Abstract
Since insect mostly developed on decaying matter and contaminated fruits, they are constantly ingesting bacteria. The insect model, Drosophila, is therefore well adapted to study the interactions that take place between the gut epithelia and either resident or infectious bacteria. In order to provide an ad hoc immune response, gut epithelial cells must detect the presence of bacteria. In a recent report, Bosco-Drayon et al. identify the main receptors by which Drosophila sense gut associated bacteria and analyze how this bacteria-receptor interaction translate into gene activation.
Introduction
In multicellular animals, the presence of a multilayer array of defenses against surrounding microbes is the rule. In most cases, the first protective screen is a physical barrier such as the skin in mammals or the exoskeleton in insects. Once this layer of defense has been breached, various cells are responsible for the elimination of the invaders and hence the containment of the infection. In Drosophila, hemocytes are the bulwark of the cellular response as by phagocytizing them, they clear the circulating hemolymph (blood of insects) of contaminating microorganisms. These “blood cells” are assisted by the fat body cells (analogous to liver cells) which are the main producers of antimicrobial peptides. Host cells are also in permanent contact with microorganisms in various epithelia. Regardless of the route by which they enter the host and the type of cells by which they are eliminated, invasive microbes must first be recognized by the host immune sentinels. Previous works have taught us that the Drosophila genome encodes proteins whose function is specifically dedicated to microbial recognition.Citation1 We shall below restrict our topic to the molecules and mechanisms implicated in bacteria recognition. The best-characterized Drosophila pattern recognition receptors constitute a family of 19 proteins named peptidoglycan recognition proteins (PGRPs). In contrast to PGRPs from other taxa which exhibit broad range of binding abilities (for example camel PGRP-S binds LPS, LTA, PGN, Fatty acid..),Citation2 Drosophila PGRPs affinity seems to be restricted to PGN. Interactions between these sentinel proteins and bacterial PGN are the main triggers that initiate immune pathway activation in flies.Citation3 How this recognition step is organized and what are the consequences for both the host and the bacteria following this PGN-PGRP interaction is the focus of this addendum.
The Drosophila Immune Pathways
Once a bacteria has entered the body cavity of the fly, it can preferentially trigger either the Toll or the IMD pathway, two NFκB dependent signaling cascades. The nature of the PGN that composes the cell wall of the incoming bacteria dictates the pathways that are activated in fat body cells.Citation4 If the PGN is of Lys-type (as in most Gram positive bacterial cell walls), it will be sensed by the blood-borne PGRP-SA receptor.Citation5 Direct binding between Lys-Type PGN and PGRP-SA will lead to Toll pathway activation by its ligand Spätzle. Hence, in contrast to mammalian cells where Toll-Like-Receptors act as pattern recognition receptors (PRRs), this function is provided by the circulating PGRP-SA protein that is genetically upstream of the Toll transmembrane protein itself. If the hemolymph is now contaminated with Gram-negative bacteria or bacilli (whose cell wall is made of DAP-type PGN), the fat body cell will activate the IMD pathway.Citation6 In this case, the PGN-PGRP interaction does not take place in the circulating blood but rather at fat body cell surface through the PGRP-LC transmembrane receptor.Citation7-Citation9 Since PGRP-LC combines the functions of PRR and of signaling receptor, its interaction with DAP-type PGN activates the downstream NFκB dependent IMD signaling.Citation10 Thus, the ability of Drosophila to discriminate between DAP-type and Lys-Type PGN containing bacteria and to translate this dichotomous recognition into ad hoc responses, relies on the recognition of different forms of peptidoglycan by specific members of the PGRP family. It should be noted that not all PGRP family members act as PRR upstream of signaling pathways. Among the 19 Drosophila PGRP proteins, 5 have enzymatic activity that allows them to cleave pro-inflammatory peptidoglycan into non-stimulatory muropeptides.Citation11 By cleaving PGN into non-active fragments, PGRP with amidases enzymatic activity (such as PGRP-SC and PGRP-LB) reduce the amount of immune elicitor and, in turns, tuned-downed the immune response.Citation11-Citation16
In addition to inducing a systemic immune response when present in the hemolymph, bacteria can also trigger responses when in contact with intestinal epithelia. These so-called “local responses” can be either immunological if elimination of the bacteria from the gut is required or tolerigenic to favor the maintenance of microbiota that are beneficial for the host.Citation3 Indeed, recent works have shown that although Drosophila microbiota is much simpler that the mammalian gut community, some gut associated bacterial species impact essential physiological parameters such as the control of larval growth control or adult mating preference.Citation17-Citation19 It is therefore essential that enterocytes distinguish between infectious and commensal bacteria in order to either eliminate or tolerate them. How the Drosophila gut cells detect the presence of bacteria and how this is translated into gene activation in the gut epithelium was the focus of a recent study.Citation19
Immune Response Can Be Activated in Gut Domains that are Devoid of Bacteria
In this work, axenic flies were fed with a commensal bacteria, Lactobacillus plantarum (L. p.) which colonize within the gut and persist in the lumen, or with the phytopathogenic Erwinia carotovora carotorova (E.c.c.) which are rapidly eliminated from the gut. Using this experimental set-up, we first monitored bacteria localization within the midgut following bacteria ingestion. As the Drosophila midgut can be functionally subdivided into four domains along the anteroposterior axis (proventriculus, ventriculus, acidic copper cell region and posterior midgut) we tested whether bacteria would equally colonize the different domains (). This was indeed not the case since both E.c.c. and L.p. were mainly concentrated in anteriormost regions (proventriculus, ventriculus) and completely absent from the posterior midgut. This prompted us to test the consequences of this patchy bacteria distribution on immune activation. For this, we monitored IMD target gene transcription along the infected guts. To our surprise, we noticed that IMD targets were upregulated in all midgut domains, even those which were bacteria free. This unexpected result could be potentially explained if anteriorly localized bacteria were releasing PGN fragments that are transported along the gut lumen and eventually reach the posterior domains. This hypothesis was corroborated by further experiments in which the ingested bacteria were substituted by purified muropeptides such as TCT (tracheal cytotoxin, minimal elicitor of IMD pathway activation). Flies fed with TCT showed a gut immune response that was very similar to that obtained with live bacteria. It is highly probable that PGN degrading enzymes such as lysozyme and PGRPs with enzymatic activities play an important role in transforming bacterial PGN into smaller muropeptides that are then transported along the gut.
Figure 1. Model for bacteria detection by the Drosophila midgut. Ingested bacteria pass through the foregut (pharynx and esophagus) and reach the midgut that is made of four domains from anterior to posterior, the proventriculus, the ventriculus, the Copper cells region and posterior midgut. Bacteria are concentrated in the anterior domains but are able to release PGN fragments that can reach more posterior regions. It is unclear whether cell wall PGN or PGN released by bacteria are detected by the same mechanisms in the gut. One could imagine that one is detected by PGRP-LC while the other is a PGRP-LE ligand. The immune response in the proventriculus is fully PGRP-LC dependent and antagonized by PGRP-LE. It mainly consists of the production of antimicrobial peptides and is thought to regulate bacterial entry into the gut lumen. In the ventriculus, both PGRP-LE and –LC are required for IMD pathway activation. The more posterior domains that are devoid of bacteria are PGRP-LE dependent as far as IMD pathway activation is concerned. Activation of the NFkB pathway in these domains mainly leads to the production of IMD pathway negative regulators that by dampening the signaling protect the host from excessive immune response and prevent spreading of the immune response to remote tissues such as the fat body. This is a simplified model as AMP and INR are produced in all domains but at different levels. Some unanswered questions remain: (1) How does PGN reach or activate intracellular PGRP-LE? (2) How does PGRP-LE antagonize IMD pathway? (3) Why are both PGRP-LC and –LE required to activate IMD signaling in the ventriculus? (4) Why are AMPs mainly produced in anterior domains and INR in posterior domains? Does PGRP-LC and PGRP-LE activation lead to same transcriptional output?
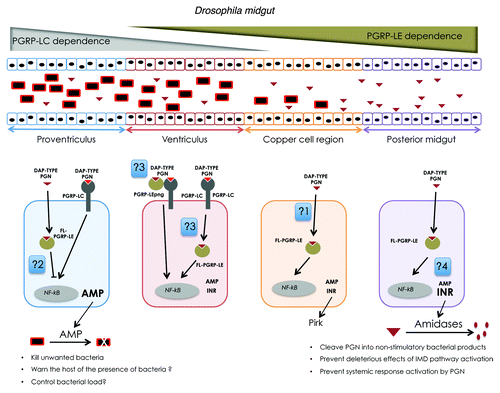
Gut IMD Pathway Activation in Not Fully PGRP-LC Dependent and Requires PGRP-LE
The fact that live bacteria in anterior domains and putative muropeptides in posterior ones, can both activate immune pathways could imply that different recognition mechanisms are required to detect bacteria along the gut length. Based on results obtained in fat body cells, PGRP-LC was the first receptor whose putative implication in gut PGN detection was tested. Infected guts from wild type, NFκb and PGRP-LC mutants were dissected into domains in which IMD target gene transcription was measured. The results show that in contrast to what is happening in fat body cells, IMD pathway activation is clearly not fully PGRP-LC dependent in the gut. While anteriormost domains (proventriculus, ventriculus) are fully PGRP-LC dependent as far as IMD pathway activation is concerned, PGRP-LC inactivation had no effect on bacteria-induced immune response in the Copper cell region and the posterior midgut. We then tested whether PGRP-LE could function as an alternative receptor in the PGRP-LC independent domains. It has been previously shown that, as for PGRP-LC, PGRP-LE is able to accommodate DAP-type PGN and its overexpression is sufficient to promote IMD downstream signaling.Citation20-Citation22 Using PGRP-LE null mutant flies, we could demonstrate that PGRP-LE is the only required bacteria receptor in both the Copper cell region and the posterior midgut. The situation is however complicated by the fact that some regions depend on both receptors for PGN detection. Indeed, elimination of either PGRP-LC or PGRP-LE renders the ventriculus insensitive to bacteria. Such an observation, which implies that the absence of one receptor cannot be compensated by the other, is compatible with a model in which PGRP-LE and PGRP-LC are two subunits of a heterodimeric receptor. It should be mentioned experiments in cell culture have shown that a truncated form of PGRP-LE can act as a PGRP-LC co-receptor for PGN detection.Citation20 Whether such a scenario takes place in the ventriculus will need further investigation. The co-requirement of PGRP-LE and -LC could also be interpreted as reflecting the epistatic relationship between the two receptors. The respective roles of PGRP-LE and -LC in the proventriculus were also at odds with data from the literature. If IMD pathway activation in the proventriculus was positively regulated by PGRP-LC, it was antagonized by PGRP-LE. This latter function of PGRP-LE as a negative regulator has not yet been described in other immune-competent tissues and neither its mechanism nor its role are yet understood. These results indicate that each gut domain responds specifically to bacteria and that the responses are differentially transduced by PGRP-LE or PGRP-LC, depending on the domain. While PGRP-LC seems essential in anterior domains, it becomes progressively dispensable in more posterior regions. In contrast, PGRP-LE that is acting as a negative regulator in the anteriormost domain is a required bacteria sensor in the posterior part of the midgut. There seems to be a reciprocal gradient of PGRP-LE and -LC requirement for PGN detection along the anteroposterior axis of the gut (). The requirement of both PGRP-LC and PGRP-LE in bacteria sensing by the gut epithelium was also reported by Neyen et al.Citation23
Why are Gut Cells Using Two Distinct PGN Receptors to Detect Bacteria?
The need for using two different receptors to sense bacteria is intriguing especially since PGRP-LE and PGRP-LC activation are supposed to both lead to IMD pathway activation. Before speculating on the respective functions of these two PGN sentinels, it is important to remind that PGRP-LC and PGRP-LE, while activating the same transducing pathway, have different subcellular localizations. While PGRP-LC is a membrane-associated protein, the full length PGRP-LE protein is an intracytoplasmic molecule whose precise subcellular localization remains unknown. It should also be mentioned that if IMD pathway activation leads to the expression of immune effectors such as antimicrobial peptides that will kill bacteria, it also triggers the production of negative regulators (INR: IMD pathway negative regulators) that will dampen IMD pathway activation and protect the host from an excessive immune activation.Citation11,Citation12,Citation15 Among these negative regulators are the PGRPs with enzymatic activities and the Pirk protein that regulates PGRP-LC subcellular distribution.Citation24-Citation26 By integrating different parameters such as bacteria localization of the gut, requirement of PGRPs along the A/P axis (PGRP-LC in anterior domains and -LE in posterior domains) and the type of IMD target genes that are induced in the different gut domains, we would like to propose the following model for gut immune response to bacteria. Since the proventriculus and ventriculus are the first domains to come in contact with bacteria streaming down from the esophagus, it is expected that they are equipped with very rapid and sensitive mechanisms of bacteria detection. Using a membrane-associated receptor such as PGRP-LC whose extracellular PGRP domain will be in direct contact with PGN circulating in the gut lumen, is certainly the most sensitive and rapid system to inform the host of the presence of bacteria. This sensitive and rapid response will mainly function to warn the host of their presence, preparing it to eliminate unwanted bacteria and to control bacterial load. Accordingly, using fluorescent constructs reporting IMD signaling activation, we could show that the proventriculus is the gut domain in which the IMD pathway is the most frequently activated gut domain under normal (non-infected) conditions. As one could expect, this primary response, mainly PGRP-LC dependent, should favor the production of AMP rather that immune pathway negative regulators. This is indeed the case. What then happens is this first line of defense is not efficient enough? We hypothesize that IMD activation by PGRP-LE dependent cells will be less rapid (since the PGN itself or a secondary messenger has to enter the cell in order to activate PGRP-LE) and probably less sensitive and require more PGN input to activate the pathway. In our model, IMD pathway, in more posterior domains, will only be activated by high levels of PGN. This could be reached after a severe infection or in response to the persistence of bacteria in the gut lumen, thus when the anteriormost domains has been overwhelmed. As previously shown, not all bacteria are able to survive in the gut, but commensal bacteria such as L. p. persist for days in the fly gut. Consistent with the proposed model, we could show that axenic flies whose gut is recolonized with L. p.Citation23 exhibit a strong response in the posterior midgut. Very interestingly, this response which is as expected, fully PGRP-LE dependent, consist mainly of the transcription of genes encoding negative regulators of the IMD pathway such as PGRP-LB or PGRP-SC1. This suggests that PGN sensing by the posterior midgut through PGRP-LE will induce proteins whose long-term function is rather to establish tolerance toward endogenous microbiota rather that to kill bacteria. In the case of mild bacterial infection, they will be taken care of by the anterior domains. Killed bacteria will have no time to proliferate and PGN fragments will not reach more posterior domains, thus will not activate PGRP-LE in posterior midgut. In the case of severe infection, the anterior immune response will be overflowed and PGN will reach posterior domains. PGRP-LE sensing will induce the production of INR to prevent host damage due to excessive immune response. As far as commensal are concerned, these are probably ignored or tolerated by the PGRP-LC response. However, their constant presence leads to PGRP-LE dependent production of negative regulators which facilitate immune tolerance and prevent damage to the host.
Although this model is built on data from multiple studies, answers to the following questions are still required to refine it and even probably to correct it. Are PGRP-LE and PGRP-LC activated by the same PGN elicitors? How can PGRP-LE and -LC both be equally required in the ventriculus region? How does PGRP-LE antagonize IMD pathway activation in the proventriculus? Why is IMD pathway activation in the different gut domains not necessarily leading to the same transcriptional output? How does extracellular PGN reach intracellular PGRP-LE? How is bacteria localization regionalized in the gut? How does the gut distinguish between infectious and commensal bacteria? Much more work is yet required to shed light on these important issues that will allow us to better appreciate the mechanisms by which Drosophila gut senses bacteria and tolerates only some of them.
Acknowledgments
We thank Marie Meister for critical comments on the manuscript. Julien Royet's research is supported by CNRS, ANR, FRM and AFM.
Disclosure of Potential Conflicts of Interest
No potential conflict was disclosed.
References
- Royet J, Gupta D, Dziarski R. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat Rev Immunol 2011; 11:837 - 51; PMID: 22076558
- Sharma P, Dube D, Sinha M, Yadav S, Kaur P, Sharma S, et al. Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid. PLoS One 2013; 8:e53756; http://dx.doi.org/10.1371/journal.pone.0053756; PMID: 23326499
- Ferrandon D. The complementary facets of epithelial host defenses in the genetic model organism Drosophila melanogaster: from resistance to resilience. Curr Opin Immunol 2013; 25:59 - 70; http://dx.doi.org/10.1016/j.coi.2012.11.008; PMID: 23228366
- Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol 2003; 4:478 - 84; http://dx.doi.org/10.1038/ni922; PMID: 12692550
- Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 2001; 414:756 - 9; http://dx.doi.org/10.1038/414756a; PMID: 11742401
- Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, et al. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 2004; 20:637 - 49; http://dx.doi.org/10.1016/S1074-7613(04)00104-9; PMID: 15142531
- Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, et al. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 2002; 416:640 - 4; http://dx.doi.org/10.1038/nature734; PMID: 11912488
- Choe KM, Werner T, Stöven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 2002; 296:359 - 62; http://dx.doi.org/10.1126/science.1070216; PMID: 11872802
- Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 2002; 416:644 - 8; http://dx.doi.org/10.1038/nature735; PMID: 11912489
- Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci U S A 2005; 102:1122 - 6; http://dx.doi.org/10.1073/pnas.0404952102; PMID: 15657141
- Mellroth P, Karlsson J, Steiner H. A scavenger function for a Drosophila peptidoglycan recognition protein. J Biol Chem 2003; 278:7059 - 64; http://dx.doi.org/10.1074/jbc.M208900200; PMID: 12496260
- Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog 2006; 2:e14; http://dx.doi.org/10.1371/journal.ppat.0020014; PMID: 16518472
- Paredes JC, Welchman DP, Poidevin M, Lemaitre B. Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 2011; 35:770 - 9; http://dx.doi.org/10.1016/j.immuni.2011.09.018; PMID: 22118526
- Zaidman-Rémy A, Poidevin M, Hervé M, Welchman DP, Paredes JC, Fahlander C, et al. Drosophila immunity: analysis of PGRP-SB1 expression, enzymatic activity and function. PLoS One 2011; 6:e17231; http://dx.doi.org/10.1371/journal.pone.0017231; PMID: 21364998
- Zaidman-Rémy A, Hervé M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 2006; 24:463 - 73; http://dx.doi.org/10.1016/j.immuni.2006.02.012; PMID: 16618604
- Bosco-Drayon V, Poidevin M, Boneca IG, Narbonne-Reveau K, Royet J, Charroux B. Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe 2012; 12:153 - 65; http://dx.doi.org/10.1016/j.chom.2012.06.002; PMID: 22901536
- Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab 2011; 14:403 - 14; http://dx.doi.org/10.1016/j.cmet.2011.07.012; PMID: 21907145
- Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, et al. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 2011; 334:670 - 4; http://dx.doi.org/10.1126/science.1212782; PMID: 22053049
- Sharon G, Segal D, Zilber-Rosenberg I, Rosenberg E. Symbiotic bacteria are responsible for diet-induced mating preference in Drosophila melanogaster, providing support for the hologenome concept of evolution. Gut Microbes 2011; 2:190 - 2; http://dx.doi.org/10.4161/gmic.2.3.16103; PMID: 21804354
- Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol 2006; 7:715 - 23; http://dx.doi.org/10.1038/ni1356; PMID: 16767093
- Takehana A, Yano T, Mita S, Kotani A, Oshima Y, Kurata S. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J 2004; 23:4690 - 700; http://dx.doi.org/10.1038/sj.emboj.7600466; PMID: 15538387
- Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol 2008; 9:908 - 16; http://dx.doi.org/10.1038/ni.1634; PMID: 18604211
- Neyen C, Poidevin M, Roussel A, Lemaitre B. Tissue- and ligand-specific sensing of gram-negative infection in drosophila by PGRP-LC isoforms and PGRP-LE. J Immunol 2012; 189:1886 - 97; http://dx.doi.org/10.4049/jimmunol.1201022; PMID: 22772451
- Kleino A, Myllymäki H, Kallio J, Vanha-aho LM, Oksanen K, Ulvila J, et al. Pirk is a negative regulator of the Drosophila Imd pathway. J Immunol 2008; 180:5413 - 22; PMID: 18390723
- Aggarwal K, Rus F, Vriesema-Magnuson C, Ertürk-Hasdemir D, Paquette N, Silverman N. Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog 2008; 4:e1000120; http://dx.doi.org/10.1371/journal.ppat.1000120; PMID: 18688280
- Lhocine N, Ribeiro PS, Buchon N, Wepf A, Wilson R, Tenev T, et al. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe 2008; 4:147 - 58; http://dx.doi.org/10.1016/j.chom.2008.07.004; PMID: 18692774