Abstract
Chromatin immunoprecipitation (ChIP) is widely used for mapping DNA-protein interactions across eukaryotic genomes in cells, tissues or even whole organisms. Critical to this procedure is the efficient cross-linking of chromatin-associated proteins to DNA sequences that are in close proximity. Since the mid-nineties formaldehyde fixation has been the method of choice. However, some protein-DNA complexes cannot be successfully captured for ChIP using formaldehyde. One such formaldehyde refractory complex is the developmentally regulated insulator factor, Elba. Here we describe a new embryo fixation procedure using the bi-functional cross-linking reagents DSG (disuccinimidyl glutarate) and DSP (dithiobis[succinimidyl propionate). We show that unlike standard formaldehyde fixation protocols, it is possible to capture Elba association with insulator elements in 2–5 h embryos using this new cross-linking procedure. We show that this new cross-linking procedure can also be applied to localize nuclear proteins that are amenable to ChIP using standard formaldehyde cross-linking protocols, and that in the cases tested the enrichment was generally superior to that achieved using formaldehyde cross-linking.
Introduction
Chromatin immunoprecipitation (ChIP) is a widely used procedure for directly mapping DNA-protein interactions across the genome in cells, tissues or even whole organisms. In this method, proteins are first covalently cross-linked to genomic DNA in intact cells/tissues or isolated nuclei, and then, after fragmentation, the protein-coupled DNA sequences are immunoprecipitated using antibodies directed against specific nuclear proteins. DNA sequences associated with these nuclear proteins are then identified by quantitative PCR (qPCR), DNA microarray or high-throughput sequencing.Citation1,Citation2 UV (UV) irradiation was used to cross-link proteins to DNA in some of the earliest ChIP experimentsCitation3-Citation5; however, UV cross-linking has several drawbacks. In addition to being inefficient, many proteins are not readily cross-linked to DNA by UV and the technique is limited to cell suspensions. For these reasons, the use of formaldehyde rather UV as the cross-linking agent soon became the standard method in ChIP experiments.Citation6-Citation8 Cells, tissues or even entire organisms (fly embryos) are first permeablized, and then fixed with the addition of formaldehyde. After cross-linking the cells, tissues, or organisms are disrupted to isolate the chromatin, which is then fragmented by sonication. The covalently linked protein-DNA complexes are subsequently isolated by immunoprecipitation with antibodies directed against the chromatin protein of interest and the associated sequences are identified.
While formaldehyde cross-linking has become the method of choice in studies on Drosophila cells and embryos, we recently discovered that a developmentally regulated insulator protein complex, called Elba, cannot be readily localized to sites in the genome using this cross-linking reagent. The Elba complex consists of 3 ~40 kD proteins, Elba1, Elba2, and Elba3.Citation9,Citation10 In vitro reconstitution experiments demonstrate that all 3 Elba proteins are required for Elba factor binding to a probe from the Bithorax complex Fab-7 insulator containing the asymmetric recognition sequence, CCAATAAG. The Fab-7 insulator is in the Abdominal-B (Abd-B) transcriptional regulatory associated domain (TARD) and it separates and ensures the functional autonomy of 2 parasegment specific cis-regulatory domains iab-6 and iab-7 that regulate Abd-B expression in parasegments PS11 and PS12 respectively.Citation11 Both Elba1 and Elba2 have a conserved C-terminal BEN domain. This domain is responsible for sequence specific DNA binding activity while sequences in N-terminal domains of Elba1 and Elba2 interact with Elba3 bringing the 3 proteins together in a tripartite complex. It is possible to dispense with Elba3 by fusing the C-terminal BEN domains of Elba1 and Elba2 to a heterologous dimerization domain like GST.
Although we were unable to detect Elba association with its recognition sequence in Fab-7 using rabbit polyclonal antibodies against the 2 BEN domain Elba proteins and standard formaldehyde ChIP protocols, there is good evidence that the tripartite Elba complex binds to Fab-7 in early embryos and is important for its insulator activity during this stage of development. First, Elba DNA binding activity is detected in nuclear extracts prepared from staged 0–6 h embryos, while it is not present later in development. Second, gel mobility supershift experiments using 2 independent polyclonal antibodies against each of the Elba proteins argue that the tripartite Elba complex is responsible for the DNA binding activity detected in 0–6 h nuclear extracts. Third, mutations in the Elba recognition sequence together with RNAi knockdowns argue that the association of the Elba complex with Fab-7 confers insulator activity in early embryos.
Since our polyclonal antibodies supershifted the Elba-DNA complex, we hypothesized that our failure to reliably detect Elba-Fab-7 association in ChIP experiment might be due to the cross-linking reagent. Formaldehyde cross-linking of proteins to DNA requires the close juxtaposition of a primary amino group in the protein and reactive nitrogen in the DNA. Although the BEN domains of Elba1 and Elba2 directly contact DNA, it seemed possible that potentially reactive primary amino groups in these 2 proteins are not in close enough proximity and/or favorably oriented in space to be efficiently cross-linked to Fab-7 DNA sequences by formaldehyde. In fact, there are proteins that are known to bind directly to DNA but are nevertheless refractory to in vivo formaldehyde cross-linking. For example, Nowak et al. have shown that formaldehyde doesn’t reproducibly cross-link NFκB to its in vivo binding sites.Citation12 A further indication that Elba proteins may be refractory to formaldehyde cross-linking because of their primary sequence, rather than some unusual feature of the BEN DNA binding domain are studies on another fly BEN domain protein, Insensitive (Insv), which is encoded by a gene located next to Elba2. Structural analysis of Insv-DNA complexes indicates that the Insv BEN domain binds directly to its recognition sequence, the palindrome CCAATTGG, as a dimer.Citation13 Insv seems to differ from Elba1 and Elba2 in that it can readily form homodimers and bind DNA without a heterologous partner. It also differs from Elba in that its genome wide distribution can be readily detected by using standard formaldehyde ChIP protocols.Citation13
For these reasons we decided to explore the use of bi-functional cross-linking reagents that could potentially link primary amino groups in the Elba proteins to reactive nitrogens in the Fab-7 DNA. Bi-functional cross-linking reagents have been successfully used for not only for formaldehyde refractory sequence specific DNA binding proteins like NFκB, but also for proteins that are linked to DNA indirectly like transcriptional co-activators/co-repressors.Citation14-Citation16 These studies indicated that combining homo bi-functional imido-esters or NHS (N-hydroxysuccinimide)-esters with formaldehyde fixation greatly improved the detection of specific protein-DNA complexes. However, in all of these studies, the bi-functional reagents were used for mapping protein-DNA complexes in single cells (tissue culture or yeast) not whole organisms. Here we have developed a method for using homo bi-functional NHS chemicals to cross-linking staged Drosophila embryos for ChIP analysis. Unlike standard formaldehyde cross-linking procedures, we show that it is possible to capture specific Elba-DNA complexes using these bi-functional reagents. We also show that this new procedure enhances the ChIP enrichment over standard formaldehyde procedures for other chromatin-associated proteins.
Results
Formaldehyde cross-linking fails to capture Elba binding to its target sequence in Fab-7
Both Elba DNA binding activity in nuclear extracts and Elba insulator activity are restricted to early embryogenesis.Citation9,Citation10 For these reasons, we used staged 2–5 h old embryos for our ChIP experiments. In previous studies we were able to show that the d-Myc protein localized to the sex-specific switch element in the Sex-lethal (Sxl) establishment promoter, Sxl-Pe, by fixing isolated nuclei from similarly staged Drosophila embryos with 1.9% formaldehyde.Citation17 However, when we used the same procedure for the 2 Elba BEN domain proteins, we were unable to reliably detect an association between either of them and the Elba recognition sequence in Fab-7. As shown in for Elba1 the enrichment (immune/pre-immune ratio) for Fab-7 was only between 1–2-fold. Moreover, the enrichment was also indistinguishable from 2 controls, 1 from the Sxl gene and the other from the BX-C iab-7 cis-regulatory domain, which do not have sequences resembling the Elba recognition motif. Similar results were obtained for a second Elba1 rabbit antibody and for an Elba2 antibody (data not shown).
Figure 1. DSG or DSP cross-linking of embryo improves the chromatin IP of Elba1. (A) The Elba1 ChIP from formaldehyde (FA) cross-linked embryo did not give significant enrichment of the Fab-7 Elba recognition sequence. 2–5 h old Oregon R embryos were treated as indicated with formaldehyde, and the isolated nuclei were subjected to the ChIP processes with anti-Elba1 #2 antiserum of ref. Citation9. The relative amounts of the DNA fragments were measured by SYBR-green qPCR and the ratio of 'Immune serum IP/Pre-immune serum IP' (fold-enrichment) was calculated using the ∆∆Ct method. Each experiment was repeated more than 3 times to obtain the standard deviation shown as an error bar. (B) The cross-linking of embryo with NHS-esters improved the enrichment of Elba1 ChIP at the target site. The 2–5 h old Oregon R embryos were first cross-linked with indicated concentration of DSG or DSP in the PBS/heptane for one hour followed by 4% FA treatment for 15 min. The ChIP experiments and data analyses were applied as described above.
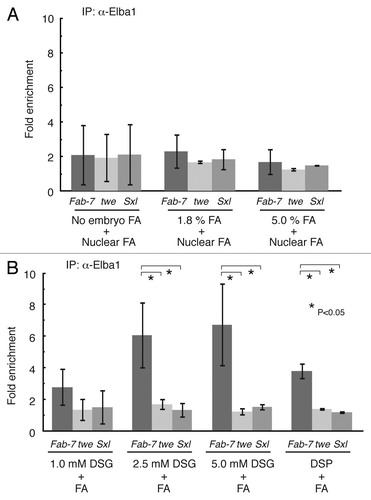
It was possible that unlike d-Myc, most if not all the Elba complex dissociates from chromatin during the isolation of the nuclei. For this reason we switched to a 2-step formaldehyde fixation procedure. In the first step, 2–5 h embryos were cross-linked with 1.8% formaldehyde in the presence of heptane.Citation1 Nuclei were isolated from these fixed embryos and subjected to a second formaldehyde fixation.Citation17 In spite of this double fixation, there was not an appreciable improvement in the immune/pre-immune signal (). Moreover, the Fab-7 specific enrichment was also still indistinguishable from the 2 controls. Similar results were obtained for the second Elba1 antibody and for the Elba2 antibody (not shown).
As it seemed possible that insufficient amounts of formaldehyde penetrated the embryonic membranes, we switched to the 5% formaldehyde/hexane procedure of Li and Biggin.Citation2 However, if anything this procedure decreased rather than increased the enrichment of the Fab-7 sequences in the immune vs. the pre-immune immunoprecipitation (). Likewise, there were still no significant differences between Fab-7 and the 2 control sequences.
Elba association with the Fab-7 insulator can be captured stabilized by bi-functional NHS esters
There are 2 kinds of bi-functional cross-linking reagents that have been used for ChIP experiments with cell suspensions: (1) Bi-functional imido-estersCitation14 and (2) Bi-functional NHS-esters.Citation12,Citation14-Citation16 We selected the NHS-esters DSG (disuccinimidyl glutarate) and DSP (dithiobis[succinimidyl propionate]) for our experiments because they are relatively hydrophobicCitation16 and we thought they would better suited for cross-linking chromatin proteins to DNA in heptane permeabilized Drosophila embryos.
We treated embryos with increasing concentrations of DSG (1.0–5 mM) in the presence of an equal volume of heptane. After vigorously shaking the DSG-embryo suspension for 1 h, formaldehyde was added so that the final concentration in the water phase would be 4%. After a 15 min incubation with formaldehyde, the embryos were processed following the protocol of Kappes et. al.,Citation17 including the nuclear formaldehyde cross-linking step. shows that the DSG-formaldehyde fixation is much more effective in capturing Elba association with Fab-7 in vivo than formaldehyde alone. The Elba1 antibody ChIP for both the 5.0 mM and 2.5 mM DSG-treated embryos showed an appreciable enrichment (6–7 fold) of Fab-7 compared with the pre-immune control. The pull down of Fab-7 sequences appears to be specific as the 2 control loci, twe and Sxl, showed only a 1–1.5 fold enrichment. While there wasn’t much difference between the 5.0 mM and the 2.5 mM DGS fixation, having a sufficiently high concentration of this cross-linking reagent does seem to be important, as there was only a limited enrichment with 1.0 mM DSG cross-linking.
DSP has a longer spacer arm with a disulfide bond between the 2 reactive groups, but is significantly less soluble than DSG. At saturating concentrations (~2mM) of DSP also showed significant enrichment of Fab-7 sequences compared with the pre-immune sera and the 2 control sequences.
Similar results for both DSG and DSP were obtained in ChIP with a second Elba1 antibody and antibody for Elba2 (data not shown; see Figure 8 of ref. Citation10). We also investigated the effects of varying the sonication conditions. As shown in Figure S1, it was possible to further enhance the yield of the Fab-7 Elba1 sequence in the Elba1 ChIP by optimizing the sonication conditions.
Elba binds to a probe from the BX-C Fab-8 insulator in 0–6 h nuclear extracts
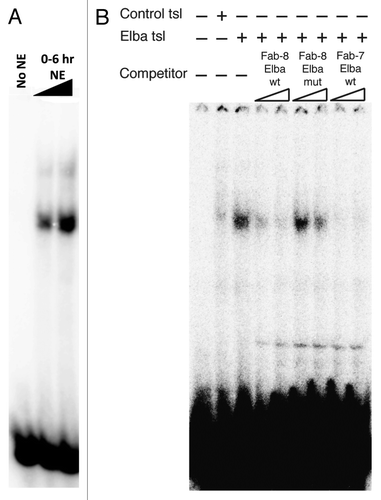
Like Fab-7, the Fab-8 insulator ensures that 2 neighboring Abd-B parasegment cis-regulatory domains, iab-7 (PS12) and iab-8 (PS13) can function autonomously.Citation18 With the aim of identifying proteins besides CTCFCitation19,Citation20 that confer Fab-8 insulator activity, we used a series of overlapping 70–90 bp probes spanning the ~0.8 kb Fab-8 insulator in EMSA (electrophoresis mobility shift assay) experiments with nuclear extracts prepared from staged 0–6 h embryos for DNA binding activity. We found that a 79 bp probe from the proximal side of the Fab-8 insulator gave a shift with 0–6 h nuclear extracts that closely resembled the shift detected with probes containing the Elba sequence from Fab-7. Like the Elba shift in Fab-7, this activity was not detected in older (6–12 h) nuclear extracts (not shown). By deletion mapping we were able to localize the Elba-like shift to a 28 bp probe that contains a sequence, CGAATAAG, which has a one base mismatch to the Elba sequence in Fab-7, CCAATAAG (see shift in ). Competition with excess cold Fab-7 Elba DNA probes, supershifts experiments with polyclonal antibodies against the 3 Elba proteins indicate that the shift seen in nuclear extracts is generated by the tripartite Elba complex (not shown). To confirm that Elba binds to this 28 bp Fab-8 probe, we in vitro translated a mixture of mRNAs encoding the 3 Elba proteins in a rabbit reticulocyte extract. As shown in , this mixture of Elba1, Elba2 and Elba 3 mRNAs primes the synthesis of a factor that specifically shifts the Fab-8 probe. The Fab-8 shift is competed by excess cold Fab-7 and Fab-8 Elba DNA, but not by cold Fab-8 DNA in which the putative Elba recognition sequence was mutated.
Figure 2. Elba complex binds to a semi-consensus sequence in Fab-8 in vitro. The 28 base pair DNA fragment from Fab-8 that contains the 'CGAATAAG' sequence was end-labeled with 32P and subjected to the EMSA (Electrophoresis Mobility Shift Assay) with either A) increasing amounts of nuclear extract (NE) from 0–6 h embryos or B) with the products of a rabbit reticulocyte in vitro-translation primed with a control mRNA ('Control tsl', lane 2) or with a mixture of mRNAs encoding the 3 Elba protein ('Elba tsl', lanes 3–9) as described in ref. Citation9. In panel B, lanes 4–9, 50-fold (lanes 4, 6, and 8) or 100-fold (lanes 5, 7, and 9) excess amounts of unlabeled DNA fragments as indicated were added as cold competitors. Competitors. Fab-8 Elba wt: 28 bp wild-type DNA from Fab-8. Fab-8 Elba mut: 28 bp Fab-8 DNA fragment that has 8 bp alteration of 'ATCCGCCT ' instead of the semi-consensus sequence. Fab-7 Elba wt: 27 bp Fab-7 fragment that spans the original Elba site (See ref. Citation9 and Citation10).
Elba association with the BX-C Fab-8 insulator can be captured with the bi-functional cross-linker DSG
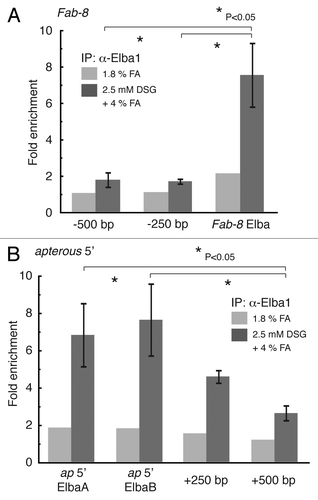
An expectation of these in vitro EMSA experiments is that the Elba complex will be associated with the Fab-8 insulator in early embryos. To determine if this Elba-Fab-8 DNA complex can be captured by DSG we assayed the Elba1 ChIP samples prepared using the 2.5 mM DSG-formaldehyde combination procedure. In the experiment shown in , we tested for enrichment of sequences spanning the 28 bp Fab-8 probe that is shifted by the Elba complex, as well as sequences that are located either 250 bp or 500 bp centromere proximal to this Fab-8 probe (on the distal edge of the iab-7 cis-regulatory domain). We also compared the enrichment using DSG-formaldehyde with the 1.8% formaldehyde fixation procedure. For sequences spanning the 28 bp fragment containing the Fab-8 Elba binding site, we observed an enrichment of about 7 fold in the Elba1 immunoprecipitate compared with the pre-immune control, while sequences 250 or 500 bp away give an enrichment of only about 1.5 fold. As was seen for the Fab-7 Elba binding site, the enrichment of sequences spanning the Fab-8 Elba site using the 1.8% formaldehyde fixation procedure (in one trial) was only about 2 fold.
Figure 3. It is possible to capture Elba complexes in Fab-8 and in the apterous boundary using the DSG-formaldehyde combination procedure. 2–5 h Oregon R embryos were cross-linked with either 1.8% formaldehyde (FA) or 2.5 mM DSG + 4% formaldehyde (FA) and then ChIP’d with Elba1 antibody (antiserum #1 in the ref. Citation9). The DNA recovered with the pre-immune and immune sera was analyzed by qPCR with primer pairs spanning the 28 fragment containing the Fab-8 Elba semi-consensus site (A) or 2 consensus (CCAATAAG) sequences at the far 5′ end of the apterous locus close to the lethal (2) 09851 gene (B). The ChIP enrichment was calculated as described in . We also tested the enrichment of sequences 250 bp and 500 bp proximal (in iab-7) to the Fab-8 Elba site and sequences 250 bp and 500 bp from the first apterous Elba consensus sequence (closest to the apterous gene). For comparison, we have included the extent of enrichment at each site obtained using the 1.8% formaldehyde (FA) procedure.
Elba1 is associated with an insulator on the distal side of the apterous locus
As a further test of the efficacy of the DSG-formaldehyde fixation procedure in capturing Elba-DNA complexes, we assayed for Elba1 association with sequences spanning 2 Elba CCAATAAG recognition motif located in an insulator element at the distal edge of the apterous (ap) locus just upstream of the lethal (2) 09851gene (Daryl Gohl and Martin Muller, pers comm.). As can be seen in , sequences spanning the 2 Elba recognition motifs are enriched 7–8 fold in the Elba1 ChIP prepared using the 2.5 mM DSG-formaldehyde procedure, while neither sequences is appreciably enriched in Elba1 immunoprecipitates of chromatin samples prepared using the 1.8% formaldehyde fixation.
DSG-formaldehyde fixation can be used to capture other protein-DNA complexes
To determine if the DSG-formaldehyde fixation procedure can be used to capture other protein-DNA complexes we tested 2 factors that, unlike Elba, can be readily detected using standard formaldehyde fixation procedures, the BEN domain protein InsvCitation13 and the large subunit of RNA polymerase II.
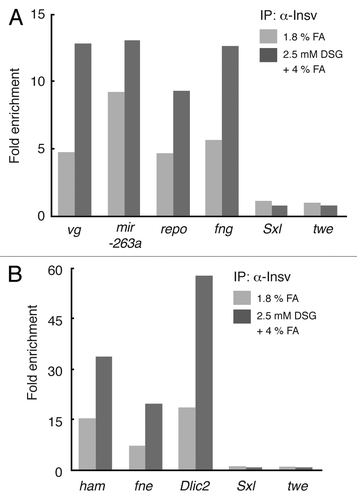
For Insv we tested whether Insv association with seven chromosomal sites previously identified in ChIP-Seq experiments using formaldehyde fixation could also be detected using the 2.5 mM DSG-formaldehyde fixation procedure. These sites were associated with the Dynein light intermediate chain2 (dlic2), found in neurons (fne), fringe (fng), hamlet (ham), mira-263a, reversed polarity (repo), and vestigal (vg) genes.Citation13 As the rabbit polyclonal antibody, staging of the embryos (2–6.5 h vs. 2–5 h) and formaldehyde fixation conditions used in these studies are different from than in Dai et al.,Citation13 we also determined the enrichment for these same Insv associated sequences in chromatin samples prepared following the 1.8% formaldehyde fixation procedure we used here.
As can be seen in , all of the previously identified Insv in vivo binding sites are significantly enriched in ChIPs of 2.5 mM DSG-formaldehyde fixed chromatin, whereas the 2 control sequences from the twe and Sxl genes are not. In this figure we subdivided the in vivo Insv associated sites into 2 categories based on the extent of enrichment observed with the 2.5 mM DSG-formaldehyde procedure. Insv sites in the first category () include those associated with the vg, mira-263a, repo, and fng genes. In this category the extent of enrichment of the Insv associated chromosomal sites was less than 15-fold. The Insv sites in the second category () were associated with the ham, fne and dlic2 genes and for these sites the enrichment was greater than 15-fold. As was seen for Elba, the extent of enrichment compared with the pre-immune control was greater at all of the Insv sites tested for chromatin fixed using the 2.5 mM DSG-formaldehyde procedure than it was with 1.8% formaldehyde. Interestingly, it would also appear that there are site-specific variations in the relative efficacy of DSG-formaldehyde and formaldehyde alone fixations. For example, the ratio of 2.5 mM DSG-formaldehyde/1.8% formaldehyde was 1.4 for the mir-263a Insv site, while it was 3.1 for the dlic2 site.
Figure 4. ChIP enrichment of Insensitive (Insv) protein is improved by using DSG-formaldehyde combination procedure. 2–5 h old Oregon R embryos were cross-linked with either 1.8% formaldehyde or 2.5 mM DSG + 4% formaldehyde and subjected to the ChIP with rabbit pre-immune serum and Insv serum as described in and . We used primers spanning 7 published Insv in vivo binding sitesCitation13 as well as 2 control loci Sex lethal (Sxl) and twine (twe) for qPCR. In vivo Insv sites showing < 15-fold enrichment with the DSG-formaldehyde combination procedure are presented in panel A, while those showing > 15-fold enrichment are presented in panel B. For comparison, we have included the extent of enrichment at each site obtained using the 1.8% formaldehyde (FA) procedure. The previously identified in vivo Insv binding sites are as follows: vg: vestigial. mir-263a: microRNA-263a or bereft. repo: reversed polarity. fng: fringe. ham: hamlet. fne: found in neurons. Dlic2: Dynein light intermediate chain 2.
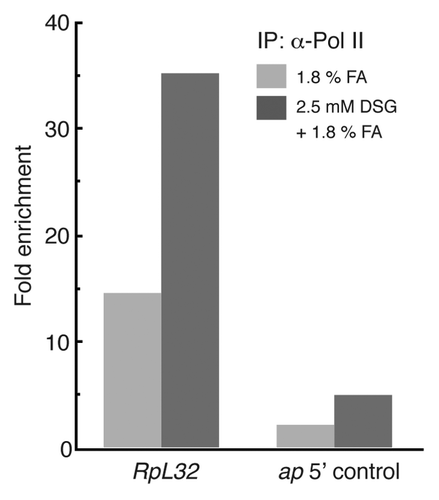
For the large subunit of Pol II we selected sequences spanning the promoter of the ribosomal protein gene, rp-L32. shows that a ~15 fold enrichment for the large Pol II subunit is detected at the promoter of the rp-L32 gene in 1.8% formaldehyde cross-linked chromatin. For the DSG-formaldehyde combination, the enrichment at the promoter is ~35 fold. The control sequence from upstream of the apterous transcription unit is enriched ~1.5 fold for formaldehyde while it is enriched ~3.5 fold in the DSG-formaldehyde combination.
Figure 5. ChIP enrichment of RNA polymerase II (pol II) is improved at some promoters by the DSG-treatment of embryo. Ten - 12 h embryos were cross-linked with either 1.8% formaldehyde only or with 2.5 mM DSG followed by 1.8% formaldehyde. The embryos were disrupted by sonication and resulting chromatin was subjected to IP with either anti-pol II CTD (C-terminal domain) or control IgG. The relative amounts of specific genome sites were measured by qPCR, and the ChIP enrichments were calculated from the ratio of Pol II IP/control IgG IP. The results of promoter of RpL32 (Ribosomal protein L32) is shown here as the representative of the Pol II-bound promoters in this developmental stage. A region upstream of apterous transcription unit was used as a negative control (ap 5′ control).
Discussion
We describe here a new procedure, using 2 bi-functional cross-linking reagents DSG and DSP, for detecting protein-DNA complexes in staged Drosophila embryos by ChIP. One advantage that these bi-functional reagents offer over standard formaldehyde fixation is that it possible to capture protein-DNA complexes that cannot be readily detected using formaldehyde fixation. In spite of compelling evidence for a direct association between the Elba factor and the Fab-7 insulator, several standard formaldehyde cross-linking procedures failed to capture immunoprecipitable protein-DNA complexes containing Elba1 (see above) or Elba2 (not shown). In contrast, Elba1 (and Elba2: see ref. Citation10) binding to the Elba site in Fab-7 could be readily detected in 2–5 h embryos using a combination of DSG or DSP and formaldehyde. Importantly, under the same fixation conditions control sequences from the twe and Sxl genes that are not expected to interact with the Elba factor show little enrichment in Elba1 (or Elba2: see ref. Citation10) ChIPs. The DSG-formaldehyde is also able to capture Elba1-DNA complexes at 2 other fly insulators, the BX-C insulator, Fab-8, and an insulator that separates apterous from the neighboring gene, l(2)09851. For Fab-8, experiments with nuclear extracts and recombinant proteins demonstrate that Elba binds to probes spanning an Elba-like recognition sequence on the proximal side of the insulator. Consistent with these biochemical studies, Elba complexes in this region of Fab-8 are readily detected in staged embryos using the DSG-formaldehyde procedure. For the apterous boundary, Elba binding was predicted from the presence of 2 CCAATAAG motifs and the fact a DNA fragment spanning these 2 sites has insulator activity in transgene assays. As expected, we can detect Elba complexes associated with both apterous sites using the DSG-formaldehyde cross-linking procedure. We also present data showing that our DSG-formaldehyde procedure can be used to recover other protein-DNA complexes in Drosophila embryos and that for at least 2 such complexes, Insv and Pol II, the enrichment is superior to that obtained using a standard formaldehyde cross-linking procedure.
Our results with the Elba complex and the related BEN domain protein Insv also point to potential problems with experiments such as genome-wide ChIPs and Chromatin Conformation Capture that depend upon fixing proteins to DNA and other proteins. In these experiments it is generally assumed that there is no inherent protein/sequence or protein-protein dependent bias in the efficiency of cross-linking (or the recovery of the cross-linked complexes) by the cross-linking reagent. However, this is clearly not the case for Elba and Insv. Even though a conserved BEN domain mediates DNA binding by both the Elba complex and Insv, only the Insv-DNA complex can be captured using formaldehyde as the cross-linking reagent. While the Elba complex might be an extreme example, it seems clear that the efficiency of cross-linking by formaldehyde or other reagents like DSG/DSP will be different from one protein to the next. Another possible complication is suggested by comparing the relative enrichment of Insv-DNA complexes using DSG-formaldehyde vs. formaldehyde at known Insv binding sites. Instead of being more or less constant, the relative enrichment with the 2 protocols varies from one in vivo Insv site to the next. While we’ve only tested one protein and our sample size is very small, this variation in relative enrichment raises the possibility that cross-linking efficiency isn’t just determined by the identity of the protein, but might also be dependent upon the specific chromosomal context. Clearly it will be important to undertake a much more exhaustive comparative study to determine to what extent and for which proteins chromosomal context can affect cross-linking efficiency.
Methods
Embryo preparation
Population cages were used embryo collections as described in ref. Citation10. The appropriately stage embryos were washed off apple juice-agar plates, dechorionated with 50% bleach (2.6% sodium hypochloride) for 3 min, and rinsed with 0.12 M NaCl/0.04% Triton X-100 and then 0.12 M NaCl. For the original ChIP protocol by ref. Citation17, embryos were frozen in liquid nitrogen and stored at -80 after dechorionation. For the other fixation methods described here the embryos were processed immediately after dechorionation.
Cross-linking of embryos
For fixation of embryos with 1.8% formaldehyde in water or 5% formaldehyde in hexane we adapted, as outlined below, protocols described in references Citation21 and Citation22 respectively. For fixation of embryos with the bi-functional cross-linkers DSG and DSP we devised the protocol described in C) after several optimization trials.
A) Cross-linking with 1.8% formaldehyde
For fixation with 1.8% formaldehyde, freshly dechorionated embryos (0.2 - 1.0 g) were transferred to a 50 ml conical tube and suspended in 10 ml of a water-based cross-linking solution consisting of 1.8% formaldehyde / 50 mM HEPES (pH 8.0) / 1 mM EDTA / 0.5 mM EGTA / 100 mM NaCl. (The formaldehyde was freshly added from a 37% stock solution.) We then added 30 ml of n-heptane and the tube was shaken vigorously at room temperature for 15 min. The embryos were collected by centrifugation at 500 × g for 1 min and the heptane phase was carefully removed from the top. The cross-linking reaction was then terminated by adding ~30 ml of the PBS (Phosphate buffered saline, 10 mM Na2HPO4 / 1.76 mM KH2PO4 / 137 mM NaCl / 2.7 mM KCl, pH 7.4) supplemented with 0.125 M glycine and 0.01% Triton X-100. After a brief incubation, the embryos were collected by centrifugation, and washed with ~30 ml of the PBS / 0.125 M glycine / 0.01% Triton X-100. This was followed by 2 washes with ~30 ml of PBST (PBS with 0.05% Triton X-100). The cross-linked embryos were either used directly for ChIP experiments or quick frozen in the liquid nitrogen for long-term storage at -80°C.
B) Cross-linking with 5% formaldehyde
For fixation of embryos with with 5% formaldehyde, the cross-linking was performed in hexane. Before beginning the dechorionation of the embryos, the formadehyde/hexane mixture was first prepared by adding 0.13 volume of 10 × PBS and 0.175 volume of 37% formaldehyde to 1 volume of hexane (~20 ml for 0.5–2.0 g of embryo) in a 50 ml conical tube. The mixture was then shaken for at least 30 min so that the formaldehyde would saturate the hexane in organic phase. The dechorionated embryos were collected in a plastic sieve “Cell Strainer” (70 µm mesh, Corning-Falcon 352350) and then briefly soaked in a tray containing 2-propanl to remove residual water from the surface of embryos. After removing the 2-propanol by blotting the sieve on paper towels, the embryos were transferred to a 50 ml conical tube and weighed. An appropriate volume of hexane (saturated with formaldehyde) was added to the tube and then the mixture was shaken vigorously for 5 min. The embryos were allowed to settle to the bottom of the formaldehyde/hexane mixture. Once the embryos had settled, the formaldehyde/hexane mixture was carefully decanted. The embryos were then washed 2 times by suspending with ~20 ml of PBS containing 0.5% Triton X-100 and by centrifugation at 500 × g for 1 min. The cross-linked embryos were either used directly the ChIP experiments or quick frozen for the storage at -80 °C as described above.
C) Fixation with a NHS-cross-linker and 4% formaldehyde
1) After dechorionation, the embryos were weighed and transferred to a 50 ml conical tube. If there was more than 2 g of embryos, we split the sample into multiple tubes.
2) The bi-functional NHS-esters were dissolved in DMSO (Dimethyl sulfoxide). In the case of DSG (Proteochem c1104), we dissolved 20 mg of DSG powder in 108 µl of DMSO to give ~120 µl of a 500mM DSG stock solution. In the case of DSP (Thermo Scientific Pierce 22585), we dissolved 20 mg of powder in 85 µl of DMSO to give ~99 µl of 500 mM DSP stock solution. The NHS-ester/DMSO solutions can be stored for 2~3 d by freezing and keeping at 4 °C. The NHS-esters have a short shelf life and should be discarded 3 mo after purchase.
3) The fixation mix was prepared by adding the NHS-ester/DMSO solution to phosphate-buffered saline (PBS, 150 mM sodium chloride (NaCl) / 10 mM sodium phosphate (pH 7.6)). The optimal concentration of NHS-esters may differ between the target proteins. The maximum concentrations of DSG and DSP in aqueous solution are 5 mM and 2 mM, respectively. For less than 0.5 g of embryo, prepare 5 ml of solution. Increase the volume by 5 ml for every 0.5 g of additional amount of embryo. Because the NHS-esters are unstable in aqueous solutions, this fixation mix should be prepared immediately before use.
4) Pour the fixation mix into the conical tube containing the dechorionated embryos. Add an equal volume of heptane.
5) Shake the tube vigorously for 1 h.
6) Add 550 µl of 37% formaldehyde per 5 ml of aqueous phase so that the final concentration of formaldehyde 4%. Shake vigorously for additional 15 min.
7) Centrifuge the tube in swinging bucket rotor at 500 × g for one minute to pellet the fixed embryo. Remove the heptane phase by pipetting.
8) Add an equal volume of the “stop” solution (PBS with 125 mM Glycine and 0.1% Triton X-100) to the aqueous phase. Mix well, let stand for 2 min and then pellet the fixed embryos by centrifuging a 500 × g for 1 min.
8) Remove the supernatant by pipetting. Suspend the embryos in 25 ml of stop solution and after a 2 min incubation collect the embryos by centrifugation. Spin again in the same condition.
9) Remove the supernatant, and wash the embryos by suspending in 25 ml of PBST (PBS + 0.05% Triton X-100) and spin again in the same condition. Repeat this step 2 times.
10) The fixed embryos can either be immediately processed for ChIP or transferred to a 1.5ml tube, frozen in liquid nitrogen and then stored at -80 °C until use.
Isolation of nuclei
The processed of the NHS-ester fixed embryos for ChIP was as described in ref. Citation17 with several modifications. ~100 mg of embryos was used for immunoprecipitation each antiserum. The embryos were first suspended in 100µl / 100 mg embryos of Nuclear Isolation Buffer (NIB, 50mM Hepes pH 7.6 / 60mM KCl / 250mM Sucrose)+ 1x Protease Arrest (PA) (Millipore 539124) and disrupted with motorized Teflon-glass homogenizer. After homogenization, the glass cylinder was washed with NIB + 1 × PA (100µl / 100 mg embryos) to recover the homogenate that remained in the homogenizer. The combined homogenate was then centrifuged at 500 × g for 6 min to pellet any large incompletely homogenized embryonic debris. Under these conditions the cross-linked nuclei remain in the supernatant. In the case of the pol II experiment in , the isolated nuclei were recovered by further centrifugation at 3000 × g and directly subjected to the sonication. In all other experiments, the nuclei were first fixed with 1% formaldehyde as follows: A 1/9th volume of 10% formaldehyde was added to nuclei in the supernatant so that the final concentration was 1%. The 10% formaldehyde solution was prepared from 5x Fixation Buffer (250mM Hepes PH 8.0 / 5mM EDTA / 500mM NaCl) and a 37% formaldehyde stock solution. After addition of the formaldehyde, sample was rotated at room temperature for 15 min. Cross-linking was stopped by the addition of 1/3 volume of 1.0 M Glycine. The cross-linked nuclei were recovered by centrifugation at 3000 × g and washed 3 times with cold PBS (using the same centrifugation conditions).
Sonication of chromatin
The isolated nuclei were suspended in 480 µl / 100 mg embryo of Sonication Buffer (10mM Tris pH 8.0 / 0.1% SDS / 10mM EDTA pH 8.0) supplemented with 2 × concentration of PA. The samples were then divided into 1.5 ml tubes so that the volume in each tube would be 250 ~320 µl. The conditions used to shear the chromatin are discussed in detail in the section “Optimization of sonication condition” in the supplemental materials. In the ChIPs of Elba1 and Insv, a Branson Sonifier 450 with the small tip was used, whereas a Branson Digital Sonifier 450 was used for the pol II ChIPs.
Immunoprecipitation (IP)
The sonicated chromatin was first supplemented with × 0.11 volume of 10% Triton X-100, × 0.01 volume of 10% sodium deoxycholate (Na-DOC), and × 0.033 volume of 5M NaCl so that the salt / detergent concentrations would be the same as those of RIPA Buffer (10mM Hepes pH 7.6 / 1mM EDTA / 0.5mM EGTA / 0.1% SDS / 1% Triton X-100 / 0.1% Na-Deoxycholate 140mM NaCl). The sample was then centrifuged at 10,000 × g to remove debris. To reduce non-specific background, the sonicated chromatin was first incubated with 40 µl Protein A-agarose beads (Santa Cruz Biotechnology sc-2001) /100 mg of starting embryos for 1 h on a rotating mixer at room temperature. After the Protein A-agarose beads were removed by centrifugation, samples were split so that each immunoprecipitation would correspond to approximately 100 mg of embryos. An aliquot corresponding to 1/100 of one IP was kept at 4 °C as the '1/100 input' sample. For each IP sample of Elba1 and Insv, 5 µl of pre-immune or immune serum was added and incubated for overnight (> 10 h) at 4 °C. In the case of pol II, Pol II CTD 8WG16 antibody (Millipore 05–952) or a control mouse IgG was used. Following the overnight incubation, 40 µl of a 50% slurry of Protein A agarose beads was added to each sample and incubated for 3 h to allow the Protein A on the beads to interact with the antibody. The beads were collected by centrifugation at 700 × g for 1 min and then washed with ~1 ml of RIPA buffer 3 times. In the original procedure,Citation17 the beads were washed with successively higher salt concentrations. However, we omitted this step because we found that the high-salt washes tended to give more variable results for the control (pre-immune) ChIPs.
Reversing the cross-links and recovery of immunoprecipitated DNA fragments from the Protein A agarose beads
The precipitated beads were first suspended in 500 µl of TE (10 mM Tris pH7.5 / 1 mM EDTA) with 50 mM NaCl. Five micro litters of 10 mg/ml RNase A was added to each sample followed by a 15 min incubation at room temperature. After removing the supernatant by centrifugation, the beads were re-suspended in 500 µl of TE containing 0.5% SDS. The '1/100 input' sample was also adjusted to 500 µl using the same buffer. Five micro litter of 20 mg/ml proteinase K was added to each tube and the samples were incubated for one hour at 37°C. At the end of the incubation, 60 µl of 10% SDS and 36 µl of 5M NaCl was added to each tube and tubes were incubated at 65°C overnight. The supernatant, which contained the DNA fragments, was recovered by centrifugation, extracted with phenol/chloroform 2 times and then precipitated by adding 1/10 volume of 3 M sodium acetate, 1 µl of precipitation carrier 'Ethachin-mate' (Wako chemicals 312–01791) and an equal volume of 2-propanol. After a thorough mixing, the precipitated DNA was pelleted by centrifugation at 14,000 × g for 10 min. The precipitates were washed with 70% ethanol, air-dried and re-suspended in 30 µl of water.
Quantitative PCR (qPCR)
The qPCR was performed with SYBR-Green reagent (Applied Biosystems 4367669) by using either Agilent Mx3000p or Applied Biosystems StepOnePlus real-time PCR system. The fold-enrichment (Immune IP/Pre-immune IP ratio or Anti-pol II IP/control IgG IP ratio) was calculated by comparative Ct method (∆∆CT method). The sequences of primer used in qPCR are listed in the Table S1.
Polyclonal antibody production for Insensitive
The procedure for the production of the Insv rabbit polyclonal antiserum as that used to produce antibodies against the Elba proteins (see ref. Citation9). Six × His-T7 tagged and 6 × His-HA tagged Insv proteins were expressed in bacteria, purified by SDS-PAGE, and then electro-eluted from the gels. 2 rabbits were used for immunization with these Insv proteins. One of the 2 resulting sera was used in this work.
Electrophoresis Mobility Shift Assay (EMSA)
The preparation of nuclear extracts and in vitro translated Elba complex was as described in ref. Citation9, while the labeling of probe and electrophoresis of EMSA is described in ref. Citation10. The oligo DNA sequences of probes/competitors are listed in Table S1.
Additional material
Download Zip (847 KB)Acknowledgments
This research was supported by grants from NIH to P.S. (GM043432) and E.C.L. (NS074037). P.S. would also like to acknowledge support from a grant to the Gene Biology Institute by the Russian Federation Ministry of Education and Science (14.B25.31.0022). E.C.L. would like to acknowledge support from the Burroughs Wellcome Fund. We would like to thank Jamila Horabin, Attilio Pane, and Shelby Blythe for useful discussions and suggestions and Linda E Alger at Thermo Scientific Pierce for her valuable advice on the use of bifunctional cross-linkers. We would also like to thank our colleagues in the Lai, Stern and Schedl lab for helpful discussions and Gordon Grey for preparing fly food.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
Supplementary material may be found here: www.landesbioscience.com/journals/fly/article/26805.
References
- Ghavi-Helm Y, Furlong EE. Analyzing transcription factor occupancy during embryo development using ChIP-seq. Methods Mol Biol 2012; 786:229 - 45; http://dx.doi.org/10.1007/978-1-61779-292-2_14; PMID: 21938630
- Li XY, Biggin MD. Genome-wide in vivo cross-linking of sequence-specific transcription factors. Methods Mol Biol 2012; 809:3 - 26; http://dx.doi.org/10.1007/978-1-61779-376-9_1; PMID: 22113265
- Gilmour DS, Lis JT. Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes. Proc Natl Acad Sci U S A 1984; 81:4275 - 9; http://dx.doi.org/10.1073/pnas.81.14.4275; PMID: 6379641
- Gilmour DS, Lis JT. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol Cell Biol 1985; 5:2009 - 18; PMID: 3018544
- Gilmour DS, Rougvie AE, Lis JT. Protein-DNA cross-linking as a means to determine the distribution of proteins on DNA in vivo. Methods Cell Biol 1991; 35:369 - 81; http://dx.doi.org/10.1016/S0091-679X(08)60580-4; PMID: 1664030
- Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 1988; 53:937 - 47; http://dx.doi.org/10.1016/S0092-8674(88)90469-2; PMID: 2454748
- Dedon PC, Soults JA, Allis CD, Gorovsky MA. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal Biochem 1991; 197:83 - 90; http://dx.doi.org/10.1016/0003-2697(91)90359-2; PMID: 1952079
- Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 1993; 75:1187 - 98; http://dx.doi.org/10.1016/0092-8674(93)90328-N; PMID: 7903220
- Aoki T, Sarkeshik A, Yates J, Schedl P. Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. Elife 2012; 1:e00171; http://dx.doi.org/10.7554/eLife.00171; PMID: 23240086
- Aoki T, Schweinsberg S, Manasson J, Schedl P. A stage-specific factor confers Fab-7 boundary activity during early embryogenesis in Drosophila. Mol Cell Biol 2008; 28:1047 - 60; http://dx.doi.org/10.1128/MCB.01622-07; PMID: 18039839
- Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 1997; 124:1809 - 20; PMID: 9165128
- Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques 2005; 39:715 - 25; http://dx.doi.org/10.2144/000112014; PMID: 16315372
- Dai Q, Ren A, Westholm JO, Serganov AA, Patel DJ, Lai EC. The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev 2013; 27:602 - 14; http://dx.doi.org/10.1101/gad.213314.113; PMID: 23468431
- Kurdistani SK, Grunstein M. In vivo protein-protein and protein-DNA crosslinking for genomewide binding microarray. Methods 2003; 31:90 - 5; http://dx.doi.org/10.1016/S1046-2023(03)00092-6; PMID: 12893178
- Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques 2006; 41:694 - , 696, 698; http://dx.doi.org/10.2144/000112297; PMID: 17191611
- Fujita N, Wade PA. Use of bifunctional cross-linking reagents in mapping genomic distribution of chromatin remodeling complexes. Methods 2004; 33:81 - 5; http://dx.doi.org/10.1016/j.ymeth.2003.10.022; PMID: 15039090
- Kappes G, Deshpande G, Mulvey BB, Horabin JI, Schedl P. The Drosophila Myc gene, diminutive, is a positive regulator of the Sex-lethal establishment promoter, Sxl-Pe. Proc Natl Acad Sci U S A 2011; 108:1543 - 8; http://dx.doi.org/10.1073/pnas.1017006108; PMID: 21220321
- Barges S, Mihaly J, Galloni M, Hagstrom K, Müller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 2000; 127:779 - 90; PMID: 10648236
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet 2007; 3:e112; http://dx.doi.org/10.1371/journal.pgen.0030112; PMID: 17616980
- Ciavatta D, Rogers S, Magnuson T. Drosophila CTCF is required for Fab-8 enhancer blocking activity in S2 cells. J Mol Biol 2007; 373:233 - 9; http://dx.doi.org/10.1016/j.jmb.2007.07.065; PMID: 17825318
- Cavalli G, Orlando V, Paro R. Mapping DNA target sites of chromatin associated proteins by formaldehyde cross-linking in Drosophila embryos. In: Bickmore WA, ed. Chromosome Structural Analysis: A Practical Approach. New York: Oxford University Press, 1999:20–37.
- Toth J, Biggin MD. The specificity of protein-DNA crosslinking by formaldehyde: in vitro and in drosophila embryos. Nucleic Acids Res 2000; 28:e4; http://dx.doi.org/10.1093/nar/28.2.e4; PMID: 10606672
