Abstract
In this Extra View we comment on our recent work on Sudestada1 (Sud1), a Drosophila 2-oxoglutarate (2OG)-dependent dioxygenase that belongs to the Ribosomal Oxygenase (ROX) subfamily. Sud1 is required for normal growth in Drosophila, and is conserved in yeast and mammals. We reported that Sud1 hydroxylates the ribosomal protein S23 (RPS23), and that its loss of function restricts growth and provokes activation of the unfolded protein response, apoptosis and autophagy. In this Extra View we speculate on the role that RPS23 hydroxylation might play in stop codon recognition and on the possible link between Sud1 loss-of-function and activation of the Unfolded Protein Response, Stress Granules formation and growth impairment.
2OG-Dependent Dioxygenases and Cell Growth
2-oxoglutarate (2OG) dependent dioxygenases comprise a large superfamily of enzymes that catalyze different oxidative reactions, employing molecular oxygen and 2OG as co-substrates, and ferrous iron (Fe2+) as a co-factor. Dioxygenases couple the two-electron oxidation of their substrates to the decarboxylation of 2OG, yielding succinate and carbon dioxide.Citation1 In animals 2OG dioxygenases mediate the hydroxylation and demethylation, via hydroxylation, of proteins, DNA, or RNAs.Citation2,3
Among the most extensively characterized 2OG dependent dioxygenases, are the oxygen sensing prolyl hydroxylase domain (PHD) enzymes that modify the Hypoxia Inducible Factor-α subunits (HIF-α).Citation4,5 At atmospheric oxygen levels (21%) HIF-α is post-translationally hydroxylated by PHDs at specific prolyl residues located in a central oxygen-dependent-degradation domain (ODDD). This hydroxylation enables recognition by VHL, the substrate recognition component of an E3 ubiquitin ligase that promotes HIF-α degradation. In hypoxia PHD activity is reduced, allowing HIF-α stabilization and the activation of HIF dependent transcriptional responses. In Drosophila, Sima and Fatiga are the homologs of HIF-α and PHDs, respectively.Citation6 Our lab has previously reported that Fatiga regulates cell growth in a Sima dependent manner.Citation7 However, another report has shown that Fatiga can also promote cell growth in a Sima-independent manner, through the activation of the Cyclin D/Cdk4 complex,Citation8 suggesting that Sima dependent and Sima independent mechanisms are involved in Fatiga-mediated growth regulation.
Given the effect of Fatiga/PHD on cell growth, we wondered if other 2OG dioxygenases are required for normal cell or organ growth. To this end, we performed an RNAi screen targeting all (n = 48) predicted or demonstrated Drosophila 2OG dioxygenases, looking for those required for normal growth.Citation9 Nine 2OG dioxygenase genes scored as positives in the screen, displaying reduced growth after RNAi-mediated silencing. One of the strongest hits was the gene CG44254, which we have named sudestada1 (sud1) after a wind that blows across the south eastern coastline of South America. Further analysis of the sud1 growth phenotype revealed that sud1 silencing provokes both reduction of cell size and reduction in the number of cells that form the affected organ, the latter due to increased apoptosis.
Sud1 is distantly related to the oxygen sensing PHD enzymesCitation10 and extensively conserved in evolution. Using molecular and biochemical techniques, our work along with two other studies performed in yeast and mammalian cells and published simultaneously demonstrated that Sud1 and its orthologs hydroxylate the small ribosomal subunit protein RPS23 at a highly conserved prolyl residue.Citation9,11,12 Whereas in mammalian cells and flies the key prolyl residue of RPS23 is mono-hydroxylated, in yeast the analogous proline is di-hydroxylated by the Sud1 homolog Tpa1p. Thus, di-hydroxylation of the target proline in RPS23 is an evolutionary primitive feature of mysterious function, apparently lost in more derived phylogenetic groups. In Drosophila, sud1 silencing provokes defects in protein synthesis and strong activation of the unfolded protein response (UPR), along with increased eIF2α phosphorylation and concomitant stress granules formation.
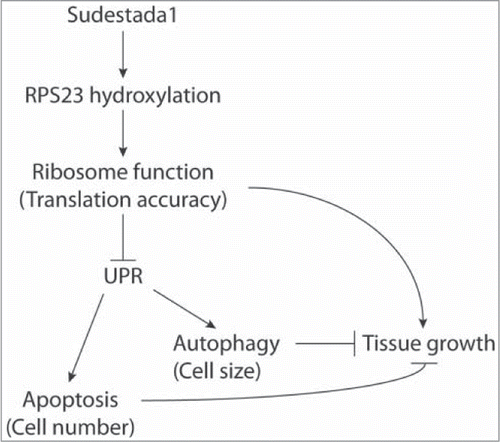
Given that sud1 silencing provokes translational stress and considering that Sud1 mediates hydroxylation of a ribosomal protein, we sought to investigate why UPR is triggered, so we directed our attention to the process of protein translation. It has been extensively documented in Drosophila that mutations that affect components of the protein synthesis machinery -the so called minute genes- provoke growth defects.Citation13 So in this context, it is not surprising that failure to introduce a post-translational modification to a ribosomal protein elicits a growth defect. What is the molecular consequence of the lack of hydroxylation of a single proline of RPS23? Does it affect ribosomal function? Why does lack of RPS23 prolyl hydroxylation impinge on cell or organ growth? We don't have definitive answers for these questions. One possibility is that failure to hydroxylate RPS23 affects translation fidelity resulting in the accumulation of mutated proteins, which in turn may lead to formation of protein aggregates and consequent activation of UPR (). Noteworthy, UPR activation following Sud1 silencing is, at least in part, responsible for growth impairment, since the Sud1 growth phenotype is partially suppressed when sud1 RNAi is co-expressed with an RNAi against Perk, a key effector kinase of UPR that targets eIF2α. Citation14 It remains to be investigated to what extent Sud1 is involved in hypoxia-triggered UPR.
Figure 1. Speculative model of the role of Sudestada1 (Sud 1) in cell physiology and organ growth. Sud1 mediates the hydroxylation of RPS23, thereby affecting ribosomal function (translation fidelity). Sud1 inhibition might provoke defects in stop-codon recognition that in turn leads to protein misfolding, accumulation of protein aggregates and consequent UPR activation. UPR in turn induces autophagy and apoptosis, thereby affecting cell and tissue growth.
The parallel studies performed in yeast,Citation12 and to a lesser extent in mammalian cells,Citation11 suggest that RPS23 hydroxylation by the Sud1 homologs –Tpa1p in yeast, and OGFOD1 in mammals- might be necessary for mRNA translation accuracy. Before speculating on possible links between impaired translation accuracy and activation of UPR in Drosophila, we will briefly discuss how accuracy is attained, and how might hydroxylation of RPS23 impinge on it.
Protein Synthesis and Translation Accuracy
Protein synthesis is one of the most energy consuming processes in the cell, and can be divided into 4 steps: 1. Initiation, 2. Elongation, 3. Termination and 4. Ribosome recycling. Initiation is regulated at the level of pre-initiation complex assembly. This complex is composed of the 40S small subunit of the ribosome, the initial fMet-tRNA, and several translation initiation factors. The pre-initiation complex recognizes and binds to the 5′-terminal m7G structure of the mRNA (m7G CAP) to initiate 5′ UTR scanning until the first AUG codon is found. Once the initiation codon is recognized, the large subunit of the ribosome assembles with the small subunit and the elongation phase begins.Citation15,16 After the fMet-tRNA occupies the P site of the ribosome, the A site receives the second aminoacyl-tRNA. The correct aminoacyl tRNA is selected by the ribosome in a process called translation decoding that occurs in the so-called decoding center of the ribosome. During mRNA translation, each amino acid is added to the nascent polypeptide chain, in a cycle composed by three steps: 1. Selection of the cognate aminoacyl tRNA; 2. Formation of the peptide bond; and 3. Shift of the ribosome to the next codon of the mRNA. This cycle is repeated until the ribosome reaches a stop codon and the termination phase takes place. In the termination phase, different termination factors contribute to the separation of ribosome subunits from the mRNA and to the release of the newly synthetized polypeptide. Defects in codon recognition by the ribosome can lead either to incorporation of wrong aminoacyl-tRNAs at the decoding center during the elongation phase, or to stop codon read-through in the termination phase.
Pioneering studies in the field demonstrated that the three-dimensional structure of the decoding center is crucial for translation fidelity.Citation17,18 Studies in yeast and bacteria revealed that mutations in RPS28 or RPS12 (RPS23 homologs in S. cerevisiae and E. coli respectively) provoke increased stop codon read-through, while other mutations in the same proteins provoke reduced stop codon read-through.Citation19-21 Crystallographic studies of mammalian and yeast ribosomes indicate that the prolyl residue of RPS23 that is hydroxylated by Sud1/OGFOD1/Tpa1p forms the apex of a conserved loop that projects toward the decoding center.Citation22,23
Interestingly, studies in yeast revealed that Tpa1p catalyzed hydroxylation of RPS23 is important to ensure proper stop codon recognition. However, in contrast to previously characterized translational accuracy modulators such as antibiotics or the translation termination factor eRF3, hydroxylation of RPS23 can either increase or decrease stop codon read-through, depending on the sequence context that surrounds the stop codon.Citation12 These results support the notion that the conformation of the decoding center might affect translation termination efficiency in a bidirectional manner. On the other hand, mammalian OGFOD1 hydroxylates proline 64 of RPS23, homologous to yeast Pro62. In this case, a role of the dioxygenase in translation accuracy is not so evident. Analysis of stop codon read-through revealed no effect or a modest increase in Ogfod1 knockout MEFs or U2O2 cells expressing Ogfod1 shRNA.Citation11 These results suggest that it is unlikely that RPS23 hydroxylation is necessary for general translation fidelity in higher eukaryotes. We cannot rule out however, that RPS23 hydroxylation is important for translation accuracy of specific mRNAs or for general translation fidelity under certain stress conditions.
From the above observations, one possibility is that in Drosophila RPS23 hydroxylation contributes to translation fidelity of certain transcript. In this line, one can postulate that inhibition of RPS23 hydroxylation following sud1 silencing leads to synthesis of mutated proteins, thereby provoking ER stress and subsequent UPR activation. Preliminary assessment of stop codon read-through using a transgenic bicistronic reporter seems to support this notion: sud1 knock-down larvae exhibited a small but consistent increase in UGA stop codon read-through, but only in certain points of the Drosophila life cycle.
Concluding Remarks and Perspectives
Recently, a subfamily of 2OG dependent dioxygenases, the so-called Ribosomal Oxygenases (ROXs), has been defined and found to modulate protein synthesis through the hydroxylation of ribosomal proteins and tRNAs.Citation24-26 Feng T. et al. (2014)Citation27 reported recently that optimal translation termination depends on the hydroxylation of the termination factor eRF1 by the 2OG-dependent dioxygenase Jmjd4. These works highlight the importance of 2OG dependent dioxygenases in protein synthesis regulation. Our work in Drosophila, along with the two papers published simultaneouslyCitation9,11,12 has strengthened the emerging paradigm that ROXs play an important role in the regulation of translation through the modification of different components of the protein synthesis machinery. An exciting possibility that deserves thorough investigation is as whether or not the activity of ROXs or other 2OG dioxygenases can be modulated by 2OG and/or O2 availability in a physiological context. If this were the case, the cellular environment could affect protein synthesis fidelity or other functions fulfilled by 2OG dioxygenases.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Matthew Cockman for critical reading of this manuscript.
Funding
This work was funded by Wellcome Trust grant WT087675MA and ANPCyT grant PICT 2011 Nº 0090.
References
- Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci 2011; 36:7-18; PMID:20728359; http://dx.doi.org/10.1016/j.tibs.2010.07.002
- Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol 2008; 4:152-6; PMID:18277970; http://dx.doi.org/10.1038/nchembio0308-152
- McDonough MA, Loenarz C, Chowdhury R, Clifton IJ, Schofield CJ. Structural studies on human 2-oxoglutarate dependent oxygenases. Curr Opin Struct Biol 2010; 20:659-72; PMID:20888218; http://dx.doi.org/10.1016/j.sbi.2010.08.006
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 2001; 294:1337-40; PMID:11598268; http://dx.doi.org/10.1126/science.1066373
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 2001; 107:43-54; PMID:11595184; http://dx.doi.org/10.1016/S0092-8674(01)00507-4
- Lavista-Llanos S, Centanin L, Irisarri M, Russo DM, Gleadle JM, Bocca SN, Muzzopappa M, Ratcliffe PJ, Wappner P. Control of the hypoxic response in Drosophila melanogaster by the basic helix-loop-helix PAS protein similar. Mol Cell Biol 2002; 22:6842-53; PMID:12215541; http://dx.doi.org/10.1128/MCB.22.19.6842-6853.2002
- Centanin L, Ratcliffe PJ, Wappner P. Reversion of lethality and growth defects in Fatiga oxygen-sensor mutant flies by loss of hypoxia-inducible factor-alpha/Sima. EMBO Rep 2005; 6:1070-5; PMID:16179946; http://dx.doi.org/10.1038/sj.embor.7400528
- Frei C, Edgar BA. Drosophila cyclin D/Cdk4 requires Hif-1 prolyl hydroxylase to drive cell growth. Dev Cell 2004; 6:241-51; PMID:14960278; http://dx.doi.org/10.1016/S1534-5807(03)00409-X
- Katz MJ, Acevedo JM, Loenarz C, Galagovsky D, Liu-Yi P, Pérez-Pepe M, Thalhammer A, Sekirnik R, Ge W, Melani M, et al. Sudestada1, a Drosophila ribosomal prolyl-hydroxylase required for mRNA translation, cell homeostasis, and organ growth. Proc Natl Acad Sci U S A 2014; 111:4025-30; PMID:24550463; http://dx.doi.org/10.1073/pnas.1314485111
- McDonough MA, Li V, Flashman E, Chowdhury R, Mohr C, Liénard BM, Zondlo J, Oldham NJ, Clifton IJ, Lewis J, et al. Cellular oxygen sensing: Crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2). Proc Natl Acad Sci U S A 2006; 103:9814-9; PMID:16782814; http://dx.doi.org/10.1073/pnas.0601283103
- Singleton RS, Liu-Yi P, Formenti F, Ge W, Sekirnik R, Fischer R, Adam J, Pollard PJ, Wolf A, Thalhammer A, et al. OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc Natl Acad Sci U S A 2014; 111:4031-6; PMID:24550447; http://dx.doi.org/10.1073/pnas.1314482111
- Loenarz C, Sekirnik R, Thalhammer A, Ge W, Spivakovsky E, Mackeen MM, McDonough MA, Cockman ME, Kessler BM, Ratcliffe PJ, et al. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc Natl Acad Sci U S A 2014; 111:4019-24; PMID:24550462; http://dx.doi.org/10.1073/pnas.1311750111
- Morata G, Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol 1975; 42:211-21; PMID:1116643; http://dx.doi.org/10.1016/0012-1606(75)90330-9
- Pomar N, Berlanga JJ, Campuzano S, Hernández G, Elías M, de Haro C. Functional characterization of Drosophila melanogaster PERK eukaryotic initiation factor 2alpha (eIF2alpha) kinase. Eur J Biochem 2003; 270:293-306; PMID:12605680; http://dx.doi.org/10.1046/j.1432-1033.2003.03383.x
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731-45; PMID:19239892; http://dx.doi.org/10.1016/j.cell.2009.01.042
- Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol 2010; 10:24-35; PMID:20029446; http://dx.doi.org/10.1038/nri2685
- Alksne LE, Anthony RA, Liebman SW, Warner JR. An accuracy center in the ribosome conserved over 2 billion years. Proc Natl Acad Sci U S A 1993; 90:9538-41; PMID:8415737; http://dx.doi.org/10.1073/pnas.90.20.9538
- Lodmell JS, Dahlberg AE. A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science 1997; 277:1262-7; PMID:9271564; http://dx.doi.org/10.1126/science.277.5330.1262
- Murgola EJ, Adelberg EA. Streptomycin-suppressible lethal mutations in Escherichia coli. J Bacteriol 1970; 103:20-6; PMID:4912524
- Gorini L, Kataja E. STREPTOMYCIN-INDUCED OVERSUPPRESSION IN E. COLI. Proc Natl Acad Sci U S A 1964; 51:995-1001; PMID:14215657; http://dx.doi.org/10.1073/pnas.51.6.995
- Sharma D, Cukras AR, Rogers EJ, Southworth DR, Green R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J Mol Biol 2007; 374:1065-76; PMID:17967466; http://dx.doi.org/10.1016/j.jmb.2007.10.003
- Chandramouli P, Topf M, Ménétret JF, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure 2008; 16:535-48; PMID:18400176; http://dx.doi.org/10.1016/j.str.2008.01.007
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae–tRNA-ribosome and subunit-subunit interactions. Cell 2001; 107:373-86; PMID:11701127; http://dx.doi.org/10.1016/S0092-8674(01)00539-6
- Ge W, Wolf A, Feng T, Ho CH, Sekirnik R, Zayer A, Granatino N, Cockman ME, Loenarz C, Loik ND, et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat Chem Biol 2012; 8:960-2; PMID:23103944; http://dx.doi.org/10.1038/nchembio.1093
- Noma A, Ishitani R, Kato M, Nagao A, Nureki O, Suzuki T. Expanding role of the jumonji C domain as an RNA hydroxylase. J Biol Chem 2010; 285:34503-7; PMID:20739293; http://dx.doi.org/10.1074/jbc.M110.156398
- Chowdhury R, Sekirnik R, Brissett NC, Krojer T, Ho CH, Ng SS, Clifton IJ, Ge W, Kershaw NJ, Fox GC, et al. Ribosomal oxygenases are structurally conserved from prokaryotes to humans. Nature 2014; 510:422-6; PMID:24814345
- Feng T, Yamamoto A, Wilkins SE, Sokolova E, Yates LA, Münzel M, Singh P, Hopkinson RJ, Fischer R, Cockman ME, et al. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol Cell 2014; 53:645-54; PMID:24486019; http://dx.doi.org/10.1016/j.molcel.2013.12.028
