Abstract
Clostridium difficile infection (CDI) is a major healthcare-associated disease worldwide. Recurring infections and increasing antibiotic resistance have complicated treatment of CDI. While C. difficile spores are important for transmission and persistence of CDI, other factors such as gut colonization and formation of bacterial communities in the gut may also contribute to pathogenesis and persistence, but have not been well investigated. Recently, we reported that important clinical C. difficile strains are able to form composite biofilms in vitro. C. difficile biofilm formation is a complex process, modulated by several different factors, including cell surface components and regulators. We also reported that bacteria within biofilms are more resistant to high concentrations of vancomycin, the antibiotic of choice for treatment of CDI. Here we summarize our recent findings and discuss the implications of biofilm formation by this anaerobic gut pathogen in disease pathogenesis and treatment.
Introduction
Clostridium difficile is a spore-forming, gram-positive anaerobic bacillus which can cause severe gastrointestinal infections in humans.Citation1C. difficile infection (CDI), one of the predominant nosocomial infections worldwide, usually occurs when the normal intestinal flora is damaged or absent, commonly after the use of antibiotics. The clinical symptoms can range from mild or severe diarrhea to serious inflammatory conditions, including pseudomembranous colitis.Citation1 Recently it was demonstrated that transmission of clostridial disease occurs through spores.Citation2,Citation3 The best-characterized virulence factors of C. difficile are two large clostridial toxins, toxin A (TcdA) and B (TcdB).Citation4-Citation6C. difficile toxins cause disorganization of the cell actin cytoskeleton and tight junctions, induction of apoptosis, fluid accumulation, and destruction of the epithelium.Citation6 Although the toxins are crucial for virulence, in recent years, attention has been focused on bacterial colonization of the gut, especially due to increased instances of recurrent CDI. The adhesin, fibronectin-binding protein A, was shown to play a role in C. difficile colonization,Citation7 and the high- and low-molecular-weight surface layer proteins (SLPs) were predicted to be involved in adherence of C. difficile to host cells during the infection.Citation8,Citation9 The cell wall proteins (CWPs) Cwp66 and Cwp84 were shown to be important in the adherence and degradation of the extracellular matrix.Citation10,Citation11
For several pathogens recurrent infections have been associated with the ability to form sessile surface-associated microbial communities or “biofilms”.Citation12 Bacteria within biofilms are protected and more resistant to different environmental stresses, like antibiotic or oxygen stress.Citation13 Common diseases such as dental caries and periodontitis are caused by bacteria in biofilm and biofilm formation has been connected with persistent tissue infections such as chronic otitis media, chronic rhinosinositis, recurrent urinary tract infections, endocarditis and cystic fibrosis-associated lung infections.Citation14 Recently, biofilms were also associated with chronic inflammatory diseases as Crohn diseaseCitation15 and acute bacterial infections.Citation16,Citation17 Moreover, biofilms represent a big problem when formed on artificial devices used in medicine, such as catheters, stents, orthopedic implants, contact lenses and implantable electronic devices.Citation14,Citation18 Resistance of bacteria within biofilms to antimicrobials makes treatment of disease difficult and unsuccessful. Furthermore, mature biofilms are highly resilient to the action of the innate and adaptive immune defense systems.Citation12
Biofilm formation by individual gut species, particularly anaerobic species, has not been well characterized. In addition to spore formation, a known means of adaptation to stress, it is likely that C. difficile forms microcolonies in vivo to survive the unfavorable environment of the human gut. As biofilm formation by C. difficile, especially by clinically important strains, was not previously characterized, we sought to develop in vitro assays to study C. difficile biofilm formation. Our recent paper in the Journal of Bacteriology describes biofilm formation by clinical C. difficile strains, bacterial proteins and regulators involved in this process and the effects of antibiotics on biofilms.Citation19 We summarize below the findings of this paper and discuss the biological and clinical implications of our findings.
C. difficile Forms Complex, Structured Biofilms In Vitro
We studied two strains, C. difficile strain 630 and strain B1/NAP1/027 R20291, isolated from the Stoke Mandeville outbreak in 2004 and 2005 (R20291). Multiple techniques were employed (confocal microscopy, crystal violet (CV) assay and CFU counts) to demonstrate and quantitate biofilms (). We find that in vitro strain R20291 () forms more biofilm as compared with strain 630 (). Both strains need rich medium for maximum biofilm formation and the addition of glucose increases biofilm formation by 630.Citation19 While the strain R20291 forms maximum biofilm after incubation for 1 day by CV assays and CFU counts, strain 630 has the highest number of CFU on day 1, but when stained with CV shows the maximum biofilm on day 5. This suggests that strain 630 behaves differently to R20291, and may accumulate more biofilm matrix upon longer incubation.Citation19 These in vitro data indicate that the strains may have different behaviors in vivo in terms of biofilm or microcolony formation, which may in turn influence their abilities to persist in the gut.
Figure 1. C. difficile biofilm formation in vitro. (A) Confocal microscopy analysis of biofilms formed by C. difficile R20291. Live/Dead staining shows dead bacteria red and live green, (propidium iodide and Syto 9, respectively). Biofilm was incubated for 3 days. 3D images of biofilms (right panel) depicting biofilm thickness in micrometers. Time course for biofilm formation by strain 630 (B) and R20291 (C) measured by CV staining (bars) and colony counts (CFU/ml, line). The results are presented in log scale, and the error bars represent standard deviations (P < 0.05).The data are representative of at least three independent experiments, each performed in triplicates. (D) Characterization of C. difficile biofilm matrix: 3D confocal microscopy images of R20291 biofilms stained with murine anti-R20291 after incubation for 3 days (E) Biofilms stained with antibodies to a synthetic C. difficile PSII polysaccharide (red) and DAPI (blue), which stains the bacterial DNA.
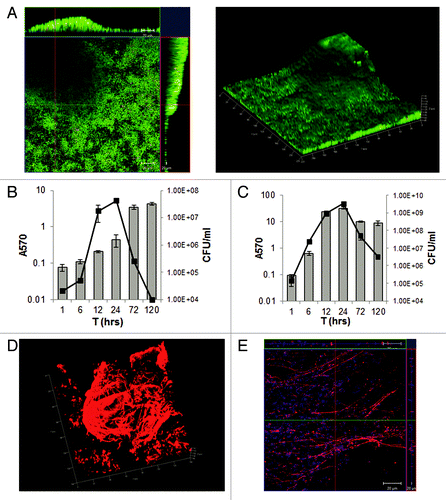
The self-produced biofilm matrix is known to protect bacteria by providing an enclosed environment.Citation20,Citation21 Biofilm matrices are typically made up of extracellular polymeric substance (EPS), which comprises primarily of proteins, DNA and polysaccharide. When C. difficile R20291 was stained with antibody against total bacteria we observed a complex biofilm matrix with just few stained individual bacteria (). This suggested that the compact biofilm may comprise surface-associated or secreted bacterial components and may be impenetrable to antibodies. Proteinase K and DNase I treatments inhibited the formation of biofilms and caused disassembly of pre-formed biofilms. This further showed that proteins as well as external DNA are part of the biofilm matrix.Citation19 Additionally, staining biofilm with a synthetic derivative of the C. difficile surface PSII polysaccharide (), which was reported to be immunogenic recently,Citation22 revealed the presence of polysaccharide components in the biofilm matrix. In all, the data clearly show that C. difficile biofilms are composed of a thick multi-component biofilm matrix. Biofilm impenetrability has been proposed as a feature of bacterial biofilms that contributes to escape of immune responses during the infection, as well as antibiotic resistance in vivo.Citation20,Citation21
C. difficile Biofilm Formation is Multifactorial
We examined selected known virulence or regulatory factors of C. difficile for biofilm formation. Cwp84 is a cysteine protease, which is responsible for maturation of S-layer proteins (SLPs).Citation23,Citation24 A mutant in cwp84, which shows similar growth rates as wildtype in broth culture, is unable to form biofilm, indicating that a mature S-layer, which also hosts several other CWPs, is needed for C. difficile biofilm formation.Citation19 We suppose that the S-layer and/or CWP’s may be involved in early steps of biofilm formation such as adhesion. A C. difficile mutant in flagellin, a principal component of flagella, affects biofilm formation in vitro.Citation19 However, flagella appear to be important at later points of biofilm formation, as the mutant does not display defects in in vitro assays at earlier time points. The role of flagella in biofilm development by motile bacteria varies between species.Citation25,Citation26 Flagella may be involved in the maturation of C. difficile biofilms, as seen for other bacteria such as Pseudomonas sp, where flagella contribute to the architecture of mature biofilms.Citation27 Studies on the functions of clostridial flagella in biofilms need to be performed under conditions of flow, where development and maturation of biofilms can be monitored.
Al-2, one of the major modulators of quorum sensing which is largely conserved across bacterial species,Citation28 has an important role in bacterial biofilm formation.Citation29,Citation30 The C. difficile genome carries a 453-bp gene that encodes a protein which shares 40% identity to the V. harveyi LuxS protein and is responsible for autoinducer-2 (AI-2) production.Citation31 We found that C. difficile LuxS mutant is unable to form biofilm in vitro, suggesting that a luxS-mediated quorum-sensing system may be important for biofilm formation.Citation19 Further work needs to be done to clarify the mechanisms involved in the luxS-mediated control of biofilm formation.
All the mutants were genetically complemented either episomally or chromosomally, and biofilm formation was completely or partly restored.
Master Regulator for Sporulation, Spo0A, is Involved in Clostridium difficile Biofilm Formation
Sporulation and biofilm formation have been linked in other bacteria. The main regulator that controls entry into sporulation is Spo0A, which is well conserved in Bacillus and Clostridium species.Citation32,Citation33 In addition to control of entry into sporulation, Spo0A controls a range of other regulatory factors, including pathways unrelated to sporulation.Citation34 The relation between Spo0A and biofilm formation has been well studied in Bacillus subtilis. Among more than 100 genes that are regulated by Spo0A are genes involved in biofilm matrix expression.Citation35 In the case of B. subtilis, the switch between biofilm formation and sporulation depends on the concentration of phosphorylated Spo0A (Spo0A-P). Intermediate levels of Spo0A-P induce expression of genes for biofilm matrix formation, while high levels induce sporulation in the cell. Initial phosphorylation induces biofilm formation as result of matrix gene expression; once biofilm matures, Spo0A-P accumulates in some cells and activates sporulation.Citation35
Our data, and recent data reported by Dawson et al.,Citation36 demonstrate the importance of spo0A for biofilm formation by C. difficile. Under our in vitro conditions, where biofilms are not mature, we detected very low number of spores. Yet, we have defective biofilm formation for a spo0A mutant.Citation19 Our data indicate that the lack of spo0A may result in decreased adhesion to surfaces in early stages of biofilm formation. Spo0A has also been previously implicated in toxin production in C. difficile, although the exact relationship between the two is currently unclear.Citation33,Citation37 Thus, spo0A appears to control multiple stress-induced pathways in C. difficile. We hypothesize that this regulator may act as a switch between different pathways such as sporulation, biofilm formation, and toxin production, selectively inducing one or more of these depending on the local environment. Examining the spo0A regulon in C. difficile under different stress conditions could provide valuable information about the role/s of this regulator in C. difficile physiology. Indeed, during infection, C. difficile spores may be a major part of mature biofilms, where spore formation is in response to nutrient starvation, as seen in other bacteria.Citation38
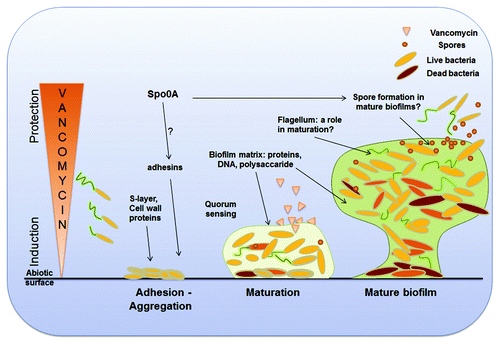
Based on our results, we have summarized the roles of C. difficile factors in a hypothetical model of biofilm development (). Mechanistic studies on how these different proteins mediate biofilm formation and how their functions are coordinated will shed more light on this complex process.
Figure 2. A hypothetical model for C. difficile biofilm development. Bacteria first recognize specific or nonspecific attachment sites on an appropriate surface and adhere to it. An intact S-layer and/or adhesins are important for this initial step of biofilm formation. The regulator of sporulation, Spo0A, controls this step, likely by upregulating expression of adhesins. Quorum sensing mediated by luxS is crucial during the early and late maturation phases when cells start to produce a biofilm matrix composed of proteins, extracellular DNA and polysaccharides. C. difficile flagella have a role in maturation of biofilms and perhaps in the early steps of biofilm formation, in directing bacteria to the right attachment sites. Spores are part of C. difficile biofilms, although numbers of spores may be maximal in mature biofilms where conditions of nutrient stress are likely. C. difficile biofilms can protect bacteria within from the antibiotic vancomycin, while at low concentrations (sub-inhibitory and inhibitory concentrations) biofilm formation is induced.
C. difficile Biofilms and Antibiotics: A Complex Relationship?
One of the main functions of a biofilm is to protect bacteria within from unfavorable conditions such as antibiotics, particularly during infection. A role for bacterial biofilms in resisting antibiotics has been well demonstrated for many pathogenic species e.g., methicillin-resistant S. aureus.Citation39 Resistance to antibiotics in biofilm can increase from 10- to 1,000-fold more compared with planktonic bacteria.Citation40 An increase in the numbers of recurring clostridial infectionsCitation41 and a rise in C. difficile strains resistant to drugs currently used for treatment such as metronidazole and rifampicin have made management of CDI difficultCitation42,Citation43 Antibiotics at minimal concentrations, on the other hand, could also act as stress signals and biofilm formation can be a defensive reaction to the presence of antibiotics, as shown for gram negative bacteria Escherichia coli and Pseudomonas aeruginosa.Citation44,Citation45 Induction of biofilms in presence of sub-inhibitory concentrations of antibiotics has been previously attributed to alterations in the level of c-di-GMP.Citation45 Such induction in vivo could be clinically relevant when there is exposure to low doses of antibiotics, like at the beginning or end of antibiotic therapy, which could perhaps explain ineffective treatment.Citation44
We examined effects of a range of concentrations of vancomycin, an antibiotic used to effectively treat C. difficile infections. Bacteria within biofilms were found to be more resistant to high concentrations of vancomycin (20 μg/mL), and C. difficile biofilm formation was induced when bacteria were exposed to sub-inhibitory and inhibitory concentrations of vancomycin (0.25 μg/mL and 0.5 μg/mL, respectively).Citation19 Thus, in the case of C. difficile, biofilms may have multiple roles depending on the antibiotic concentrations in the environment. In vivo, C. difficile may form biofilms to escape antibiotics used for treatment, resulting in recurrent infections.
Several mechanisms are known to mediate antibiotic resistance of bacteria within biofilms. The biofilm matrix, a main player, can act as an initial physical barrier that affects penetration of antimicrobial agents.Citation40 Other more complex attributes such as the physiological state of the bacterium, like the presence of persistent cells in population, can also contribute to resistance.Citation46 Biofilm matrix has the role of a protective barrier within which bacteria are more tolerant to antibiotics. Concentration gradients are formed across the biofilm matrix decreasing the effective concentration of antibiotics reaching the bacteria within.Citation40 Persistent cells are phenotypical variants of the wildtype in a dormant state and are more resistant to the antibiotics. Upon re-inoculation, persistent bacteria display similar level of tolerance to antibiotics.Citation47 Indeed, genetic mutations could also occur at low frequencies within bacteria in biofilm environments, contributing to antibiotic resistance.Citation48 Genetic changes mediated by systems such as stress responses and slow growth could be responsible for the increased tolerance to the antibiotics.Citation40 In our initial studies on mechanisms involved in vancomycin resistance, we disrupted biofilms and treated them with antibiotics. These bacteria were not resistant to vancomycin anymore, indicating that vancomycin resistance does not involve genetic or inherited changes in the bacteria.Citation19 As seen in other bacterial species, our data indicate that antibiotic resistance of clostridial biofilms may be mediated by the thick biofilm matrix and/or the physiological state of bacteria within biofilms.Citation49 Clearly, much remains to be understood with regard to how clostridial biofilms may resistant antibiotics.
C. difficile Biofilms: A Way to Survive the Gut?
The human large intestine represents a huge variety of bacterial species, constituting an extremely complex and metabolically active site.Citation50 Most studies on the colonic microbiota have been done on planktonic bacteria found in faeces, but the sessile bacteria that form biofilms in the mucus layer of the gut are likely to play a fundamental role in gut health and disease.Citation51 Biofilms formed by anaerobic bacteria from the human intestinal tract have been poorly characterized. One of the reasons for this is perhaps the difficulties associated in cultivation and standardizing conditions for biofilm formation in vitro. Recently the ability to form mono- and duo- species biofilms for several gut bacteria such as Bacteroides oralis, various Clostridium species including C. difficile, Finegoldia magna, and Fusobacterium necrophorum was shown in vitro.Citation52 C. difficile colonization of the gut and mechanisms involved in this process in vivo are poorly understood, but it is likely that formation of large microcolonies or biofilm communities have a key role in gut colonization and bacterial survival. It is also possible that microcolony formation precedes toxin production in the colon. Formation of bacterial mats has been reported previously in murine C. difficile infections.Citation2 Such biofilm or multicellular structures could potentially protect bacterium from cellular immune responses invoked by toxin production and from antibiotics used for the treatment of CDI. In addition to spores, which have been linked to persistence of clostridial disease,Citation2,Citation3 biofilm formation in vivo could be another factor contributing to recurrence of CDI. Investigating biofilm development by C. difficile during infection in vivo is therefore essential for a better understanding of clostridial pathogenesis. In vivo studies with mutants that are selectively defective for sporulation or biofilm formation may help understand the contribution of spores and biofilms to C. difficile persistence. Screening for inhibitors or designing vaccines against proteins crucial for biofilm development could provide effective solutions for treatment or prophylaxis of CDI.
Acknowledgments
The research leading to these results received funding from the European Community's Seventh Framework Programmes “CLOSTNET” (PEOPLE-ITN-2008–237942). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Submitted
04/30/2013
Revised
07/15/2013
Accepted
07/23/2013
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 2009; 7:526 - 36; http://dx.doi.org/10.1038/nrmicro2164; PMID: 19528959
- Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun 2009; 77:3661 - 9; http://dx.doi.org/10.1128/IAI.00558-09; PMID: 19564382
- Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 2012; 80:2704 - 11; http://dx.doi.org/10.1128/IAI.00147-12; PMID: 22615253
- Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. The role of toxin A and toxin B in Clostridium difficile infection. Nature 2010; 467:711 - 3; http://dx.doi.org/10.1038/nature09397; PMID: 20844489
- Lyras D, O’Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, et al. Toxin B is essential for virulence of Clostridium difficile.. Nature 2009; 458:1176 - 9; http://dx.doi.org/10.1038/nature07822; PMID: 19252482
- Voth DE, Ballard JD. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 2005; 18:247 - 63; http://dx.doi.org/10.1128/CMR.18.2.247-263.2005; PMID: 15831824
- Barketi-Klai A, Hoys S, Lambert-Bordes S, Collignon A, Kansau I. Role of fibronectin-binding protein A in Clostridium difficile intestinal colonization. J Med Microbiol 2011; 60:1155 - 61; http://dx.doi.org/10.1099/jmm.0.029553-0; PMID: 21349990
- Calabi E, Calabi F, Phillips AD, Fairweather NF. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect Immun 2002; 70:5770 - 8; http://dx.doi.org/10.1128/IAI.70.10.5770-5778.2002; PMID: 12228307
- Karjalainen T, Waligora-Dupriet AJ, Cerquetti M, Spigaglia P, Maggioni A, Mauri P, et al. Molecular and genomic analysis of genes encoding surface-anchored proteins from Clostridium difficile.. Infect Immun 2001; 69:3442 - 6; http://dx.doi.org/10.1128/IAI.69.5.3442-3446.2001; PMID: 11292772
- Waligora AJ, Hennequin C, Mullany P, Bourlioux P, Collignon A, Karjalainen T. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect Immun 2001; 69:2144 - 53; http://dx.doi.org/10.1128/IAI.69.4.2144-2153.2001; PMID: 11254569
- Janoir C, Péchiné S, Grosdidier C, Collignon A. Cwp84, a surface-associated protein of Clostridium difficile, is a cysteine protease with degrading activity on extracellular matrix proteins. J Bacteriol 2007; 189:7174 - 80; http://dx.doi.org/10.1128/JB.00578-07; PMID: 17693508
- Römling U, Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med 2012; 272:541 - 61; http://dx.doi.org/10.1111/joim.12004; PMID: 23025745
- Davey ME, O’toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 2000; 64:847 - 67; http://dx.doi.org/10.1128/MMBR.64.4.847-867.2000; PMID: 11104821
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science 1999; 284:1318 - 22; http://dx.doi.org/10.1126/science.284.5418.1318; PMID: 10334980
- Claret L, Miquel S, Vieille N, Ryjenkov DA, Gomelsky M, Darfeuille-Michaud A. The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway. J Biol Chem 2007; 282:33275 - 83; http://dx.doi.org/10.1074/jbc.M702800200; PMID: 17827157
- Hannan TJ, Totsika M, Mansfield KJ, Moore KH, Schembri MA, Hultgren SJ. Host-pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic Escherichia coli bladder infection. FEMS Microbiol Rev 2012; 36:616 - 48; http://dx.doi.org/10.1111/j.1574-6976.2012.00339.x; PMID: 22404313
- Kumagai Y, Matsuo J, Cheng Z, Hayakawa Y, Rikihisa Y. Cyclic dimeric GMP signaling regulates intracellular aggregation, sessility, and growth of Ehrlichia chaffeensis.. Infect Immun 2011; 79:3905 - 12; http://dx.doi.org/10.1128/IAI.05320-11; PMID: 21788390
- Lynch AS, Robertson GT. Bacterial and fungal biofilm infections. Annu Rev Med 2008; 59:415 - 28; http://dx.doi.org/10.1146/annurev.med.59.110106.132000; PMID: 17937586
- Ðapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, et al. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile.. J Bacteriol 2013; 195:545 - 55; http://dx.doi.org/10.1128/JB.01980-12; PMID: 23175653
- Beloin C, Roux A, Ghigo JM. Escherichia coli biofilms. Curr Top Microbiol Immunol 2008; 322:249 - 89; http://dx.doi.org/10.1007/978-3-540-75418-3_12; PMID: 18453280
- Watnick PI, Lauriano CM, Klose KE, Croal L, Kolter R. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol Microbiol 2001; 39:223 - 35; http://dx.doi.org/10.1046/j.1365-2958.2001.02195.x; PMID: 11136445
- Adamo R, Romano MR, Berti F, Leuzzi R, Tontini M, Danieli E, et al. Phosphorylation of the synthetic hexasaccharide repeating unit is essential for the induction of antibodies to Clostridium difficile PSII cell wall polysaccharide. ACS Chem Biol 2012; 7:1420 - 8; http://dx.doi.org/10.1021/cb300221f; PMID: 22620974
- Calabi E, Ward S, Wren B, Paxton T, Panico M, Morris H, et al. Molecular characterization of the surface layer proteins from Clostridium difficile.. Mol Microbiol 2001; 40:1187 - 99; http://dx.doi.org/10.1046/j.1365-2958.2001.02461.x; PMID: 11401722
- Kirby JM, Ahern H, Roberts AK, Kumar V, Freeman Z, Acharya KR, et al. Cwp84, a surface-associated cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile.. J Biol Chem 2009; 284:34666 - 73; http://dx.doi.org/10.1074/jbc.M109.051177; PMID: 19808679
- Kobayashi K. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis.. Mol Microbiol 2007; 66:395 - 409; http://dx.doi.org/10.1111/j.1365-2958.2007.05923.x; PMID: 17850253
- Lemon KP, Freitag NE, Kolter R. The virulence regulator PrfA promotes biofilm formation by Listeria monocytogenes.. J Bacteriol 2010; 192:3969 - 76; http://dx.doi.org/10.1128/JB.00179-10; PMID: 20511507
- Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol 2008; 10:2331 - 43; http://dx.doi.org/10.1111/j.1462-2920.2008.01658.x; PMID: 18485000
- Hardie KR, Heurlier K. Establishing bacterial communities by ‘word of mouth’: LuxS and autoinducer 2 in biofilm development. Nat Rev Microbiol 2008; 6:635 - 43; http://dx.doi.org/10.1038/nrmicro1916; PMID: 18536728
- Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol 2005; 3:383 - 96; http://dx.doi.org/10.1038/nrmicro1146; PMID: 15864263
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae.. Mol Microbiol 2003; 50:101 - 4; http://dx.doi.org/10.1046/j.1365-2958.2003.03688.x; PMID: 14507367
- Carter GP, Purdy D, Williams P, Minton NP. Quorum sensing in Clostridium difficile: analysis of a luxS-type signalling system. J Med Microbiol 2005; 54:119 - 27; http://dx.doi.org/10.1099/jmm.0.45817-0; PMID: 15673504
- López D, Kolter R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis.. FEMS Microbiol Rev 2010; 34:134 - 49; http://dx.doi.org/10.1111/j.1574-6976.2009.00199.x; PMID: 20030732
- Underwood S, Guan S, Vijayasubhash V, Baines SD, Graham L, Lewis RJ, et al. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J Bacteriol 2009; 191:7296 - 305; http://dx.doi.org/10.1128/JB.00882-09; PMID: 19783633
- Molle V, Fujita M, Jensen ST, Eichenberger P, González-Pastor JE, Liu JS, et al. The Spo0A regulon of Bacillus subtilis.. Mol Microbiol 2003; 50:1683 - 701; http://dx.doi.org/10.1046/j.1365-2958.2003.03818.x; PMID: 14651647
- Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 2013; 11:157 - 68; http://dx.doi.org/10.1038/nrmicro2960; PMID: 23353768
- Dawson LF, Valiente E, Faulds-Pain A, Donahue EH, Wren BW. Characterisation of Clostridium difficile biofilm formation, a role for Spo0A. PLoS One 2012; 7:e50527; http://dx.doi.org/10.1371/journal.pone.0050527; PMID: 23236376
- Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 2012; 80:2704 - 11; http://dx.doi.org/10.1128/IAI.00147-12; PMID: 22615253
- Piggot PJ. Spore development in Bacillus subtilis.. Curr Opin Genet Dev 1996; 6:531 - 7; http://dx.doi.org/10.1016/S0959-437X(96)80080-3; PMID: 8939730
- Olson KM, Starks CM, Williams RB, O’Neil-Johnson M, Huang Z, Ellis M, et al. Novel pentadecenyl tetrazole enhances susceptibility of methicillin-resistant Staphylococcus aureus biofilms to gentamicin. Antimicrob Agents Chemother 2011; 55:3691 - 5; http://dx.doi.org/10.1128/AAC.00302-11; PMID: 21646481
- Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 2001; 9:34 - 9; http://dx.doi.org/10.1016/S0966-842X(00)01913-2; PMID: 11166241
- Kelly CP. Can we identify patients at high risk of recurrent Clostridium difficile infection?. Clin Microbiol Infect 2012; 18:Suppl 6 21 - 7; http://dx.doi.org/10.1111/1469-0691.12046; PMID: 23121551
- Carman RJ, Boone JH, Grover H, Wickham KN, Chen L. In vivo selection of rifamycin-resistant Clostridium difficile during rifaximin therapy. Antimicrob Agents Chemother 2012; 56:6019 - 20; http://dx.doi.org/10.1128/AAC.00974-12; PMID: 22908175
- Surawicz CM, Alexander J. Treatment of refractory and recurrent Clostridium difficile infection. Nat Rev Gastroenterol Hepatol 2011; 8:330 - 9; http://dx.doi.org/10.1038/nrgastro.2011.59; PMID: 21502971
- Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, et al. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol 2009; 72:1500 - 16; http://dx.doi.org/10.1111/j.1365-2958.2009.06739.x; PMID: 19460094
- Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 2005; 436:1171 - 5; http://dx.doi.org/10.1038/nature03912; PMID: 16121184
- Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. Persisters: a distinct physiological state of E. coli.. BMC Microbiol 2006; 6:53; http://dx.doi.org/10.1186/1471-2180-6-53; PMID: 16768798
- Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 2004; 230:13 - 8; http://dx.doi.org/10.1016/S0378-1097(03)00856-5; PMID: 14734160
- Tyerman JG, Ponciano JM, Joyce P, Forney LJ, Harmon LJ. The evolution of antibiotic susceptibility and resistance during the formation of Escherichia coli biofilms in the absence of antibiotics. BMC Evol Biol 2013; 13:22; http://dx.doi.org/10.1186/1471-2148-13-22; PMID: 23356665
- Lewis K. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol 2008; 322:107 - 31; http://dx.doi.org/10.1007/978-3-540-75418-3_6; PMID: 18453274
- Probert HM, Gibson GR. Bacterial biofilms in the human gastrointestinal tract. Curr Issues Intest Microbiol 2002; 3:23 - 7; PMID: 12400635
- Croucher SC, Houston AP, Bayliss CE, Turner RJ. Bacterial populations associated with different regions of the human colon wall. Appl Environ Microbiol 1983; 45:1025 - 33; PMID: 6847178
- Donelli G, Vuotto C, Cardines R, Mastrantonio P. Biofilm-growing intestinal anaerobic bacteria. FEMS Immunol Med Microbiol 2012; 65:318 - 25; http://dx.doi.org/10.1111/j.1574-695X.2012.00962.x; PMID: 22444687