Abstract
Genetically modified (GM) mustard line (V4) with increased carotenoid content was compared with native mustard to find the difference in allergenic potential, if any. Simulated gastric fluid (SGF) digestibility of crude protein extract from GM as well as its native counterpart mustard crop was envisaged to understand the intended or unintended changes in GM crop along with IgE immunoblotting. BALB/c mice were used as model for allergenicity studies for monitoring total and specific IgE, specific IgG1, histamine level, histopathology, and systemic anaphylaxis score. Allergenicity of mustard was checked in humans by clinical history, skin prick test and IgE levels. Similar results were evident by significant increase in total IgE, specific IgE, IgG1, histamine levels, in GM and native mustard in comparison to control group. Prominent anaphylactic symptoms (score 2: 60%; score 3: 20%; score 4: 20% in native mustard and score 2: 40%; score 3: 40%; score 4: 20% in GM mustard) and eruptive histopathological changes were observed in both GM and native mustard when compared with controls. One protein of approximately 16 kDa was found stable up to 1 h in both GM as well as non GM mustard. IgE immunoblotting detected three protein components of approximately 29, 24 and 16 kDa in both GM and non GM varieties. Collectively, our data demonstrate substantially equivalent allergic responses against GM as well as its native counterpart. Therefore, the GM mustard may be as safe as its native counterpart with reference to allergenic responses.
Abstract
Introduction
Food allergy is an important health concern and getting global attention. It is an immune provocation induced by innocuous food in susceptible individuals.Citation1 Food allergy appears to be increasing in its prevalence and is estimated to affect four to eight per cent of the population.Citation2 As many as 29,000 cases of anaphylactic reactions to different foods are treated in the emergency departments of US hospitals each year and 200 deaths occur every year due to food induced anaphylaxis reactions.Citation3 Advancement in biotechnology has resulted in an increasing number of genetically modified food crops with improved characteristics as compared with the native variety. These GM crops may have better insect, herbicide tolerance properties, or have more nutritional value. However, there is increasing perception that these GM crops should be evaluated for various attributes including allergenic responses. For example, it has been reported that GM soybean containing a Brazil nut protein has improved protein content but retained its allergenicity.Citation4 Also, GM corn with cry9c gene has allergenic properties, but no immunoglobulin E (IgE) was found in patients’ sera.Citation5 Thus it is becoming increasingly essential to assess the allergenic potential of novel gene product according to standard guidelines to ensure safety of GM crop before their commercialization.Citation6,Citation7
Mustard (Brassica juncea) is one of the most widespread crops, mainly grown in Canada, United States, Great Britain, Hungary, Africa, Pakistan and India. It is one of the widely used spices and condiments all over the world. Mustard seeds are rich source of oil (48%), protein (43%), also rich in selenium and omega-3-fatty acid.Citation8 Mustard (rapeseed) oil is the third largest produce among edible oils and accounts for approximately 12% of the total world's edible oil production.Citation9,Citation10 India is the world’s fourth-largest edible oil producer after the USA, China and Brazil.Citation11 Recently, genetically modified mustard (GM Mustard) lines have been developed by overexpression of phytoene synthase (psy) from Zea mays and phytoene desaturase (crtI) from Erwinia uredovora driven by a seed-specific napin promoter, using Agrobacterium-mediated transformation to increase carotenoid content.Citation12-Citation14 However, comprehensive safety evaluation is required to assess the allergenic potential of GM mustard.
We assessed the allergenic potential of GM mustard expressing psy and crtI gene and compared with its native counterpart to identify intended/unintended changes due to genetic modification.
Using murine model, we have compared the immunological response obtained both with native and GM mustard crop, to assess the allergenicity of GM variety with respect to its native counterpart. We used different approaches including bioinformatics study, pepsin digestibility assay, IgE-immunoblotting, estimation of different immunoglobulin levels and histopathological analysis of lung, intestine and spleen to unravel the allergenic potential of GM mustard and compared with its native counterpart. Additionally, human studies were also performed to observe prevalence of native mustard allergenicity. The present study provides valuable information regarding the mustard allergy in consumer group and may be helpful for allergenicity assessment of other newly expressed gene products.
Results
Prevalence of mustard allergy in humans
Skin tests were performed in 300 patients with allergic symptoms and out of these, 225 patients (75%) showed sensitivity to various common allergens such as pollens, fungi, house dust and mites. Eight patients (3.5%) showed SPT positive results against mustard. In mustard sensitized patients’ total IgE level ranged between 876 to 2600 IU/mL, while in healthy individuals it ranged between 66 and 700 IU/mL. Specific IgE level against allergenic mustard proteins in mustard sensitized individuals ranged between 0.91 to 1.92 OD (492 nm), while in healthy individuals it was in the range of 0.065–0.092 ().
Table 1. Details of allergic patients demonstrating mustard allergy
Bioinformatics analysis of inserted psy and crtI genes
FASTA computer algorithm was used for the sequence alignment studies. Comparison of inserted gene product with well-known allergens listed in AllergenOnline database was done over a sliding window of 80 amino acids. Gene product of psy and crtI genes did not show sequence homology with any known allergen. Similarly, the identification of exact matches of any eight amino acid long sequence of psy gene and crtI product with known allergens were done by AllergenOnline database and no matches were found with known allergens ().
Table 2. Bioinformatics analysis of inserted gene psy and crtI product
SGF Assay and densitometry analysis of native as well as transgenic mustard
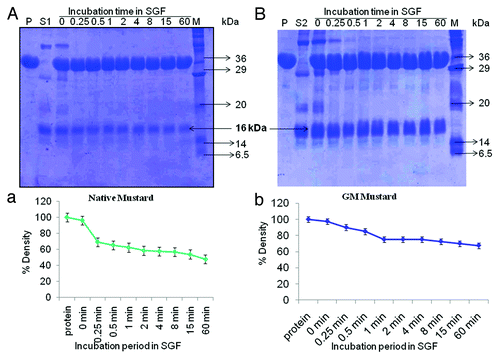
One protein of molecular weight 16 kDa was found to be pepsin resistant in simulated gastric fluid both in native as well as transgenic mustard (). Densitometry analysis of SDS-PAGE of SGF assay results revealed a similar pattern in both native as well as GM mustard and their densities were more than 50% after 1 h ().
IgE immunoblot of native and transgenic mustard CPE
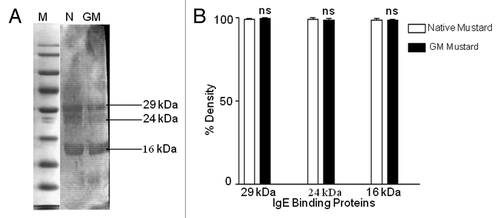
Three proteins of mol wt 29, 24 and 16 kDa have shown binding to IgE immunoglobulin (). The densitometry analyses of IgE immunoblots have shown bands with similar density in GM and native mustard (). Protein extracts of mustard were further tested for allergenicity using native and GM mustard sensitized sera. Immunoblotting of SDS-PAGE separated mustard proteins with sensitized mice sera showed three IgE binding proteins of mol wt 29, 24 and 16 kDa. The SGF stable mustard protein of mol wt 16 kDa showed IgE binding both to native as well as GM mustard.
Figure 2. IgE Immunoblotting and densitometry analysis: (A) native mustard (indicated by N) and GM mustard (indicated by GM) using mice sera showed three protein bands of mol wt 29, 24 and 16 kDa. M is denoted for mol wt marker. (B) Densitometry analysis of IgE immunoblot of both native and GM mustard. Results are expressed as mean ± SEM from three separate experiments (ns = non significant).
Total /specific IgE and specific IgG1 measurement
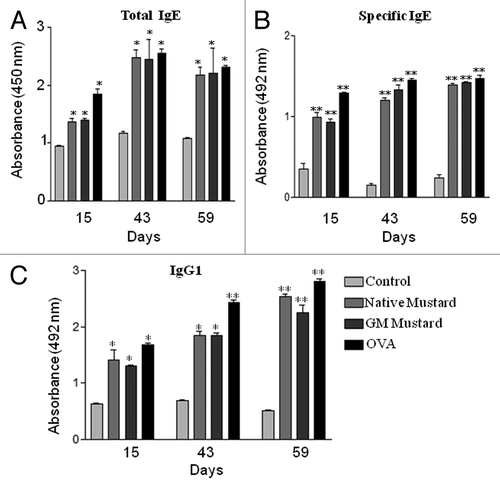
As shown in , total IgE levels in both native and GM mustard CPE (p < 0.05) sensitized groups were higher as compared with control group (PBS treated). The native and GM mustard CPE elicited high (p < 0.01) specific IgE response on day 15, 43 and 59 when compared with control (). A similar trend was observed for protein specific IgG1 levels with p value < 0.05 ().
Systemic anaphylaxis score and histamine level
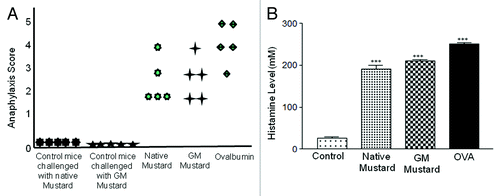
Hypersensitive symptoms became evident within 20 to 30 min after protein challenge (native, GM mustard protein/or OVA in respective sensitized groups, separately). The severe anaphylactic symptoms as expected were observed in positive control group of OVA sensitized mice with 40% mortality, whereas, mice sensitized separately with native and GM mustard showed lesser reaction without any mortality. In native and GM mustard, 60% mice showed score 3, 20% score 2 and 20% score 1. However, control mice did not show any hypersensitivity symptoms either with native or GM mustard when challenged on day 60 (). Histamine, a prominent mediator of anaphylaxis, was investigated in plasma of sensitized mice after challenge. A 5-fold increase in histamine level was found is native as well as mice treated with GM mustard as compared with control group after challenge ().
Figure 4. Anaphylaxis score and histamine level: Mice challenged on day 60. Anaphylaxis score and histamine levels were measured after 30 min (A) Anaphylaxis score in control, native mustard, GM mustard, Ovalbumin. (B) Histamine level in native mustard, GM mustard and OVA. Results are expressed as mean ± SEM from three separate experiments (***p < 0.001) when compared with control.
Histopathological studies
Histopathological analysis indicated effect of native vs GM mustard on lungs, spleen and intestine. HE (hematoxylin - eosin) stained lung section of mice exposed to native and GM mustard revealed distortion of epithelium and thickening of alveolar septa. Peribronchial and perivascular lymphoid proliferation in addition to narrowing of bronchial lumen was evident in native as well as GM mustard treated group ( and ). Cellular infiltration and lumen narrowing were more prominent in lung sections of OVA treated mice (). The presence of lymphoid hyperplasia, which is a characteristic histological feature of the spleen in response to an allergen, reflected massive proliferation and differentiation of T lymphocytes.
Figure 5. Histopathology of lungs, spleen and intestine of control group and sensitized mice with different proteins: (A) lung of control mice; (B) spleen of control mice; (C) intestine of control mice; (D) lung of native mustard; (E) spleen of native mustard; (F) intestine of native mustard sensitized mice; (G) lung of GM mustard; (H) spleen of GM mustard; (I) intestine of GM mustard sensitized mice; (J) lung of OVA; (K) spleen of OVA; (L) intestine of OVA sensitized mice.
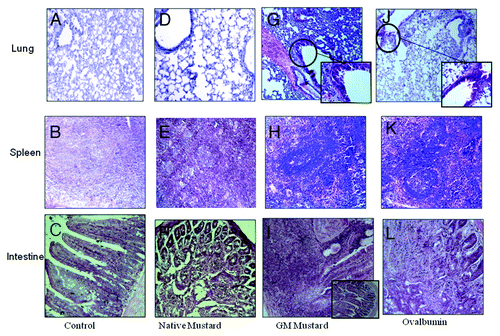
The lung section from PBS treated mice showed normal airway epithelia with occasional infiltrates (). Histopathology of spleen from native and GM mustard sensitized/ challenged group showed a few large megakaryocytes throughout the parenchyma ( and ), whereas, positive control treated with OVA showed lymphoid hyperplasia, large megakaryocytes and fibrous tissue proliferation (). Normal histological structure with distinct red and white pulp was observed in spleen of control animals (). Histopathology of mice intestine in these animals treated with native and GM mustard showed mucosal exfoliation and goblet cell hyperplasia ( and ), while animals treated with OVA showed mild hypertrophy of sub-mucosal glands (). The control animals had normal intestine histology ().
Discussion
In the present study, we have assessed the allergenic potential of both GM and native mustard by comparing the allergenic responses elicited by them in animals. Various international agencies like Codex, OECD, FAO, WHO have given guidelines for safety assessment of genetically modified crops and their products.
Prevalence of mustard allergy among allergic patients was evident by elevated specific IgE levels, positive skin prick tests and higher level of total IgE in these patients. These data are supported by earlier reports regarding several food allergies including mustard allergy.Citation15,Citation16
The Sequence homology with known allergenic proteins for newly expressed gene product using in silico approach to identify allergenicity is an initial requirement. Search for 80 amino acids alignments by FASTA, 8 amino acids exact matches and search for full-length alignments by FASTA have showed no matching with any allergen indicated that chances of allergic reactions are very minimum. In silico evaluation of a protein by the FASTA algorithm helps to predict about the clinically relevant cross reactive allergen. Use of other softwares such as Algpred and/or ADFS may also be helpful as they will reduce false allergenic prediction to a great extent.Citation17 Therefore, allergenicity prediction should be confirmed by in vivo and in vitro laboratory methods for allergenicity testing.
The stability to pepsin digestion is one of the important factors contributing to the allergenicity of food proteins.Citation18,Citation19 Our earlier studies also indicated that proteins stable in SGF for > 2 min were able to bind to specific IgE of allergic patient’s sera in case of soybean and chickpea.Citation20,Citation21 According to DBT, India guidelines (2008), proteins with more than 10% stainable full-length protein band remaining at > 30 to 60 min may be considered stable.Citation22 In both native and GM mustard the density of 16 kDa pepsin resistant protein was more than 50% even after 1 h incubation. This stability may be important for both sensitization of the immune system after reaching mucosa and, for the elicitation of gastrointestinal symptoms of food allergy.
To confirm the allergenicity of mustard, in vivo studies were carried-out using BALB/c mice as this mouse strain has been reported to be susceptible to allergic reactions.Citation23,Citation24 In this study, mice were sensitized through intraperitoneal route, as it is one of the most appropriate routes for allergenicity assessment of food allergens.Citation23 The levels of total IgE, specific IgE and IgG1 observed in native and GM mustard treated sera were comparable and higher than control. IgE plays an important role in mediating immediate-type food allergy both in humans as well as animal models.Citation25,Citation26 Th2 cells help in production of IgE and IgG1 antibodies that are responsible for induction of allergic reactions, while Th1 cells antagonize these responses by inducing the production of IgG2a antibody.Citation21 Therefore, allergic responses are characterized by elevated IgE and IgG1 levels. These data put together demonstrate comparable ability of native and GM mustard to elicit allergenic responses.
In the present study, mice pre-sensitized with native and GM mustard on challenge showed visible signs of anaphylaxis. These systemic anaphylactic symptoms involved multiple target organs, including gut and respiratory system. The level of histamine in plasma was higher in both native and GM mustard sensitized/challenge mice as compared with control. Histamine plays a major role in the pathogenesis of allergy process.Citation27 Upon allergen mediated cross linking of mast cells bound IgE; histamine and other mediators are released and contribute to the symptoms of anaphylaxis. However, histamine is reported to be best characterized mediator among the other inflammatory substances released from the mast cells in the acute phase of immediate type reactions. Taken together, these results indicate comparable allergenicity inducing potential of both native and GM mustard.
Histopathological examination of the intestine confirmed that there was an anaphylactic response in the gut after mice were challenged with native as well as GM mustard, as revealed by mucosal exfoliation which is a characteristic feature of diarrhea commonly reported in case of anaphylaxis. These histological changes underpin the modulated levels of cytokine and immunoglobulin, proposing toward allergenic potential of GM mustard which is comparable to that of native counterpart. These symptoms are very common in allergic reaction reported by several authors.Citation21,Citation25,Citation26
This study is an effort to evaluate the allergenic potential of GM mustard proteins. Both native and GM mustard demonstrated comparable allergenic responses in mice. Presence of only one pepsin resistant protein of 16 kDa in both GM and native crop may also be the reason for inducing similar allergic responses in mice. Comparable induction of IgE, IgG1, histamine levels, histopathological changes and anaphylaxis scores in mice for both native and GM mustard also supports substantially equivalent responses. Taken together, our findings confirmed that native and GM mustard elicit equal allergenic responses in mice. Hence, GM mustard with psy and crtI gene products did not demonstrate any un-intended effects in mice when compared with its native counterpart. However, efforts are underway to understand the mechanism of allergic reaction and to pin point the role played by individual allergenic proteins of native and GM mustard in causing allergenicity.
Materials and Methods
Materials
All the chemicals were of highest grade purity available. Pepsin from porcine gastric mucosa (Cat No. P-6887), O-phenylenediamine (OPD), bovine serum albumin (BSA), reagents for sodium dodecyl sulfate– PAGE (SDS–PAGE) and goat anti-human IgE peroxidase conjugate (Cat No. A9667) were purchased from Sigma Chemical Company, St. Louis, USA. The anti-human IgE antibody preparation is specific for human IgE when tested against purified human IgG, by manufacturer using Ouchterlony Double Diffusion (ODD) and immunoelectrophoresis (IEP). Goat anti-mouse IgG1 and IgE-HRP linked were obtained from Southern Biotech, Birmingham, USA. All the other chemicals used were of the highest purity available from other commercial sources. GM Mustard seeds were provided by The Energy and Resources Institute, New Delhi, India and the same lot was used throughout the study. The dry seeds were stored at 5–8°C and 25–30% relative humidity. Seeds were put in a water proof container with a thin layer of colored silica gel desiccant as indicator of dryness.
Screening of mustard sensitive patients
Human study was performed to unravel the prevalence of allergy to mustard in susceptible individuals according to previously reported method.Citation25 Skin prick tests (SPT) were performed with allergen kit (Alicit India Private Limited) on nasobronchial allergic patients (n = 300, 225 males and 75 females) at the Department of Pulmonary Medicine, Chhatrapati Shahuji Maharaj Medical University (CSMMU), Lucknow. The results were read 20 min after allergens prick. Weal diameter equal to positive control or more were considered as marked positive skin reactions.Citation28 If smaller weal as compared with positive control then +1, equal +2, larger 3 and for bigger with pseudopodia then +4. Blood was collected from mustard SPT positive patients and normal healthy volunteers. Serum was separated and stored at -80°C ().
Total IgE level in mustard sensitive patients
Serum total IgE level in patients was determined using commercially available double antibody sandwich ELISA kit following manufacturers’ instructions (Immunology Consultants Laboratory, Inc., Newberg, USA). IgE concentration was calculated using the standard curve and presented as IU/ml (1 IU/ml = 2.4 ng/ml). Total IgE levels were measured twice in triplicate and results are expressed as mean ± SEM.
Specific IgE levels in mustard sensitive patients
Specific IgE levels against native mustard were estimated by enzyme-linked immunosorbent assay (ELISA) according to the method described earlier.Citation26 Briefly, the wells of microtiter plate (Nunc, Denmark) were coated with 1 µg of native or GM mustard protein in carbonate buffer (pH 9.6) separately. After blocking, the plates were incubated with diluted sera (1:10 v/v) of healthy and mustard-hypersensitive patients overnight at 4°C. The plate was washed and incubated with HRP-conjugated goat-anti-human IgE (1:1000 v/v, Sigma Chemical Co., St. Louis, USA) for 3 h at 37°C. Color was developed using orthophenylenediamine (OPD). The reaction was stopped after 20 min by adding 5 N H2SO4 and the absorbance was read at 492 nm. Specific IgE levels were measured in triplicate and results are expressed as mean ± SEM.
Bioinformatics analysis of GM mustard
There are number of guidelines for allergenicity assessment of new proteins incorporated into the food crops.Citation29-Citation31 Bioinformatics is one of the criteria for allergenicity assessment of transgenic proteins. Sequence homology studies were performed for gene psy and crtI inserted into mustard using “Allergen Online” software < www.allergenonline.com>. Allergen Online database provides allergen list and sequence searchable database for identifying proteins that may present a potential risk of allergenic cross-reactivity and help in assessing the safety of proteins that may be introduced into foods through genetic engineering or other methods.
Search for full-length alignments of psy and crtI gene by FASTA
FASTA is commonly used bioinformatic tool for phylogenetic analysis of any protein or nucleotide sequences. If the overall FASTA alignment with identity matches greater than 50% indicates possible cross-reactivity with other allergens. Bioinformatics analysis of proteins introduced in genetically engineered crops is an important initial step to assess the potential allergenicity of the protein and sequence homology is one of the recommended methods.Citation32
Preparation of crude protein extracts (CPE)
Crude proteins from native as well as GM mustard seeds were extracted as described by Astwood et al., (1996) with slight modifications.Citation33 Briefly, seeds were powdered with grinder and defatted with n-hexane. Defatted flour was macerated for 2 min with grinder in the phosphate buffer (20mM Na2HPO4, 2 mM KH2PO4, 5.4 mM KCl, 0.5 M NaCl, pH 7.0). The mixture was agitated overnight at 4°C, centrifuged for 30 min at the 10,000 g, supernatant recovered, filtered through 0.45 μm syringe filter. Aliquots were stored at –80°C until used. Protein content of CPE was estimated according to Lowry method with BSA as standard.Citation34
BALB/c mice
Healthy 6–8 week old female BALB/c mice (22 ± 3 g) were obtained from the CSIR-IITR, Lucknow, India animal breeding colony. Mice were maintained under standard laboratory conditions in pathogen free environment and fed mustard free diet.
BALB/c mice treatment protocol
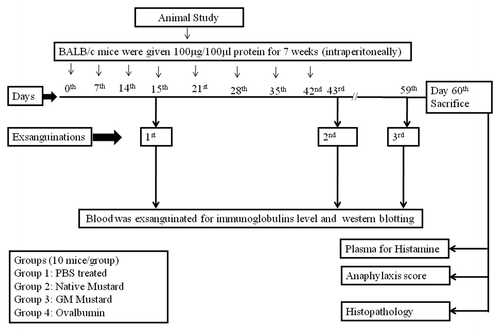
Six to eight week old BALB/c mice were randomly divided into four groups of 10 mice each. After two weeks of acclimatization period, group 1, 2, 3 and 4 were given phosphate buffer saline (PBS), native mustard, GM mustard and OVA by intraperitoneal route, respectively. Mice were given 100 μg proteins (native mustard, GM mustard and OVA) in 100 μl PBS once a week for 7 weeks according to method described earlier.Citation26 Blood samples were collected from retro-orbital sinus to measure levels of serum total / specific IgE and IgG1 antibodies at different time intervals (15, 43 and 59 d). Mice in each group were challenged on day 60 and the symptoms were scored. Challenged mice in each group were sacrificed. Lungs, intestine and spleen were taken out for histopathological analysis and fixed in 10% formalin in PBS ().
Simulated Gastric Fluid (SGF) assay and densitometry analysis
SGF was prepared as described by U. S. Pharmacopia and performed as described earlier.Citation26,Citation33,Citation35 In brief; 3.2-mg pepsin was dissolved in 1 mL of 0.03 M NaCl, pH 2.0 for preparation of SGF. Pepsin activity was expressed as “change in absorbance (ΔA280) of 0.001 per min at pH 2.0 at 37°C, measured as TCA-soluble products using hemoglobin as substrate is equivalent to one unit (of pepsin activity).” SGF (430 μl) was incubated at 37°C prior to addition of 86 μl of test protein solution. Reaction was stopped at different time points (0.25, 0.5, 1, 2, 4, 8, 15 and 60 min) by the addition of the 60 μl of the incubation mixture to 60 μl of stopping solution, that is 2× Tris-tricine SDS sample buffer (containing 200mM dithiothreitol, 4% SDS, 0.2% bromophenol blue, 20% glycerol and 100mM Tris, pH 8.8). The 15 μl/well incubation mixture of native mustard while 30 μl/well GM mustard (double amount was taken to check out any possibility of extra pepsin resistant protein in GM mustard) were subjected to SDS-PAGE gel electrophoresis.Citation36 Standard molecular weight marker (Sigma, Wide Range, M.W. 6.5–205 kDa, Cat # M-4038) were included on all gels to calculate the molecular weight of the proteins. Coommasie Brilliant Blue (G-250) staining was used to assess the extent of digestibility.
Gel image was captured using Syngene Bio Imaging System (Syngene, Cambridge, UK) and the resulting undigested protein bands were subjected to densitometry analysis for validation of SGF results using the GENETOOLS software (Syngene), in which raw volume of the bands was recorded. The raw volume was defined as a three-dimensional size of the band where intensity was the third dimension.
IgE immunoblotting
To detect IgE binding proteins immunoblotting was performed using sera of native and GM mustard treated mice according to the method described earlier.Citation26 Mustard CPE (native as well as GM mustard) were resolved onto 12% SDS-PAGE. Resolved proteins were electrophoretically transferred onto PVDF membrane with pore size 0.45 μm (Immobilon-P, Millipore Co) according to method described earlier for 2 h at 0.8 mA/ cm2 in transfer buffer at room temperature with a semidry blotting unit (Amersham Biosciences, San Francisco, USA).Citation37 After termination of the electrophoretic transfer, the blot was cut into strips and each strip was blocked with 5% defatted milk in 2 TBS-T buffer (TRIS-HCl 20 mM, 500 mM NaCl, pH 7.4) containing 0.1% Tween-20) overnight at 4°C. After washing 3× (5 min each), with TBS-T, the strips were incubated in sera of native and GM mustard treated mice overnight at 4°C (dilution 1:10 in TBST + 1% BSA). After 3 washes (5 min each) with TBS-T, the strips were incubated for 6 h at room temperature with goat anti-mouse IgE peroxidase conjugated (Sothern Biotech, dilution 1:1000 in TBST + 1% BSA). After two additional washes (5 min each), the IgE binding proteins were visualized with 3–30-diaminobenzidine tetrahydrocholride (DAB system). The densitometry analysis was performed to observe relative density of native and GM mustard immunoblot using the GENETOOLS software (Syngene).
Total IgE level
Total IgE level was estimated by using ELISA kit (BD biosciences) according to manufacturers’ instructions.
Specific IgE and IgG1 level
Serum IgE and IgG1 antibodies specific for native and GM mustard CPE were measured using ELISA method.Citation38 Briefly, the microtiter plates (Maxisorp; NuncTM Immunomodule, Roskilde, Denmark) were coated with 1 µg protein in 100 µl per well in coating buffer (carbonate buffer, pH 9.6) and left overnight at 4°C. Subsequently, the plates were washed thoroughly with PBS-T (PBS + 0.05% Tween-20), blocked with 250 µl of blocking solution (1% BSA in PBS-T, w/v) for 2 h at 37°C. After washing, the plates were incubated with diluted native and GM mustard treated mice sera for IgE (1:10) and IgG1 (1:500) estimation. Similar dilutions of serum from PBS treated group were added in separate well and run along to serve as negative controls. The plates were incubated for two hours at 37°C and then washed three times with washing buffer (PBS-T). To each well, 100 µl of either HRP-conjugated rat anti-mouse IgE or IgG1 antibodies (1:1000; Southern Biotech, Birmingham, USA) were added and incubated for 1 h at 37°C. The plates were washed with washing buffer. A 50 µl substrate solution (5 mg ortho-phenylenediamine in 10 ml substrate buffer and 10 µl H2O2) was added to each well and the plates were incubated for 30 min at 37°C in dark. The reaction was stopped by the addition of 50 µl of stopping solution (5N H2SO4) and the absorbance was taken at 492 nm in ELISA plate reader (Spectra Max, Molecular Devices, USA).
Systemic anaphylaxis score
Mice were challenged intraperitoneally with 10 mg of native/ GM mustard CPE or OVA on day 60. Anaphylactic reactions were scored after 30 min using a previously described scoring system 0, no symptoms; 1, scratching and rubbing around the snout and head; 2, puffiness around the eyes and snout, pilar erection, diarrhea, and reduced activity or standing still with an increased respiratory rate; 3, wheezing, labored respiration, and cyanosis around the mouth; 4, symptoms as in no; 3 with loss of consciousness, tremors, and/or convulsion; 5, death.Citation39
Histamine release assay
On 60th day, blood was collected in chilled ethylenediamine tetra acetic acid (EDTA) coated tubes, 30 min after challenge, for estimation of histamine level. Plasma histamine levels were determined using immunoassay kit (SPI-BIO, Montigny le Bretonneux, France) according to the manufacturers’ protocol.
Histopathology
The mice were sacrificed after challenge by cervical dislocation. Spleen, lungs and intestine were taken for histological analysis. Tissues were fixed in 10% formalin in PBS, embedded in paraffin, and cut into 3–5-μm thick sections.
Statistical Analysis
The data are expressed as mean ± SEM. All the data were analyzed using software package from Instat version 3·0 (Graphpad, San Diego, CA, USA; www.graphpad.com). Differences between values were compared by using one-way analyses of variance (ANOVA) with Bonferroni as a post hoc test. A value of p < 0.05 was considered statistically significant.
Ethical permissions
SPT and sera collection were carried-out with patient’s consent and the study protocol was approved by the human ethics committee of the CSMMU, Lucknow, India. Animal study was performed after approval of animal ethical committee of IITR, Lucknow, India.
Acknowledgments
We are grateful to Director of our Institute for his keen interest in this study. We are thankful to Monsanto for the technology to develop GM mustard. We cordially acknowledge Dr. Rajendra Prasad, Head, Department of Pulmonary Medicine, CSM Medical University, Lucknow for providing blood sample of healthy volunteers and allergic patients. Authors thank Mr. B.D. Bhattacharjee for his help in editing of the manuscript. AM, SK and AKV are thankful to ICMR and CSIR, New Delhi, respectively for their Senior Research Fellowships.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding Sources
This work is supported by supra institutional project (SIP-08) of Council of Scientific and Industrial Research (CSIR), New Delhi and United States Agency for International Development (USAID).
References
- Kumar S, Verma AK, Das M, Dwivedi PD. Allergenic diversity among plant and animal food proteins. Food Rev Int 2012; 28:277 - 298; http://dx.doi.org/10.1080/87559129.2011.635391
- Sicherer SH. Food allergy. Mt Sinai J Med 2011; 78:683 - 96; http://dx.doi.org/10.1002/msj.20292; PMID: 21913199
- Sampson HA. Anaphylaxis and emergency treatment. Pediatrics 2003; 111:1601 - 8; PMID: 12777599
- Nordlee JA, Taylor SL, Townsend JA, Thomas LA, Bush RK. Identification of a Brazil-nut allergen in transgenic soybeans. N Engl J Med 1996; 334:688 - 92; http://dx.doi.org/10.1056/NEJM199603143341103; PMID: 8594427
- Bucchini L, Goldman LR. Starlink corn: a risk analysis. Environ Health Perspect 2002; 110:5 - 13; http://dx.doi.org/10.1289/ehp.021105; PMID: 11781159
- Lack G, Chapman M, Kalsheker N, King V, Robinson C, Venables K, BSACI working party. Report on the potential allergenicity of genetically modified organisms and their products. Clin Exp Allergy 2002; 32:1131 - 43; http://dx.doi.org/10.1046/j.1365-2222.2002.01464.x; PMID: 12190648
- Kumar S, Misra A, Verma AK, Roy R, Tripathi A, Ansari KM, et al. Bt brinjal in India: a long way to go. GM Crops 2011; 2:92 - 8; http://dx.doi.org/10.4161/gmcr.2.2.16335; PMID: 21865863
- Mustard seeds, WHFoods. Retrieved on 2012-1- 24 from www.whfoods. com/ genpage.php?tname=foodspice&dbid=106.
- Edible oil, Ministry of food processing industries, retrieved Sept 14, 2012, from http://mofpi.nic.in/Content Page.aspx? Category Id=687.
- Mustard seed oil, retrieved Sept 15, 2012, from http:// www.Commodityonline.com/ commodities/oil-oilseeds/mustardseedoil.php.
- Report on GPDP project in edible oil industry in India, retrieved Sept 15, 2012, from http://www.apo-tokyo.org/gp/31pagpdp/rama/rama_tech01.pdf.
- Li F, Vallabhaneni R, Yu J, Rocheford T, Wurtzel ET. The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol 2008; 147:1334 - 46; http://dx.doi.org/10.1104/pp.108.122119; PMID: 18508954
- Cong L, Wang C, Chen L, Liu H, Yang G, He G. Expression of phytoene synthase1 and carotene desaturase crtI genes result in an increase in the total carotenoids content in transgenic elite wheat (Triticum aestivum L.). J Agric Food Chem 2009; 57:8652 - 60; http://dx.doi.org/10.1021/jf9012218; PMID: 19694433
- Al-Babili S, Hoa TT, Schaub P. Exploring the potential of the bacterial carotene desaturase CrtI to increase the beta-carotene content in Golden Rice. J Exp Bot 2006; 57:1007 - 14; http://dx.doi.org/10.1093/jxb/erj086; PMID: 16488912
- Misra A, Prasad R, Das M, Dwivedi PD. Prevalence of legume sensitization in patients with naso-bronchial allergy. Immunopharmacol Immunotoxicol 2008; 30:529 - 42; http://dx.doi.org/10.1080/08923970802135294; PMID: 18618316
- Vereda A, Sirvent S, Villalba M, Rodríguez R, Cuesta-Herranz J, Palomares O. Improvement of mustard (Sinapis alba) allergy diagnosis and management by linking clinical features and component-resolved approaches. J Allergy Clin Immunol 2011; 127:1304 - 7; http://dx.doi.org/10.1016/j.jaci.2011.01.020; PMID: 21349575
- Verma AK, Misra A, Subash S, Das M, Dwivedi PD. Computational allergenicity prediction of transgenic proteins expressed in genetically modified crops. Immunopharmacol Immunotoxicol 2011; 33:410 - 22; http://dx.doi.org/10.3109/08923973.2010.523704; PMID: 20964517
- Besler M, Steinhart H, Paschke A. Stability of food allergens and allergenicity of processed foods. J Chromatogr B Biomed Sci Appl 2001; 756:207 - 28; http://dx.doi.org/10.1016/S0378-4347(01)00110-4; PMID: 11419714
- Thomas K, Aalbers M, Bannon GA, Bartels M, Dearman RJ, Esdaile DJ, et al. A multi-laboratory evaluation of a common in vitro pepsin digestion assay protocol used in assessing the safety of novel proteins. Regul Toxicol Pharmacol 2004; 39:87 - 98; http://dx.doi.org/10.1016/j.yrtph.2003.11.003; PMID: 15041142
- Misra A, Prasad R, Das M, Dwivedi PD. Probing novel allergenic proteins of commonly consumed legumes. Immunopharmacol Immunotoxicol 2009; 31:186 - 94; http://dx.doi.org/10.1080/08923970802578782; PMID: 19514993
- Verma AK, Kumar S, Tripathi A, Chaudhari BP, Das M, Dwivedi PD. Chickpea (Cicer arietinum) proteins induce allergic responses in nasobronchial allergic patients and BALB/c mice. Toxicol Lett 2012; 210:24 - 33; http://dx.doi.org/10.1016/j.toxlet.2012.01.011; PMID: 22285431
- DBT. (2008). Protocols for food and feed safety assessment of GE crops: Department of Biotechnology, Ministry of science and technology, Government of India.
- Dearman RJ, Stone S, Caddick HT, Basketter DA, Kimber I. Evaluation of protein allergenic potential in mice: dose-response analyses. Clin Exp Allergy 2003; 33:1586 - 94; http://dx.doi.org/10.1046/j.1365-2222.2003.01793.x; PMID: 14616873
- Dearman RJ, Caddick H, Stone S, Basketter DA, Kimber I. Characterization of antibody responses induced in rodents by exposure to food proteins: influence of route of exposure. Toxicology 2001; 167:217 - 31; http://dx.doi.org/10.1016/S0300-483X(01)00462-0; PMID: 11578801
- Kumar S, Verma AK, Misra A, Tripathi A, Chaudhari BP, Prasad R, et al. Allergenic responses of red kidney bean (Phaseolus vulgaris cv chitra) polypeptides in BALB/c mice recognized by bronchial asthma and allergic rhinitis patients. Food Res Int 2011; 44:2868 - 79; http://dx.doi.org/10.1016/j.foodres.2011.06.043
- Misra A, Kumar R, Mishra V, Chaudhari BP, Raisuddin S, Das M, et al. Potential allergens of green gram (Vigna radiata L. Millsp) identified as members of cupin superfamily and seed albumin. Clin Exp Allergy 2011; 41:1157 - 68; http://dx.doi.org/10.1111/j.1365-2222.2011.03780.x; PMID: 21631610
- López-Expósito I, Song Y, Järvinen KM, Srivastava K, Li XM. Maternal peanut exposure during pregnancy and lactation reduces peanut allergy risk in offspring. J Allergy Clin Immunol 2009; 124:1039 - 46; http://dx.doi.org/10.1016/j.jaci.2009.08.024; PMID: 19895992
- Aggarwal S, Chhabra SK, Saxena RK, Agarwal MK. Heterogeneity of immune responses to various Aspergillus species in patients with allergic respiratory diseases. Indian J Chest Dis Allied Sci 2000; 42:249 - 58; PMID: 15597671
- FAO/WHO. Evaluation of allergenicity of genetically modified foods. Report of a joint FAO/WHO expert consultation on allergenicity of foods derived from biotechnology. (Food and Agriculture Organization of the United Nations (FAO), Rome, 2001.
- Codex Alimentarius Commission. Alinorm 03/34: Joint FAO/WHO Food Standard Programme, Codex Alimentarius Commission, Twenty-Fifth Session, Rome, 30 June–5 July, 2003. Appendix III, Guideline for the conduct of food safety assessment of foods derived from recombinant-DNA plants and Appendix IV, Annex on the assessment of possible allergenicity, 2003.
- Ladics GS, Bannon GA, Silvanovich A, Cressman RF. Comparison of conventional FASTA identity searches with the 80 amino acid sliding window FASTA search for the elucidation of potential identities to known allergens. Mol Nutr Food Res 2007; 51:985 - 98; http://dx.doi.org/10.1002/mnfr.200600231; PMID: 17639511
- Metcalfe DD, Astwood JD, Townsend R, Sampson HA, Taylor SL, Fuchs RL. Assessment of the allergenic potential of foods derived from genetically engineered crop plants. Crit Rev Food Sci Nutr 1996; 36:Suppl S165 - 86; http://dx.doi.org/10.1080/10408399609527763; PMID: 8959382
- Astwood JD, Leach JN, Fuchs RL. Stability of food allergens to digestion in vitro. Nat Biotechnol 1996; 14:1269 - 73; http://dx.doi.org/10.1038/nbt1096-1269; PMID: 9631091
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem 1951; 193:265 - 75; PMID: 14907713
- The United States Pharmacopoeia 23. Simulated gastric fluid, TS. In The National Formulary 18; Board of trustees, Eds. United States Pharmacopoeia Convention, Inc. Rockville, MD, 1995; pp. 2053.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227:680 - 5; http://dx.doi.org/10.1038/227680a0; PMID: 5432063
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 1979; 76:4350 - 4; http://dx.doi.org/10.1073/pnas.76.9.4350; PMID: 388439
- Misra A, Kumar R, Mishra V, Chaudhari BP, Tripathi A, Das M, et al. Partial characterization of red gram (Cajanus cajan L. Millsp) polypeptides recognized by patients exhibiting rhinitis and bronchial asthma. Food Chem Toxicol 2010; 48:2725 - 36; http://dx.doi.org/10.1016/j.fct.2010.06.047; PMID: 20600518
- Li XM, Schofield BH, Huang CK, Kleiner GI, Sampson HA. A murine model of IgE-mediated cow’s milk hypersensitivity. J Allergy Clin Immunol 1999; 103:206 - 14; http://dx.doi.org/10.1016/S0091-6749(99)70492-6; PMID: 9949309