in the closest relatives of this species.Citation10
Experimental Evaluation to Long-Standing Functional Constraints
The collinear chromosome region examined, hereafter referred to as CG15121-CG16894 after its outermost genes, ranks first in size, and includes 36 protein-coding genes and a species-specific number of non-coding (nc) RNA genes. Importantly, CG15121-CG16894 encompasses several characteristics suggestive of the presence of constraints. First, the region is enriched for male-biased genes and for genes involved in chemosensory perception, with some of them belonging to both categories. Genes of these expression neighborhoods could be coordinately regulated during a specific physiological and/or developmental stage. Additionally, CG15121-CG16894 includes four intervals with a high density of highly conserved non-coding elements (or HCNE peaks), more than any other conserved chromosome region. HCNEs are thought to act as long-range enhancers in cisCitation11 and have been found to correlate with chromosome regions showing preserved gene organization both in vertebrates and Diptera.Citation4,Citation12 Indeed, in silico promoter predictions and expression profiles throughout the Drosophila life cycle were consistent with some genes in CG15121-CG16894 being regulated by HCNEs. Remarkably, a gene arrangement including chemosensory perception genes and the gene Toll-7 was found to be conserved in A. gambiae and A. aegypti, lineages also characterized by high rates of chromosomal rearrangementsCitation13 and therefore denoting the possible presence of some kind of constraint.
Given the lack of reported naturally occurring mutations disrupting the integrity of CG15121-CG16894, we engineered a chromosomal inversion with a breakpoint within this region, using an FLP-mediated recombination system available in D. melanogaster.Citation14 The induced rearrangement separated genes that were members of the testis and chemosensory perception expression neighborhoods, as well as relocated one of the HCNE peaks to a different chromosomal location. Special care was taken to avoid generating artifactual position effects. In addition, during the course of the experiments, we generated up to three independent types of control strains carrying the region CG15121-CG16894 in its intact form to control for all possible confounding effects. A variety of assays to test for differences in viability between strains with and without the disrupted region CG15121-CG16894 did not show evidence of impaired development as a result of the disruption. In a second set of assays we sought phenotypic effects related to the function of the gene constituents of the expression neighborhoods, and to the normal homeostasis of adult individuals. Although no parameter related with male fertility or normal homeostasis appeared to be affected, we detected a modified odor response to attractant volatile compounds. Subsequent expression profiling with microarray technology of adult individuals with and without the disrupted region CG15121-CG16894 did not reveal differences in mRNA abundance directly related to the engineered disruptions in the targeted region.
Evidence for Constraints and Other Mechanisms
Comparative studies, mostly in vertebrates, have concluded that regulatory-based constraints play a crucial role in preventing the fixation of breakpoints in particular regions of the genome.Citation3,Citation5,Citation15 This mechanism would prevent gross chromosomal rearrangements that might perturb long-range regulatory elements spanning large conserved regions, which could hamper the proper regulation of gene expression. Examples supporting this model are provided by translocations affecting the genes sonic hedgehog and Sox9, and by an inversion affecting the gene TRPS1.Citation16-Citation18 However, mammalian genomes have lower rates of chromosome rearrangements than those of worms and Diptera.Citation7,Citation19 Therefore, in the absence of direct evidence for a link between a structural variant and a disease phenotype,Citation16-Citation18 or of the induction of a detrimental phenotype upon disruption of the region under study,Citation9 it remains possible that regions putatively under constraints may just reflect a common ancestry.
Our work with the region CG15121-CG16894 revealed no evidence of severe detrimental effects upon selective gene rearrangements, which was unexpected since collinearity conservation is supposed to be an effective proxy for constraints in highly malleable genomes such as those of Diptera.Citation7 One possibility could be that most of the phenotypic tests performed lacked statistical power, or alternatively that the time frame used to detect the expected detrimental effects was not appropriate.Citation20,Citation21 However, we were able to detect other phenotypic differences between the strains, albeit apparently unrelated to the disruption of CG15121-CG16894. Other possibilities are that the relevant phenotype was not examined, or that the gene expression analyses performed were not detailed enough. For example, the differences detected in odorant perception were not paralleled by significant changes in mRNA abundance between flies with the intact and disrupted region CG15121-CG16894. However, these experiments were done with material from whole bodies, and some of the genes with odorant functions present in CG15121-CG16894 are known to possess complex tissue-specific patterns of expression.Citation22,Citation23 Therefore, the alteration of these patterns could have gone unnoticed by our microarray experiments. Furthermore, there were no phenotypic effects upon disruption of the arrangement between several Odorant binding protein genes and the gene Toll-7, which has been faithfully maintained despite the extent of genome rearrangement during ~970 myr of divergence in the Diptera species compared. In addition, putative long-range interactions mediated by HCNE peaks involving genetic factors at both sides of the induced breakpoint are unlikely to be enough to justify the maintenance of the integrity of the region CG15121-CG16894 across the genus Drosophila.
Although further targeted disruptions are needed in this and other large conserved chromosome regions in order to reach solid conclusions on whether and how functional constraints impact collinearity conservation, it seems necessary to start considering alternative mechanisms or genomic features. Previous workCitation7 based on simulations of the mode of chromosome evolution under different combinations of parameters has already suggested that even in the largest conserved regions associated with constraints, some intergenic regions were not under the influence of those constraints and could thus accommodate chromosomal breakpoints. Further, when the presence of constraints was simulated, no distinction was made between regulatory-based constraints and others of a different nature.Citation7 For example, we have found that chromosomal regions including genes that interact with the B-type lamin protein, one of the constituent proteins of the nuclear periphery in D. melanogaster,Citation24 are preferentially associated with the largest degrees of collinearity conservation in the genus Drosophila.Citation25 The nature of this structural constraint (nuclear organization) is poorly understood. It might be related to a limited exposure to break-prone molecular environments (e.g., in the context of the recombination machinery) or with increased repair rates.Citation26 Given the evolutionary stability enjoyed by chromosomal regions exposed to these more benign molecular environments, an interesting possibility is that some HCNE peaks had arisen subsequently in the common ancestor to the existing Drosophila lineages. In this scenario, HCNE peaks and their long-range interactions would be the consequence, and not the initial cause, of the evolutionary stability of some chromosomal regions. Region CG15121-CG16894 is in fact known to establish contact with the nuclear peripheryCitation27 and it includes five genes known to interact with the nuclear lamina in somatic cells.Citation28 Lastly, it should be considered whether large conserved chromosomal regions are particularly depressed in sequences known to mediate chromosomal rearrangements, such as transposable elements and ncRNA genes, which can mediate nonallelic homologous recombination (NAHR) events leading to chromosomal rearrangements.Citation29-Citation32 At least in the case of ncRNA genes, which have been annotated across the genus Drosophila, we were able to confirm that the region CG15121-CG16894 was not depleted of these kinds of sequences.
Future Directions
Regulatory-based constraints continue to be a good model to explain unusually high conservation of local gene organization, as indicated by the proven relationship between naturally occurring, or induced, macromutations and detrimental phenotypes.Citation9,Citation16-Citation18 However, our recent work underscores two aspects that should be more carefully considered. The first aspect is the need to use experimental systems to properly evaluate the scope of functional constraints and to uncover the underlying responsible mechanisms. The necessary molecular tools are already in place, e.g., in DrosophilaCitation33 and mice.Citation9 The second aspect is the need to analyze in depth how the presence of other features in large conserved regions, such as local density of NAHR mediating sequences, signatures of structural constraints, and patterns of occurrence of other sources of genetic variation (e.g., nucleotide substitutions and indels), differs from chromosome regions that are not conserved across lineages. Lastly, it will be important to address the relevance for organismal fitness of non-annotated but expressed genomic features, which might be more common than previously thought based on the magnitude of ectopic expression recently documented.Citation34 Some of these non-annotated but expressed genomic features may be ncRNA genes essential for the organism, and therefore no chromosomal breakpoint would be tolerated in the region where they reside. To sum up, only by adopting a more inclusive view of the factors that can impact chromosome evolution, will we be able to effectively dissect the mechanistic basis of collinearity conservation and assess the relevance of functional constraints in shaping the organization of genes on chromosomes during the evolution of eukaryotic taxa.
| Abbreviations: | ||
| myr | = | million years |
| HCNE | = | highly conserved non-coding element |
| NAHR | = | nonallelic homologous recombination |
Acknowledgments
We thank Pavel Pevzner, Mashya Abbassi, and Tiffanie Do for critical reading of the manuscript. This work is supported by NSF grant DEB-0949365 to J.R.
References
- Ranz JM, Casals F, Ruiz A. How malleable is the eukaryotic genome? Extreme rate of chromosomal rearrangement in the genus Drosophila.. Genome Res 2001; 11:230 - 9; http://dx.doi.org/10.1101/gr.162901; PMID: 11157786
- Nadeau JH, Taylor BA. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci U S A 1984; 81:814 - 8; http://dx.doi.org/10.1073/pnas.81.3.814; PMID: 6583681
- Mackenzie A, Miller KA, Collinson JM. Is there a functional link between gene interdigitation and multi-species conservation of synteny blocks?. Bioessays 2004; 26:1217 - 24; http://dx.doi.org/10.1002/bies.20117; PMID: 15499588
- Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engström PG, Fredman D, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res 2007; 17:545 - 55; http://dx.doi.org/10.1101/gr.6086307; PMID: 17387144
- Mongin E, Dewar K, Blanchette M. Long-range regulation is a major driving force in maintaining genome integrity. BMC Evol Biol 2009; 9:203; http://dx.doi.org/10.1186/1471-2148-9-203; PMID: 19682388
- Pevzner P, Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci U S A 2003; 100:7672 - 7; http://dx.doi.org/10.1073/pnas.1330369100; PMID: 12810957
- von Grotthuss M, Ashburner M, Ranz JM. Fragile regions and not functional constraints predominate in shaping gene organization in the genus Drosophila. Genome Res 2010; 20:1084 - 96; http://dx.doi.org/10.1101/gr.103713.109; PMID: 20601587
- Díaz-Castillo C, Xia XQ, Ranz JM. Evaluation of the role of functional constraints on the integrity of an ultraconserved region in the genus Drosophila. PLoS Genet 2012; 8:e1002475; http://dx.doi.org/10.1371/journal.pgen.1002475; PMID: 22319453
- Spitz F, Herkenne C, Morris MA, Duboule D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat Genet 2005; 37:889 - 93; http://dx.doi.org/10.1038/ng1597; PMID: 15995706
- Meadows LA, Chan YS, Roote J, Russell S. Neighbourhood continuity is not required for correct testis gene expression in Drosophila. PLoS Biol 2010; 8:e1000552; http://dx.doi.org/10.1371/journal.pbio.1000552; PMID: 21151342
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature 2006; 444:499 - 502; http://dx.doi.org/10.1038/nature05295; PMID: 17086198
- Engström PG, Ho Sui SJ, Drivenes O, Becker TS, Lenhard B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res 2007; 17:1898 - 908; http://dx.doi.org/10.1101/gr.6669607; PMID: 17989259
- Sharakhov IV, Serazin AC, Grushko OG, Dana A, Lobo N, Hillenmeyer ME, et al. Inversions and gene order shuffling in Anopheles gambiae and A. funestus. Science 2002; 298:182 - 5; http://dx.doi.org/10.1126/science.1076803; PMID: 12364797
- Golic KG, Golic MM. Engineering the Drosophila genome: chromosome rearrangements by design. Genetics 1996; 144:1693 - 711; PMID: 8978056
- Becker TS, Lenhard B. The random versus fragile breakage models of chromosome evolution: a matter of resolution. Mol Genet Genomics 2007; 278:487 - 91; http://dx.doi.org/10.1007/s00438-007-0287-0; PMID: 17851692
- Jeong JY, Einhorn Z, Mathur P, Chen L, Lee S, Kawakami K, et al. Patterning the zebrafish diencephalon by the conserved zinc-finger protein Fezl. Development 2007; 134:127 - 36; http://dx.doi.org/10.1242/dev.02705; PMID: 17164418
- Leipoldt M, Erdel M, Bien-Willner GA, Smyk M, Theurl M, Yatsenko SA, et al. Two novel translocation breakpoints upstream of SOX9 define borders of the proximal and distal breakpoint cluster region in campomelic dysplasia. Clin Genet 2007; 71:67 - 75; http://dx.doi.org/10.1111/j.1399-0004.2007.00736.x; PMID: 17204049
- Fantauzzo KA, Tadin-Strapps M, You Y, Mentzer SE, Baumeister FA, Cianfarani S, et al. A position effect on TRPS1 is associated with Ambras syndrome in humans and the Koala phenotype in mice. Hum Mol Genet 2008; 17:3539 - 51; http://dx.doi.org/10.1093/hmg/ddn247; PMID: 18713754
- Hillier LW, Miller RD, Baird SE, Chinwalla A, Fulton LA, Koboldt DC, et al. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol 2007; 5:e167; http://dx.doi.org/10.1371/journal.pbio.0050167; PMID: 17608563
- Nóbrega MA, Zhu Y, Plajzer-Frick I, Afzal V, Rubin EM. Megabase deletions of gene deserts result in viable mice. Nature 2004; 431:988 - 93; http://dx.doi.org/10.1038/nature03022; PMID: 15496924
- Barbaric I, Miller G, Dear TN. Appearances can be deceiving: phenotypes of knockout mice. Brief Funct Genomic Proteomic 2007; 6:91 - 103; http://dx.doi.org/10.1093/bfgp/elm008; PMID: 17584761
- Galindo K, Smith DP. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics 2001; 159:1059 - 72; PMID: 11729153
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol 2005; 15:1535 - 47; http://dx.doi.org/10.1016/j.cub.2005.07.034; PMID: 16139208
- Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol 2010; 2:a000547; http://dx.doi.org/10.1101/cshperspect.a000547; PMID: 20826548
- Ranz JM, Díaz-Castillo C, Petersen R. Conserved gene order at the nuclear periphery in Drosophila. Mol Biol Evol 2012; 29:13 - 6; http://dx.doi.org/10.1093/molbev/msr178; PMID: 21771720
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 2008; 322:597 - 602; http://dx.doi.org/10.1126/science.1162790; PMID: 18948542
- Mathog D, Sedat JW. The three-dimensional organization of polytene nuclei in male Drosophila melanogaster with compound XY or ring X chromosomes. Genetics 1989; 121:293 - 311; PMID: 2499510
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 2006; 38:1005 - 14; http://dx.doi.org/10.1038/ng1852; PMID: 16878134
- Szankasi P, Gysler C, Zehntner U, Leupold U, Kohli J, Munz P. Mitotic recombination between dispersed but related rRNA genes of Schizosaccharomyces pombe generates a reciprocal translocation. Mol Gen Genet 1986; 202:394 - 402; http://dx.doi.org/10.1007/BF00333268
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 2003; 423:241 - 54; http://dx.doi.org/10.1038/nature01644; PMID: 12748633
- Hill CW, Gray JA. Effects of chromosomal inversion on cell fitness in Escherichia coli K-12. Genetics 1988; 119:771 - 8; PMID: 2900793
- Liu SL, Sanderson KE. Rearrangements in the genome of the bacterium Salmonella typhi. Proc Natl Acad Sci U S A 1995; 92:1018 - 22; http://dx.doi.org/10.1073/pnas.92.4.1018; PMID: 7862625
- Ryder E, Ashburner M, Bautista-Llacer R, Drummond J, Webster J, Johnson G, et al. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics 2007; 177:615 - 29; http://dx.doi.org/10.1534/genetics.107.076216; PMID: 17720900
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, et al. The reality of pervasive transcription. PLoS Biol 2011; 9 - e1000625