Abstract
Genetic, nutritional, and gut microbiota-derived factors have been proposed to play a role in the development of the whole intestine that is around 40% longer in PRM/Alf mice compared with other mouse strains. The PRM/Alf genotype explains 60% of this length difference. The remaining 40% are due to a maternal effect that could depend on the gut microbiota transmitted by the mother to their pups. Germ-free PRM/Alf mice and C3H/He mice were associated with a simplified human microbiota (SIHUMI) to study its impact on gut length. The small intestines of the SIHUMI-associated mice were 16.4% (PRM/Alf) and 9.7% (C3H/He) shorter than those of the corresponding germ-free counterparts. Temporal temperature gradient gel electrophoresis and quantitative real-time PCR revealed differences in microbiota composition between both SIHUMI-associated mouse strains. Anaerostipes caccae was one log lower in PRM/Alf mice than in C3H/He mice. Since polyamines and short-chain fatty acids (SCFAs) are important intestinal growth factors, their concentrations were explored. Cecal concentrations of putrescine, spermine, spermidine, and N-acetylspermine were 1.5-fold, 3.7-fold, 2.2-fold, and 1.4-fold higher, respectively, in the SIHUMI-C3H/He mice compared with the SIHUMI-PRM/Alf mice. In addition, cecal acetate, propionate, and butyrate concentrations in SIHUMI-C3H/He mice were 1.4-fold, 1.1-fold, and 2.1-fold higher, respectively, than in SIHUMI-PRM/Alf mice. These results indicate that polyamines and SCFAs did not promote gut lengthening in any of the two mouse strains. This suggests that as yet unknown factors provided by the SIHUMI prevented gut lengthening in the SIHUMI-associated mice compared with the germfree mice.
Introduction
The intestines of the PRM/Alf mice are around 40% longer than those of other mouse strains including C3H/He, C57BL/6J, and DBA/2J.Citation1 Both small and large intestines of the PRM/Alf mouse are proportionally lengthened. This phenotype develops between birth and weaning. Cross-adoption experiments revealed that a maternal effect explains 40% of the length difference.Citation1 Therefore, growth-promoting factors in the milk as well as factors derived from the intestinal microbiota have been considered important.Citation1,Citation2 Indeed, milk from PRM/Alf dams contains higher amounts of IgA (a 2- to 3-fold increase) than that from C57BL/6J dams. Since secretory IgA controls the survival of essential commensal bacteria,Citation3 PRM/Alf milk might influence the PRM/Alf microbiota composition. Gut bacteria influence a number of morphological parameters including cecum size, crypt depth, villus height, and development of the lamina propria.Citation4-Citation6 Bacterial metabolites contribute to such effects. For instance, direct infusion of short-chain fatty acids (SCFAs) into cecum or colon leads to increased villus height, crypt depth,Citation7,Citation8 and cell proliferation.Citation9,Citation10 Trophic effects have also been observed for polyamines, which are involved in the regulation of cell growth and cell proliferation as well as neonatal gut maturation.Citation11,Citation12
The purpose of this study was to investigate the role of the intestinal microbiota on gut lengthening in the PRM/Alf mouse model and to identify microbial factors that possibly contribute to this phenomenon. We hypothesized that higher intestinal concentrations of polyamines and/or SCFAs in the gut of PRM/Alf mice as compared with C3H/He mice may be responsible for the increased intestinal length. To exclude the influence of a different intestinal microbiota composition, we took advantage of gnotobiotic mice colonized with a simplified human microbiota (SIHUMI) consisting of nine bacterial strains.Citation13 We first investigated the intestinal morphology in the SIHUMI PRM/Alf and C3H/He mice. The SIHUMI composition was then assessed in each mouse strain, as well as intestinal concentrations of SCFAs and polyamines.
Our results indicate that the SIHUMI induces a shortening of the intestine. This shortening was more pronounced in PRM/Alf mice as compared with C3H/He mice. Mucosa thickness of the proximal intestine and colon was also decreased in PRM/Alf mice compared with C3H/He mice. The mouse genotype influenced both the microbiota composition and the concentrations of SCFAs and polyamines. Higher intestinal concentrations of SCFAs and of most polyamines did not correlate with an extended gut length. Therefore, these metabolites do not promote gut lengthening. The shorter gut observed for both SIHUMI-associated mouse strains in comparison to the corresponding germfree counterparts suggests that SIHUMI-associated factors prevented gut lengthening in the SIHUMI-associated mice.
Results
To investigate the impact of the microbiota on gut length, we compared the length of the small intestine and of the colon between SIHUMI-PRM/Alf and SIHUMI-C3H/He mice and PRM/Alf and C3H/He germ-free mice, respectively (). We have evidence that the intestinal length was identical in males and females for a given microbial status (germ-free or SIHUMI), regardless of the mouse strain (data not shown). In this study only females were included.
Figure 1. Gut length of germfree and SIHUMI associated PRM/Alf (A) and C3H/He mice (B). The length was determined in 8-wk old mice. The length of the whole intestine corresponds to the sum of small and large intestinal length. Data are expressed as mean ± standard deviation. Differences among groups were analyzed by Student’s t test, ***P ≤ 0.001; ngermfree (PRM/Alf, C3H/He) = 14, 12; nSIHUMI (PRM/Alf, C3H/He) = 12, 13. (C) Morphometric comparison of the layers of the intestinal tract between SIHUMI-PRM/Alf (n = 3) and SIHUMI-C3H/He (n = 3) mice. Data are expressed as mean ± SEM. Differences among groups were analyzed by Student’s t test *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001
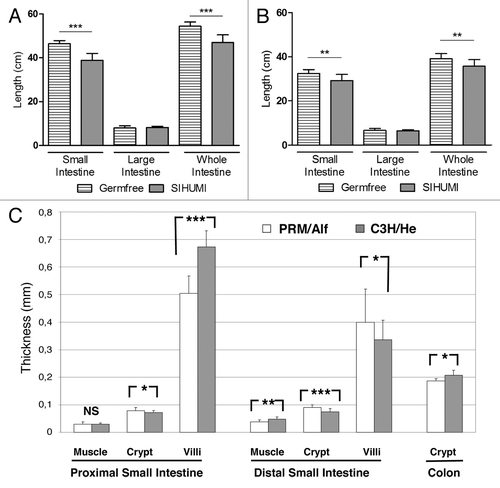
The intestine of germ-free PRM/Alf mice (54.4 ± 1.9 cm) was 39% longer than that of germ-free C3H/He mice (39.1 ± 2.3 cm, P ≤ 0.001) (). The gut length of SIHUMI-PRM/Alf mice was also 32% longer compared with SIHUMI-C3H/He mice (47.0 ± 3.5 cm vs. 35.7 ± 3.0 cm, respectively, P ≤ 0.001). The small intestine of SIHUMI-PRM/Alf and SIHUMI-C3H/He mice was 16.4% and 9.7% shorter than those of the respective germ-free counterparts, whereas the colon length did not differ ().
The intestinal walls of SIHUMI-PRM/Alf and SIHUMI-C3H/He mice were subjected to histological analysis (). The thickness of the tunica muscularis in the proximal part of the small intestine was not significantly different between SIHUMI-PRM/Alf and SIHUMI-C3H/He mice, but was decreased in the distal part of the small intestine of SIHUMI-PRM/Alf mice compared with SIHUMI-C3H/He mice (37 ± 8 µm vs. 47 ± 9 µm; P ≤ 0.01). The crypt depth of SIHUMI-PRM/Alf mice was significantly higher compared with SIHUMI-C3H/He mice in both the proximal small intestine (78 ± 11 µm and 71 ± 8 µm, respectively; P ≤ 0.05) and distal small intestine (90 ± 10 µm and 74 ± 12 µm, respectively; P ≤ 0.001). The height of the villi in the distal small intestine was higher in the SIHUMI-PRM/Alf mice compared with SIHUMI-C3H/He mice (400 ± 120 µm and 336 ± 70 µm, respectively; P ≤ 0.05). Conversely, the villi in the proximal small intestine of the SIHUMI-PRM/Alf mice were shorter than those of the SIHUMI-C3H/He mice (504 ± 63 µm and 676 ± 58 µm, respectively; P ≤ 0.001). The same applies to the thickness of the colon mucosa, which was lower in SIHUMI-PRM/Alf mice compared with SIHUMI-C3H/He mice (187 ± 8 µm and 208 ± 18 µm, respectively; P ≤ 0.05).
To find out whether differences in SIHUMI composition contributed directly to these differences, we analyzed the dominant microbiota composition in feces collected at weaning (day 22) and before killing (day 54) in both mouse strains using temporal temperature gradient gel electrophoresis (TTGE) fingerprinting and in cecal contents after killing (day 56) using quantitative real-time PCR (qPCR).
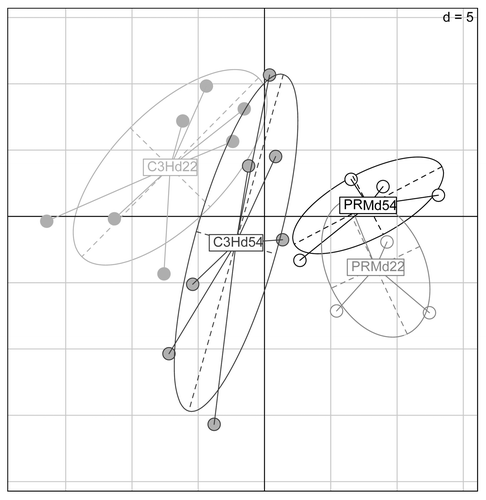
Eight of the nine bacterial members of the SIHUMI could be detected in the intestine of both mouse strains (). Only L. plantarum was never detected in the cecal content (using qPCR) at day 56 or in the feces (using TTGE fingerprinting, data not shown) at day 22 and day 54. Pearson similarity coefficient between the two strains’ microbiota, at day 22 and day 54, ranged from 91% to >95%. However, principal component analysis (PCA) data of fecal microbiota composition revealed a significant difference between SIHUMI-PRM/Alf and SIHUMI-C3H/He mice (; P = 1 × 10−4). This difference was more important at weaning than at day 54. The qPCR data from day 56 also highlighted microbiota differences in the cecum: A. caccae was one log lower in PRM/Alf mice than in C3H/He mice and those of B. thetaiotaomicron, B. longum, C. butyricum, and F. varium were also significantly lower, although these differences were numerically small (). As a consequence, the relative proportions of the community members were also modified. For example, the proportion of cecal C. ramosum was higher in PRM/Alf mice than in C3H/He mice (7.3% in PRM/Alf mice vs. 3.3% in C3H/He). All these data indicate a significant mouse strain effect on the microbiota composition.
Figure 2. Microbial cell numbers in dry cecal content of PRM/Alf (n = 12) and C3H/He (n = 13) mice, both associated with the SIHUMI consortium. Data are expressed as mean ± standard deviation. Significant differences were analyzed by Student’s t test: **P ≤ 0.01; ***P ≤ 0.001; n.d., not detected
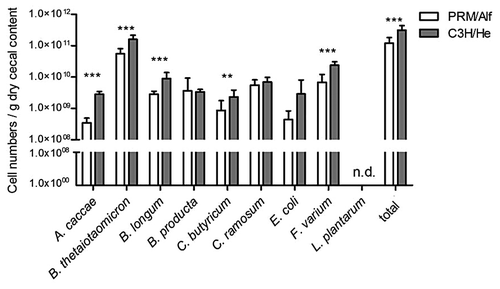
Figure 3. Profiles of dominant intestinal microbiota from PRM/Alf (n = 4) and C3H/He (n = 7) mice at day 22 and at day 54. PCA analysis highlights a significant clustering between the two mouse groups, accentuated at day 22 and to a lesser extent at day 54. The two first components of the analysis explain 28.69% of the total variance. Component 1 = 16.35%; Component 2 = 12.34%. Based on 10 000 replicates, the Monte-Carlo test of the interclass PCA highlighted a simulated P = 1 × 10−4.
Since microbial metabolites such as SCFAs and polyamines are trophic factors,Citation11,Citation14 we measured their concentrations in the cecum of SIHUMI-PRM/Alf and SIHUMI-C3H/He mice.
Concentrations of acetate, propionate, and butyrate in cecum were 1.4-fold, 1.1-fold, and 2.1-fold lower, respectively (P ≤ 0.01), in the SIHUMI-PRM/Alf mice than in the SIHUMI-C3H/He mice (). Lactate concentrations ranged between 0.39 mM and 4.39 mM (median: 2.37 mM) in SIHUMI-PRM/Alf mice and between 1.92 mM and 7.62 mM (median: 3.60 mM) in SIHUMI-C3H/He mice (data not shown, P ≤ 0.05).
Figure 4. Cecal polyamine concentrations (µM) and SCFA concentrations (mM) of PRM/Alf (n = 12) and C3H/He mice (n = 13) associated with the SIHUMI consortium. Data are expressed as medians. Whiskers show the 5th and 95th percentile. Differences among groups were analyzed by Mann Whitney. *P ≤ 0.05; **P ≤ 0.01, ***P ≤ 0.001.
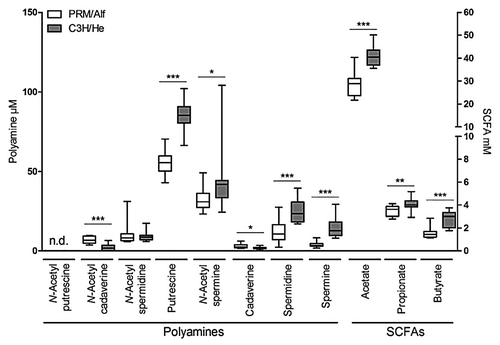
Concentrations of putrescine, spermidine, spermine, and N-acetylspermine were 1.5-fold, 2.2-fold, 3.7-fold, and 1.4-fold lower, respectively (P ≤ 0.05), in SIHUMI-PRM/Alf than in SIHUMI-C3H/He mice (), while N-acetylcadaverine and cadaverine concentrations were 3.9-fold and 1.2-fold higher, respectively (P ≤ 0.05), in the SIHUMI-PRM/Alf mice. Correlations were only found for PRM/Alf mice, whose gut length correlated positively with N-acetylcadaverine (r = 0.717, P ≤ 0.001), but negatively with putrescine (r = −0.754, P ≤ 0.001), spermidine (r = −0.619, P ≤ 0.001), and spermine (r = −0.883, P ≤ 0.001).
Discussion
Genetic and trophic factors derived from mother’s milk and/or bacterial metabolites in the intestine have been proposed to play a role in the PRM/Alf mouse model of gut lengthening.Citation1,Citation2 Our study shows that germ-free PRM/Alf mice had a longer intestine than germ-free C3H/He mice. The same has previously been observed for their conventional counterparts.Citation1 This result suggests that microbial factors are of minor importance for the development of an elongated gut in PRM/Alf mice. Interestingly, the small intestine was shorter in SIHUMI-associated mice than in germ-free mice, irrespective of genetic background, suggesting that the SIHUMI community negatively affected intestinal development. Intestinal crypt depths in SIHUMI-PRM/Alf mice were similar to those previously reported for conventional PRM/Alf mice,Citation15 suggesting that this parameter was not influenced by the type of microbiota. However, the intestinal wall was thicker in SIHUMI-associated PRM/Alf mice than in conventional PRM/Alf mice. Accordingly, villi in the proximal and distal small intestine were longer and the colon crypts were deeper in SIHUMI-PRM/Alf than in conventional PRM/Alf mice. B. longum, a member of the SIHUMI, has previously been demonstrated to cause villus lengthening in a rodent model.Citation16 Elongated intestinal villi may enhance the intestinal absorptive capacities of the SIHUMI-associated mice. SIHUMI-PRM/Alf mice displayed significantly longer small intestinal crypts than SIHUMI-C3H/He mice, but colonic crypts were shorter in these mice. Whether this discrepancy is due to differences in the distribution of the SIHUMI members between small and large intestine remains unknown since we only determined microbial cell numbers in the cecum. Former experiments with the SIHUMI model indicated that the cell numbers of some SIHUMI members measured in the small intestine with qPCR were under the detection limit (unpublished data).
SCFA and lactate concentrations of SIHUMI-C3H/He mice were between those of conventional and germfree C3H mice.Citation17 The fact that concentrations of SCFAs and of most of the analyzed polyamines were lower in SIHUMI-PRM/Alf mice than in SIHUMI-C3H/He mice might have consequences. First, since SCFAs and polyamines act as trophic factors,Citation11,Citation14 reduced concentrations of these bacterial metabolites may prevent gut lengthening in SIHUMI-PRM/Alf mice. This would support the hypothesis that these bacterial factors critically contribute to gut growth control. Second, the absorption of SCFAs and polyamines might be increased in SIHUMI-PRM/Alf mice due to an increased intestinal surface resulting from deeper small intestinal crypts and a longer intestinal length compared with that of C3H/He mice. We found a correlation between the butyrate concentration and the A. caccae cell number (r = 0.726, P ≤ 0.001). Therefore, the lower cecal butyrate concentrations observed in SIHUMI-PRM/Alf mice may be due to the lower count of the butyrate producer A. caccae detected in PRM/Alf mice.Citation18 Even though concentrations of N-acetylcadaverine and cadaverine were significantly higher in PRM/Alf mice, available data argue against a role of these compounds as growth factors. First, neither cadaverine nor N-acetylcadaverine are known to have a physiological role.Citation19 Second, N-acetylcadaverine undergoes cellular and urinary secretion and thereby plays a role in controlling the pool of free cadaverine.Citation11,Citation12 Since cadaverine may be formed by both the intestinal microbiota and mammalian tissue,Citation20 it is likely that PRM/Alf and C3H/He mice differ in their endogenous metabolism.
In summary, SIHUMI-associated PRM/Alf mice exhibited an increased intestinal length compared with SIHUMI-associated C3H/He mice. However, their intestinal length was shorter compared with their germ-free counterparts. These results provide evidence that the intestinal microbiota plays an important role in the control of the intestinal development.
Materials and Methods
Production of germ-free PRM/Alf mice
A colony of germ-free PRM/Alf mice was obtained through germ-free rederivation by aseptic hysterectomy of pregnant PRM/Alf mice from conventional rodent breeding facilities (ENVA, UMR 955 GFM) and bred in INRA germ-free rodent breeding facilities (INRA, UMR1319 Micalis, Anaxem facilities), according to established methods.Citation21
Animal care and experimentations were in accordance with guidelines of the International Guiding Principles for Biomedical Research.
Establishment of the SIHUMI in germ-free mice
Breeding colonies of PRM/Alf and C3H/He mice colonized with a simplified human microbiota composed of nine species (Anaerostipes caccae DSM 14667, Bacteroides thetaiotaomicron DSM 2079, Bifidobacterium longum NCC 2705, Blautia producta DSM 2950, Clostridium butyricum DSM 10702, Clostridium ramosum DSM 1402, Escherichia coli K-12 MG 1655, Fusobacterium varium ATCC 8501, and Lactobacillus plantarum DSM 20174) were established as described elsewhere.Citation17
Female littermates (PRM/Alf: n = 12; C3H/He: n = 13) from these SIHUMI-associated colonies were used in our experiment. In addition, germ-free PRM/Alf (n = 14) and germ-free C3H/He female mice (n = 12) were included. All mice were housed in Trexler-type isolators with free access to autoclaved, acidified water (pH = 4.0) and gamma-sterilized (45 kGy) pelleted standard chow R03-40 (Scientific Animal Food and Engineering). The rooms housing the isolators were maintained at constant temperature and humidity (21 ± 1 °C, 50 ± 5%) with a 12-h light-dark cycle.
Feces were collected at weaning (22 ± 3 d of age) and before killing (54 ± 1 d of age) for microbiota profiles assessment. Mice were killed by cervical dislocation at 56 ± 1 d of age and the gastrointestinal tract was removed. After measurement of intestinal length, cecal content was collected. Intestinal tissue was also collected for morphometrical analysis.
Morphometrical analysis
The intestines of a subgroup of PRM/Alf (n = 3) and C3H/He (n = 3) mice were dissected in three parts: the proximal half of the small intestine, the distal half of the small intestine, and the colon, using the “swiss roll” technique.Citation22 Samples were fixed for 48 h in 10% formalin in PBS, embedded in paraffin, and sectioned at 4 µm, with sections being 150 µm apart. Sections were deparaffinized in toluene, rehydrated, and stained with hematoxylin-eosin. The thickness of the muscle layer, height of the villi, and depth of the crypts were measured. Seven to ten measurements were performed on four slides. Only pictures of the highest and most perpendicular villi with respect to the muscle layer were taken into consideration. Statistical significance of differences (P ≤ 0.05) was determined with the Student’s t test.
Analysis of cecal content
Cecal bacteria were quantified using qPCR targeting the single-copy gene groELCitation23,Citation24 as described previously.Citation17 Cecal concentrations of acetate, propionate, and butyrate were determined by gas chromatography and polyamines by high-performance liquid chromatography.Citation17 Intestinal lactate concentrations were determined by an enzymatic assay (R-Biopharm) according to manufacturer’s instructions.
Statistical significance of differences (P ≤ 0.05) was determined with the Student’s t test for parametric data or the Mann Whitney test for non-parametric data. One-way ANOVA (Bonferroni) was used for comparisons between more than two groups. Correlations between bacterial metabolites and gut length were calculated using SPSS 16.0 (IBM).
DNA preparation and fingerprinting of the dominant microbiota
Total DNA was extracted from mice feces by mechanical and chemical disruption.Citation25 The variables regions V6-V8 of the bacterial 16S rRNA gene were amplified using primers (GCclamp-U968 and L1401) for TTGE fingerprinting.Citation26
Sequence-specific fingerprints were obtained with TTGE as previously described.Citation25 TTGE profiles were analyzed with Gel Compar software version 2.0 (Applied Maths). Similarity coefficients (Pearson correlation method) were calculated for each pair of profiles, yielding a similarity matrix.
Statistical analysis of microbiota profiles
Densitometric curves corresponding to each of the normalized TTGE profiles were digitized with the GelCompar software program. The resulting data matrix was used to assess the microbiota similarity between PRM/Alf and C3H/He mice associated with SIHUMI. Hierarchical clustering was applied to the data to calculate the spatial coordinates of each mouse for a Principal Components Analysis (PCA) using multivariate regression. PCAs were computed with the R software program (package ade4; http://pbil.univ-lyon1.fr/ADE-4/). In order to statistically assess the impact of the genetic background of mice on microbiota colonization, interclass PCA with genetic status as instrumental variable was computed based on the presence and abundance of specific TTGE bands. Interclass PCA with instrumental variables was further applied to determine the most discriminating variables (TTGE bands) between the microbial communities of either PRM/Alf or C3H/He mice.
Inter-class PCAs display differences in diversity between microbial communities in different individuals. This procedure maximizes the variance between the microbiota of individuals, instead of showing total variance. Thus, inter-class PCA highlights combinations of variables (TTGE bands as a surrogate for different bacterial species) that maximize variations observed between qualitative variables such as genetic background. P value of the statistical significance of interclass PCA clustering based on microbiota profiles was assessed using a Monte Carlo rank test (10 000 replicates).
| Abbreviations: | ||
| ATCC | = | American type culture collection |
| DSM | = | Deutsche Sammlung für Mikroorganismen (German culture collection) |
| NCC | = | Nestlé culture collection |
| PCA | = | principal component analysis |
| qPCR | = | quantitative real-time PCR |
| SCFA | = | short chain fatty acid |
| SIHUMI | = | simplified human microbiota |
| SIHUMI + Fv | = | simplified human microbiota supplemented with Fusobacterium varium |
| TTGE | = | temporal temperature gradient gel electrophoresis |
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
We thank P Guillaume, B Lignon-Couffon, I Grüner, and U Lehmann for excellent animal care and S Schaan and M Urbich for technical assistance. The project was supported by grant BL257/8-1 from the Deutsche Forschungsgemeinschaft and by grant ANR-08-ALIA-019 from the Agence Nationale de la Recherche.
References
- Aubin-Houzelstein G, Da Silva NR, Bellier S, Salaün P, Montagutelli X, Panthier JJ. Genetic interaction between a maternal factor and the zygotic genome controls the intestine length in PRM/Alf mice. Physiol Genomics 2003; 16:82 - 9; http://dx.doi.org/10.1152/physiolgenomics.00106.2003; PMID: 14559976
- Boumahrou N, Chevaleyre C, Berri M, Martin P, Bellier S, Salmon H. An increase in milk IgA correlates with both pIgR expression and IgA plasma cell accumulation in the lactating mammary gland of PRM/Alf mice. J Reprod Immunol 2012; 96:25 - 33; http://dx.doi.org/10.1016/j.jri.2012.08.001; PMID: 23021255
- Corthésy B. Role of secretory IgA in infection and maintenance of homeostasis. Autoimmun Rev 2013; 12:661 - 5; http://dx.doi.org/10.1016/j.autrev.2012.10.012; PMID: 23201924
- Gustafsson BE, Maunsbach AB. Ultrastructure of the enlargec cecum in germfree rats. Z Zellforsch Mikrosk Anat 1971; 120:555 - 78; http://dx.doi.org/10.1007/BF00340589; PMID: 5098551
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol 2007; 19:59 - 69; http://dx.doi.org/10.1016/j.smim.2006.10.002; PMID: 17118672
- Wostmann BS. Anatomy, morphology and function of the gastrointestinal system. Germfree and gnotobiotic animal models. Boca Raton: CRC Press, 1996:19-40.
- Frankel WL, Zhang W, Singh A, Klurfeld DM, Don S, Sakata T, Modlin I, Rombeau JL. Mediation of the trophic effects of short-chain fatty acids on the rat jejunum and colon. Gastroenterology 1994; 106:375 - 80; PMID: 8299904
- Ichikawa H, Sakata T. Stimulation of epithelial cell proliferation of isolated distal colon of rats by continuous colonic infusion of ammonia or short-chain fatty acids is nonadditive. J Nutr 1998; 128:843 - 7; PMID: 9566991
- Kissmeyer-Nielsen P, Mortensen FV, Laurberg S, Hessov I. Transmural trophic effect of short chain fatty acid infusions on atrophic, defunctioned rat colon. Dis Colon Rectum 1995; 38:946 - 51; http://dx.doi.org/10.1007/BF02049730; PMID: 7656742
- Sakata T. Stimulatory effect of short-chain fatty acids on epithelial cell proliferation in the rat intestine: a possible explanation for trophic effects of fermentable fibre, gut microbes and luminal trophic factors. Br J Nutr 1987; 58:95 - 103; http://dx.doi.org/10.1079/BJN19870073; PMID: 3620440
- Larqué E, Sabater-Molina M, Zamora S. Biological significance of dietary polyamines. Nutrition 2007; 23:87 - 95; http://dx.doi.org/10.1016/j.nut.2006.09.006; PMID: 17113752
- Seiler N, Raul F. Polyamines and the intestinal tract. Crit Rev Clin Lab Sci 2007; 44:365 - 411; http://dx.doi.org/10.1080/10408360701250016; PMID: 17558654
- Becker N, Kunath J, Loh G, Blaut M. Human intestinal microbiota: characterization of a simplified and stable gnotobiotic rat model. Gut Microbes 2011; 2:25 - 33; http://dx.doi.org/10.4161/gmic.2.1.14651; PMID: 21637015
- Scheppach W. Effects of short chain fatty acids on gut morphology and function. Gut 1994; 35:Suppl S35 - 8; http://dx.doi.org/10.1136/gut.35.1_Suppl.S35; PMID: 8125387
- Bellier S, Da Silva NR, Aubin-Houzelstein G, Elbaz C, Vanderwinden JM, Panthier JJ. Accelerated intestinal transit in inbred mice with an increased number of interstitial cells of Cajal. Am J Physiol Gastrointest Liver Physiol 2005; 288:G151 - 8; http://dx.doi.org/10.1152/ajpgi.00048.2004; PMID: 15297259
- Laparra JM, Olivares M, Gallina O, Sanz Y. Bifidobacterium longum CECT 7347 modulates immune responses in a gliadin-induced enteropathy animal model. PLoS One 2012; 7:e30744; http://dx.doi.org/10.1371/journal.pone.0030744; PMID: 22348021
- Slezak K, Hanske L, Loh G, Blaut M. Increased bacterial putrescine has no impact on gut morphology and physiology in gnotobiotic adolescent mice. Benef Microbes 2013; 4:253 - 66; http://dx.doi.org/10.3920/BM2012.0047; PMID: 23666100
- Duncan SH, Louis P, Flint HJ. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 2004; 70:5810 - 7; http://dx.doi.org/10.1128/AEM.70.10.5810-5817.2004; PMID: 15466518
- Balasundaram D, Tyagi AK. Polyamine--DNA nexus: structural ramifications and biological implications. Mol Cell Biochem 1991; 100:129 - 40; http://dx.doi.org/10.1007/BF00234162; PMID: 2008175
- Pegg A, Williams-Asheman HG. Biosynthesis of putrescine. In: Morris DR, Marton LJ, eds. Polyamines in Biology and Medicine. New York: Dekker, 1981:4-42.
- Coates ME. The germ-free animal in research. London: Academic Press, 1968.
- Moolenbeek C, Ruitenberg EJ. The “Swiss roll”: a simple technique for histological studies of the rodent intestine. Lab Anim 1981; 15:57 - 9; http://dx.doi.org/10.1258/002367781780958577; PMID: 7022018
- Hill JE, Penny SL, Crowell KG, Goh SH, Hemmingsen SM. cpnDB: a chaperonin sequence database. Genome Res 2004; 14:1669 - 75; http://dx.doi.org/10.1101/gr.2649204; PMID: 15289485
- Junick J, Blaut M. Quantification of human fecal bifidobacterium species by use of quantitative real-time PCR analysis targeting the groEL gene. Appl Environ Microbiol 2012; 78:2613 - 22; http://dx.doi.org/10.1128/AEM.07749-11; PMID: 22307308
- Lepage P, Seksik P, Sutren M, de la Cochetière MF, Jian R, Marteau P, Doré J. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm Bowel Dis 2005; 11:473 - 80; http://dx.doi.org/10.1097/01.MIB.0000159662.62651.06; PMID: 15867587
- Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol 1998; 64:3854 - 9; PMID: 9758810
