Abstract
The Drosophila R7 photoreceptor precursor is directed to its fate by signals from adjacent cells that activate its Receptor Tyrosine Kinase (RTK) and Notch (N) signaling pathways. Counter-intuitively, the N activity both promotes and inhibits the photoreceptor fate in the R7 precursor. We offer an evolutionary perspective for this in which earlier ommatidia had fewer photoreceptors and used N to inhibit the addition of any more. When additional photoreceptors were added by evolution, an RTK signal was used to overcome the N inhibition in these cells, and these new additions potently activated N in their neighboring cells, preventing them from also responding to the RTK signal. The R7 precursor also receives this block, and requires robust RTK activation for it to become a photoreceptor. This is achieved by N transcriptionally activating a new RTK, one that is potently activated in the R7 precursor and sufficing to overcome the N inhibition. The unusually high RTK signal in R7 requires additional transduction components not needed when the signal is mild; in R7 the small GTPases Ras and Rap are both required to transduce the signal, but in other photoreceptors Ras alone suffices.
Keywords: :
Introduction
Over the past 25 years, the Drosophila ommatidium has emerged as one of the key model systems for studying how short-range cellular interactions direct cell fates. Studies have examined the nature of the ligands, receptors and transduction pathways involved, and an understanding has emerged of how direct neighbor/neighbor interactions combine with diffusing signals to choreograph the specification of the various cell types of the ommatidium. The R7 photoreceptor specification has been the most intensively studied and is accordingly the best understood. Here, we review our recent work that examines a complex interplay between the N and RTK signaling pathways in the specification of the R7 photoreceptor.Citation1,Citation2
The Ommatidium Grows by Accretion
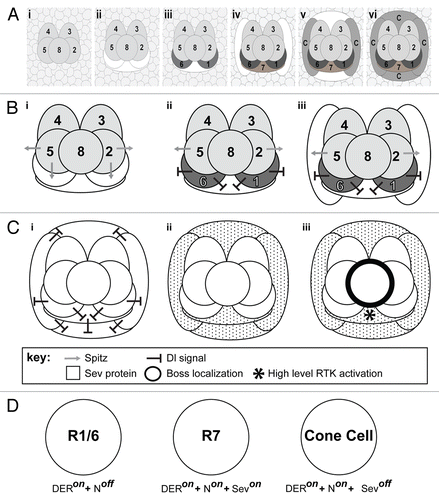
The ommatidium is constructed in two distinct phases. In the first phase, a group of cells drop out of the cell cycle, and organize together to form a five-celled unit known as the precluster.Citation3 These five cells represent the precursors of the R2,3,4,5 and 8 photoreceptors. Many aspects of precluster formation have been gleaned from many studies, but we still lack a simple narrative of how it forms. The second phase is simpler and better understood, and is the focus of our studies. During this second phase, the cells surrounding the precluster go through a final round of cell division before providing a surrounding pool of cells from which new cluster additions are recruited. As cells join the cluster they receive fate-specifying signals that direct their differentiation, and as these cells begin differentiation, they too express signals that direct the fate of the next cells to join.Citation4 The new cells are added to the cluster in a highly stereotyped manner (, i-vi). First, three cells are added to the R2/8/5 face of the cluster. These are the precursors of the R1/7/6 photoreceptors. Next, two presumptive lens secreting cone cells join onto the cluster flanks, followed by two more joining at the poles of the developing ommatidium to complete a square surrounding the photoreceptors.Citation5 Although cell recruitment continues after this stage we confine our investigations to understanding the signals that specify the R1/6/7 photoreceptors and the cone cells.
Figure 1. Cellular dynamics and signaling in growing ommatidial clusters. (A) The growth and maturation of the ommatidium, from the precluster stage to the specification of the cone cells. (i) Shows the five precluster cells surrounded by a sea of unspecified cells. (ii) Cells are systematically recruited to the unit with the first three (white shapes) incorporated along the R2/8/5 face. (iii) Two of the three begin to differentiate (R1/6 – dark gray), while the cell between them (R7 – white shape) does not. (iv) Cells are next added to flanking positions of the cluster (white shapes) as R7 begins differentiation. (v) Two more cone cell precursors are added above and below the cluster (white shapes). (vi) All seven of the newly added cells of the cluster differentiate. (B) A model for why R1/6 activate N strongly in the R7 precursor. (i) Spitz (gray arrow) is released from the R2/5 precluster cells. The R1/6 precursors directly about these cells and (ii) are rapidly specified (dark gray), and express Dl (black line shapes). The Dl expression activates N in the R7 precursor. (iii) When the flanking cone cells join the unit, they receive a potent N activation that prevents them from responding to Spitz. (C) Schematic depiction of Sev expression and activation. (i) As R7 and the flanking cone cells differentiate, they too express high levels of Dl ensuring that all four cone precurors (and R7) experience high N activation. (ii) N transcriptionally activates sev in these cells leading to a high level of Sev (speckling) in these cells. (iii) The ligand for Sev is Boss (thick black line) that is exclusively expressed on membrane of R8. The only cell expressing Sev and contacting R8 is the R7 precursor. This cell now selectively experiences high-level RTK activation (*). (D) Summary diagram depicting the fate of the photoreceptor precursor cells in response to differential activation of the N and RTK pathways. R1/6 cells are specified when DER signaling is active and there is low N activity. R7 cells are specified when N activity is high and both DER and Sev RTK pathways are engaged. The cone cells are specified when N activity is high but only the DER RTK pathway is active. Even though the cone cells express Sev, they do not contact the Boss expressing R8 cell, so the Sev RTK pathway is not engaged.
Cells Are Blind to the Cell Fate Signals Before They Join the Cluster
A key concept to this model is that there is a two-step process in cell fate specification. First, a cell joins the unit. This likely involves a change in the adhesive properties of the cell as it increases its membrane contact with the cells of the precluster. Second, once the cell has joined the precluster it then interprets the signals it receives from the precluster cells as fate-specifying directives. The non-precluster cells surrounding the unit may receive these signals, but they do not interpret them as cell fate instructions.
R1/7/6 and the Cone Cells Comprise Three Distinct Cell Types
We study the signals that specify the R1/6/7 photoreceptors and the cone cells. Within these seven cells we define three distinct cell types; R1 and R6 belong to the generic photoreceptor class, R7 is a unique photoreceptor type, and the four cone cells represent a single cell type. Thus, three distinct cell types are specified as these seven cells are added; R1/6, R7 and the cone cells. By manipulating the fate-specifying signals that these cells receive, we can switch any one cell to the fate of any other.Citation2 We therefore infer that they are all derived from an equivalence group, in which any cell entering the cluster can be specified as any one of the three fates; the cell’s fate being determined by the position it is recruited to and the signals presented at that position.
Receptor Tyrosine Kinase (RTK) Signaling
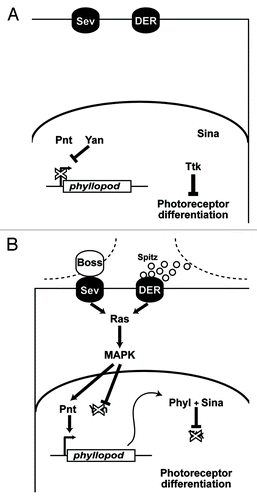
The RTK pathway plays a key role in the fate of the cells; it determines whether a cell becomes a photoreceptor or not. During ommatidial development two RTKs are involved. One is the Drosophila EGF receptor (DER) which is activated by its diffusible ligand, Spitz.Citation6,Citation7 The other RTK is Sevenless (Sev)Citation8 which, is activated by its spatially restricted ligand, Bride of Sevenless (Boss) which is only expressed on the plasma membrane of the R8 cell.Citation9 Activation of either RTK receptor results in the transduction of the Ras pathway and the subsequent phosphorylation cascade that activates MAPKCitation10-Citation12 (). Activated MAPK translocates to the nucleus where it stimulates the transcription of the phyllopod (phyl) gene.Citation13,Citation14 Phyl is an adaptor protein that targets the transcription factor Tramtrack (Ttk) for poly-ubiquitination and subsequent degradation.Citation15-Citation17 Ttk is a repressor of photoreceptor differentiation. If the RTK pathway is activated within a cell, then Ttk is degraded and the cell becomes a photoreceptor. If the RTK pathway is not activated then Ttk persists and the cell does not become a photoreceptor. Accordingly, the R1/6/7 precursors degrade Ttk whereas the cone cell precursors do not.
Figure 2. Details of the RTK transduction pathway. (A) In the absence of ligands the RTKs (Sev and DER) are inactive. The phyllopod gene is not transcribed and Ttk acts as a transcriptional repressor in the cells, and the photoreceptor fate is blocked. (B) In the presence of ligand, RTK transduction occurs via Ras leading to MAPK phosphorylation and translocation to the nucleus where it promotes phyl transcription. The resulting Phyl protein recruits the Sina E3-ubiquitin ligase to poly-ubiquitinate Ttk,Citation16,Citation17 thereby targeting it for degradation, and releasing the block on photoreceptor differentiation.
N Activation Specifies the R7 Rather Than R1/6 Photoreceptor Type
Three of the seven cells that join the precluster during the second phase of ommatidial development become photoreceptors. Yet within these three cells there are two distinct cell types; R1/6 and R7. The signal that distinguishes them is N. If a cell degrades Ttk and has a high level of N activity it becomes an R7, but if it degrades Ttk and has a low N signal it becomes an R1/6 type.Citation2 The ligand for N here is Delta (Dl), a transmembrane protein expressed by the R1/6 cells that activates N in the R7 precursor. How this N signal specifies the R7 fate remains unclear but there are a number of N target genes that have been implicated. The Enhancer of Split (E(spl)) genes encode basic helix-loop-helix transcription factors that are frequently transcriptionally activated in response to N activity. One of these genes, E(spl)mδ, is expressed in the R7 precursor (and not in the R1/6 and cone cell precursors). When it is ectopically expressed, it has a modest ability to transform R1/6 precursors into R7s.Citation18 Extramachrochaetae (emc) is another N response gene that encodes a helix-loop-helix protein that lacks a DNA binding domain. When emc is absent, R7 precursors transform into R1/6 types, and so it appears to be a critical component of the N-induced R7 specification.Citation19 Another gene, seven-up (svp), encodes a steroid receptor-like protein that inhibits R7 development in R1/6/3/4 cells,Citation20 and is transcriptionally inactivated in R7 precursors by N,Citation21 while another gene, prospero (pros), encodes a homeodomain protein that prevents R7 precursors from adopting the R8 features,Citation22 and is transcriptionally regulated by N activity.Citation23
Antagonistic Functions for N in the R7 Precursor
In addition to providing the signal that directs R7 to its appropriate photoreceptor fate, N performs two other functions within the R7 precursor.Citation2 One function inhibits the RTK pathway, so that when Spitz activates DER in the R7 precursor, the resulting RTK activation is insufficient to degrade Ttk, thereby promoting the cone cell fate. The molecular nature of this block remains unknown; all we know is that it antagonizes the ability of RTK activity to remove Ttk from the cell. The other function that N performs is to transcriptionally activate sev, resulting in a high level of Sev protein in the R7 precursor. The Sev receptor is now able to engage its unique ligand, Boss, on the adjacent R8 cell, resulting in a RTK signal with sufficient potency to overcome the N inhibition and degrade Ttk, thereby specifying the cell as a photoreceptor. These two N functions are thus antagonistic; one opposes the degradation of Ttk while the other promotes it.
An Evolutionary Perspective
In the R7 precursor, N activity serves to both inhibit and promote the photoreceptor fate. Why would one signal trigger opposing effects within the same cell? We propose that, although counter-intuitive from the developmental perspective, these opposing effects can be understood from an evolutionary perspective.
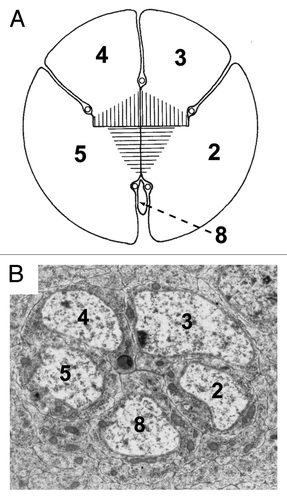
In Phyllopod crustaceans such as water beetles, ommatidia contain only five photoreceptors.Citation24 These appear to represent R2,3,4,5, 8 of the insect ommatidium, and thus the precluster represents the ancient photoreceptor grouping (). The subsequent developmental addition of the R1,6,7 photoreceptors correspond to the evolutionary acquisitions. Phyl is critically required in these cells to degrade Ttk and thereby allow them to differentiate as photoreceptors, and the phyl gene was named for its mutant atavistic phenotype in which only the five cells of the precluster are specified as photoreceptors.Citation13
Figure 3. Structural similarities between the organization of the five photoreceptor cluster of the Phyllopoda and the precluster of Drosophila. (A) Shows a drawing of the five photoreceptors of adult Leptodora kindtii adapted from.Citation24 The image has been rotated from the original, and the numbers of the photoreceptors changed to correspond with the Drosophila nomenclature. The hatching in the middle represents the rhadom of the photoreceptors. (B) Shows a TEM photograph through the Drosophila precluster. Although the two images are from different developmental stages, and the R8 cells are of dramatically different sizes, the overall topology of the cells is largely conserved.
N signaling is used frequently in developmental biology to limit the number of cells acquiring a specific fate. We propose that in the ancestral condition, once the five photoreceptors were specified, those cells expressed an N ligand to prevent subsequent cells from becoming photoreceptors. This N signal would prevent the degradation of Ttk, and thus prevent the specification of extra photoreceptors. It is this N signal, “designed” to prevent the incorporation of additional photoreceptors to the ancient ommatidium that, we propose, lies at the heart of the antagonistic effects we detect in the specification of the R7 photoreceptor.
Spitz released within the developing Drosophila precluster is critically required for the specification of the R2,3,4,5 photoreceptors, but is then subsequently needed again for the formation of R1 and R6.Citation7 The model we propose argues that Spitz signaling ceases in the five cells of the Phyllopoda, but later in evolution it reinitiates to overcome the N repression in the precursors of additional photoreceptors. However, Spitz is a diffusible ligand, which raises the question of what prevents it from promiscuously inducing new photoreceptors. We propose one explanation; once a cell is specified as a photoreceptor, it then expresses high levels of Dl which activates a strong N block to all the cells that it contacts, thereby preventing them from also responding to the Spitz signal ().
Consider the situation when the three cells destined to form R1/6/7 join the ommatidium. If R2/5 release Spitz, the R1/6 precursors will receive the signal rapidly, overcome the N induced block and begin to differentiate as photoreceptors (Fig. 1Bi,ii). The R7 precursor, which lies more distant from the source of the secreted ligand, will only receive significant levels of Spitz some time later. As the next round of cell recruitment occurs, two cells join the flanks of the cluster. These are the prospective anterior/posterior lens secreting cone cells. They directly contact the Spitz-secreting R2/5 cells – as did the R1/6 precursors a short time previously (, iii). The problem the ommatidium now faces is how to prevent the cone cell precursors from becoming photoreceptors. This, we argue, is why the R1/6 cells express high levels of Dl. This strongly activates the N pathway in the neighboring cone cell precursors, and provides a potent block to photoreceptor specification - a block that the level of RTK activation via Spitz/DER signaling is unable to overcome. This effectively solves the problem of the cone cells; it prevents them from being inappropriately specified as photoreceptors. But, in addition to contacting the cone cell precursors, R1/6 also directly contact the R7 precursor, and consequently provide the same block to photoreceptor differentiation in that cell too (, iii). Hence, when diffusing Spitz reaches the R7 precursor, the potent block is already established, and DER activation is not sufficient to specify the cell as a photoreceptor. Thus, in ensuring that the cone cell precursors do not become photoreceptors, the cell in the R7 position is concomitantly prevented from responding to the Spitz/DER signal.
At this point in the developmental/evolutionary progression, the ommatidium has incorporated two additional photoreceptors (R1/6), and ensured that the cells they contact are unable to be specified as photoreceptors by the Spitz/DER signal. This is where Sev comes in. The same N signal that is used to inhibit the formation of photoreceptors is now used to activate sev transcription. Since N activity is high, the level of sev transcription is correspondingly high resulting in an abundance of Sev in the R7 and cone cell precursors (, ii). Unlike Spitz, which diffuses, the ligand for Sev is membrane bound on the R8 precursor and can activate Sev only in cells that directly contact it. Of the R7 and cone cell precursors, only R7 contacts R8, and thus this cell alone experiences Sev activation (, iii). The high levels of Sev allow for a potent RTK signal to be transduced in the R7 precursor, a signal potent enough to overcome the block to photoreceptor specification imposed earlier by the Dl expressed in the neighboring R1/6 cells. The cell in the R7 position now degrades Ttk and begins differentiation as a photoreceptor, and the ommatidium has incorporated three additional photoreceptors to the ancestral group of five.
Transduction of the High-Level RTK Signaling in the R7 Precursor
The requirement to limit the effects of the Spitz/DER signal and prevent the specification of cone cell precursors as photoreceptors demands a high RTK activation in the R7 precursor. The R1/6 precursors, in contrast, only have mild N activity and correspondingly only a mild RTK is required for their specification. Thus the R1/6 and R7 precursors differ in the strength of RTK signal they transduce. In the former it is low, and in the latter it is high. We recently investigated whether there was any difference in which these two cell types transduce these different levels of RTK activity.Citation1
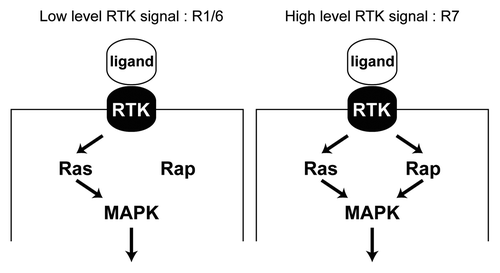
A key nexus in the RTK pathway is the activation of Ras. Ras is a small GTPase that when bound by GTP recruits Raf to the plasma membrane and thereby triggers the phosphorylation cascade that results in MAPK activation. Another member of the Ras superfamily is Rap. In the Drosophila terminal system where the Torso RTK operates, full output from the pathway requires the presence of both Rap and Ras, with both binding to Raf and triggering the phosphorylation cascade.Citation25 In the absence of either Ras or Rap, the RTK signal is significantly diminished. We therefore examined whether Rap would also be required for the high level RTK transduction that occurs in the R7 precursor. Indeed, we found that it is; when Rap levels are depleted, the R7 precursor does not degrade Ttk, and differentiates as a cone cell. The R1/6 cells in comparison do not require the presence of Rap; here the presence of Ras alone is sufficient for degradation of Ttk and the specification of the photoreceptor fate. Thus, when RTK activity is mild only Ras is required for the transduction of the pathway, but when RTK activity is high, both Ras and Rap are required (). This requirement corresponds not only with the strength of the RTK activity, but also with the strength of the N signal. In the R1/6 precursors, N signaling is mild and a correspondingly mild RTK signal requiring only Ras occurs. In the R7 precursor, N signaling is high, demanding a potent RTK signal and the concomitant need for Rap in addition to Ras.
Figure 4. Differential requirement for Rap in RTK transduction. (A) In the R1/6 precursors a mild RTK signal is transduced, where Ras alone suffices for this purpose. Rap is likely engaged, but is not required since its removal does not compromise the specification of these cells.Citation1 (B) In the R7 precursor a robust RTK signal is transduced and this requires the combined presence of both Ras and Rap. Removal of either prevents adequate transduction of the pathway and a failure of the R7 to be specified.
References
- Mavromatakis YE, Tomlinson A. The role of the small GTPase Rap in Drosophila R7 photoreceptor specification. Proc Natl Acad Sci U S A 2012; 109:3844 - 9; http://dx.doi.org/10.1073/pnas.1115108109; PMID: 22355117
- Tomlinson A, Mavromatakis YE, Struhl G. Three distinct roles for notch in Drosophila R7 photoreceptor specification. PLoS Biol 2011; 9:e1001132; http://dx.doi.org/10.1371/journal.pbio.1001132; PMID: 21886484
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol 1976; 53:217 - 40; http://dx.doi.org/10.1016/0012-1606(76)90225-6; PMID: 825400
- Tomlinson A, Ready DF. Neuronal differentiation in Drosophila ommatidium. Dev Biol 1987; 120:366 - 76; http://dx.doi.org/10.1016/0012-1606(87)90239-9; PMID: 17985475
- Tomlinson A. The cellular dynamics of pattern formation in the eye of Drosophila. J Embryol Exp Morphol 1985; 89:313 - 31; PMID: 3937883
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 1996; 87:651 - 60; http://dx.doi.org/10.1016/S0092-8674(00)81385-9; PMID: 8929534
- Kumar JP, Tio M, Hsiung F, Akopyan S, Gabay L, Seger R, et al. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development 1998; 125:3875 - 85; PMID: 9729495
- Hafen E, Basler K, Edstroem JE, Rubin GM. Sevenless, a cell-specific homeotic gene of Drosophila, encodes a putative transmembrane receptor with a tyrosine kinase domain. Science 1987; 236:55 - 63; http://dx.doi.org/10.1126/science.2882603; PMID: 2882603
- Krämer H, Cagan RL, Zipursky SL. Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature 1991; 352:207 - 12; http://dx.doi.org/10.1038/352207a0; PMID: 1857416
- Biggs WH 3rd, Zavitz KH, Dickson B, van der Straten A, Brunner D, Hafen E, et al. The Drosophila rolled locus encodes a MAP kinase required in the sevenless signal transduction pathway. EMBO J 1994; 13:1628 - 35; PMID: 8157002
- Fortini ME, Simon MA, Rubin GM. Signalling by the sevenless protein tyrosine kinase is mimicked by Ras1 activation. Nature 1992; 355:559 - 61; http://dx.doi.org/10.1038/355559a0; PMID: 1311054
- Simon MA, Bowtell DD, Dodson GS, Laverty TR, Rubin GM. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 1991; 67:701 - 16; http://dx.doi.org/10.1016/0092-8674(91)90065-7; PMID: 1934068
- Chang HC, Solomon NM, Wassarman DA, Karim FD, Therrien M, Rubin GM, et al. phyllopod functions in the fate determination of a subset of photoreceptors in Drosophila. Cell 1995; 80:463 - 72; http://dx.doi.org/10.1016/0092-8674(95)90497-2; PMID: 7888014
- Dickson BJ, Domínguez M, van der Straten A, Hafen E. Control of Drosophila photoreceptor cell fates by phyllopod, a novel nuclear protein acting downstream of the Raf kinase. Cell 1995; 80:453 - 62; http://dx.doi.org/10.1016/0092-8674(95)90496-4; PMID: 7859287
- Li S, Li Y, Carthew RW, Lai ZC. Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell 1997; 90:469 - 78; http://dx.doi.org/10.1016/S0092-8674(00)80507-3; PMID: 9267027
- Li S, Xu C, Carthew RW. Phyllopod acts as an adaptor protein to link the sina ubiquitin ligase to the substrate protein tramtrack. Mol Cell Biol 2002; 22:6854 - 65; http://dx.doi.org/10.1128/MCB.22.19.6854-6865.2002; PMID: 12215542
- Tang AH, Neufeld TP, Kwan E, Rubin GM. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell 1997; 90:459 - 67; http://dx.doi.org/10.1016/S0092-8674(00)80506-1; PMID: 9267026
- Cooper MT, Bray SJ. R7 photoreceptor specification requires Notch activity. Curr Biol 2000; 10:1507 - 10; http://dx.doi.org/10.1016/S0960-9822(00)00826-5; PMID: 11114517
- Bhattacharya A, Baker NE. The HLH protein Extramacrochaetae is required for R7 cell and cone cell fates in the Drosophila eye. Dev Biol 2009; 327:288 - 300; http://dx.doi.org/10.1016/j.ydbio.2008.11.037; PMID: 19118542
- Mlodzik M, Hiromi Y, Weber U, Goodman CS, Rubin GM. The Drosophila seven-up gene, a member of the steroid receptor gene superfamily, controls photoreceptor cell fates. Cell 1990; 60:211 - 24; http://dx.doi.org/10.1016/0092-8674(90)90737-Y; PMID: 2105166
- Miller AC, Seymour H, King C, Herman TG. Loss of seven-up from Drosophila R1/R6 photoreceptors reveals a stochastic fate choice that is normally biased by Notch. Development 2008; 135:707 - 15; http://dx.doi.org/10.1242/dev.016386; PMID: 18199577
- Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev Cell 2003; 4:853 - 64; http://dx.doi.org/10.1016/S1534-5807(03)00156-4; PMID: 12791270
- Hayashi T, Xu C, Carthew RW. Cell-type-specific transcription of prospero is controlled by combinatorial signaling in the Drosophila eye. Development 2008; 135:2787 - 96; http://dx.doi.org/10.1242/dev.006189; PMID: 18635611
- Nilsson DE, Odselius R, Elofsson R. The compound eye of Leptodora kindtii (Cladocera). An adaptation to planktonic life. Cell Tissue Res 1983; 230:401 - 10; PMID: 6850774
- Mishra S, Smolik SM, Forte MA, Stork PJ. Ras-independent activation of ERK signaling via the torso receptor tyrosine kinase is mediated by Rap1. Curr Biol 2005; 15:366 - 70; http://dx.doi.org/10.1016/j.cub.2005.02.022; PMID: 15723799