Abstract
The gut microbiota plays important roles in the development of the host immune system. We have previously shown that a combination of 46 strains of commensal Clostridia isolated from conventionally reared mice can induce the accumulation of CD4+Foxp3+ regulatory T (Treg) cells in the mouse colonic lamina propria. Subsequently, we succeeded in isolating and selecting 17 strains of Clostridia from a healthy human fecal sample that can significantly increase the number and function of colonic Treg cells in colonized rodents, thereby attenuating symptoms of experimental allergic diarrhea and colitis. Here we characterize each of the 17 strains of human-derived Clostridia in terms of sensitivity to antibiotics and ability to produce short chain fatty acids and other metabolites, and discuss their potential as biotherapeutics to correct dysbiosis and treat immune-inflammatory diseases.
Keywords: :
Seventeen Strains of Treg-Inducing Clostridia Isolated From the Human Intestine
The intestinal microbiota is composed of ~100 trillion commensal bacteria and has co-evolved with the host by participating in many essential physiologic and metabolic functions. There is also abundant evidence that the gut microbiota affects the host immune status. Immunological effects of the microbiota are not simply due to the presence of innocuous bacteria, but to the biological activities of the gut microbiota consortium.Citation1 In many cases, regulation of development and/or effector functions of different immune cell populations (such as Treg cells vs Th17 cells) depends on the activity of different members of the commensal community. The relative abundance of these different immunomodulatory members can direct the general nature of host mucosal and systemic immunity. Previously, we referred to such immunomodulatory members of the microbiota as “autobionts”.Citation2 However, currently there are relatively few specific examples of autobionts.
We and other groups have shown that segmented filamentous bacteria can potently induce interleukin-17-producing CD4+ T cells (Th17 cells) in the small intestine of mice.Citation3 We have also shown that a combination of 46 strains of Clostridia indigenous to conventionally reared mice can induce Treg cells in the mouse colonic lamina propria and thereby contribute to protecting mice against colitis and allergic responses.Citation4 In the most recent publication by our group, we identified 17 strains of human-derived Clostridia as potent inducers of Treg cells.Citation5 Starting from a complete healthy human fecal sample, a sequence of selection steps was applied to obtain Treg cell-inducing human-derived bacterial strains using gnotobiotic techniques.Citation5 We first observed full Treg cell induction in the colon of ex-germ-free mice orally inoculated with a chloroform-treated human fecal sample. Then the cecal contents from these mice were treated with chloroform, diluted, and serially transplanted into other germ-free mice, while monitoring Treg induction capability. We succeeded in obtaining colonized mice in which the complexity of the gut microbiota was greatly decreased without sacrificing Treg-inducing potency. From these mice, we cultured and selected 17 strains which, when mixed together and orally administered to gem-free mice and rats, were able to induce a significant accumulation of CD4+Foxp3+ Treg cells in the colon. Furthermore, repeated oral ingestion of the mixture of 17 strains rendered specific-pathogen-free mice resistant to experimental allergic diarrhea and trinitrobenzene sulfonic acid (TNBS)-induced colitis.Citation5 Therefore, the 17 strains have at least a prophylactic effect in mouse colitis models.
Characterization of the 17 Strains
The 17 Treg-inducing strains isolated in our study all belong to the class Clostridia.Citation5 Clostridia species are gram-positive anaerobic rods and typically can form endospores. Some Clostridia species, including Clostridium tetani, C. botulinum, and C. perfringens, are well-known pathogens that are often isolated as a singular cause of infectious disease, and C. difficile is responsible for antibiotic-associated diarrhea and colitis. On the other hand, Clostridia species are extremely heterogeneous and many of them inhabit the large intestine of human and animals as predominant symbiotic microbes. Clostridia species can be classified into 19 clusters (I to XIX),Citation6 and many of the pathogenic species listed above belong to cluster I. The 17 Treg-inducing strains fall within clusters IV, XIVa, and XVIII of Clostridia (). It has been reported that species within clusters XIVa and IV are indispensable for various physiological host functions. For instance, colonization with Clostridia clusters XIVa and IV normalizes the enlarged cecum found in germ-free mice and supports epithelial growth and turnover.Citation7 It was also shown that colonization with Clostridia cluster XIVa strains renders mice resistant to C. difficile colonization.Citation8
Figure 1. Antibiotic sensitivity of the 17 strains. The closest known species/strains for each of the 17 strains are shown. Antibiotic sensitivities are categorized as susceptible (white), intermediate (light orange) or resistant (dark orange). Antimicrobial susceptibility testing was performed using the broth microdilution method with dry plates (Eiken Chemical, Japan) according to the Clinical and Laboratory Standards institute (CLSI) guidelines M11-A8 and M100-S23. Briefly, each strain grown on Eggerth-Gagnon agar was harvested and suspended in ABCM broth (Eiken Chemical, Tokyo, Japan). The number of colony-forming units (CFU) was adjusted to 1 × 105 CFU/mL and an aliquot (100 μL) of the suspension was inoculated into each well of the plates. After incubation at 37 °C for 48 h, bacterial growth was visually assessed to determine the minimal inhibitory concentrations (MICs). The Etest (Sysmex-bioMérieux, Japan) was also employed to test for susceptibility to metronidazole and vancomycin.
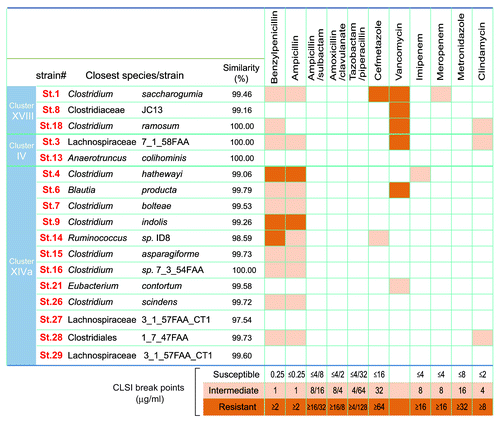
It should be noted that there have been several reports of Clostridia clusters IV, XIVa, and XVIII species (Clostridium clostridioforme, Clostridium innocuum, and Clostridium ramosum, in particular) in clinical specimens of opportunistic infections.Citation9 However they are rarely single isolates, but rather a fraction of the multiple organisms in these clinical samples. Considering that commensal Clostridia species colonize in close proximity to the gut epithelial surface,Citation10 they may translocate when the barrier is compromised, not necessarily because of specific pathogenic properties, but simply because of their local abundance. Sequencing of the genomes of the 17 strains revealed that they lack known toxins and virulence factors.Citation5 Some of the 17 strains possess genes encoding putative sialidase, hyaluronidase, flagella-related protein, and fibronectin binding protein, but with low similarity to genes found in pathogenic Clostridia species. Furthermore, ingestion of a mixture of the 17 strains of Clostridia was effective in preventing colitis induced by TNBSCitation5 and even by dextran sulfate sodium (DSS) (data not shown), which chemically disrupts the epithelial barrier. Given the lack of major virulence factors and toxins in their genomes and the lack of any toxicity upon dosing to animals, the Clostridia strains isolated in our study appear to be safe for clinical use.
To further confirm that the isolated Clostridia strains are not harmful, antibiotic sensitivity of each strain was tested. All strains were susceptible to ampicillin-sulbactam, piperacillin-tazobactam, amoxicillin-clavulanate, metronidazole, and chloramphenicole, although some strains were resistant to penicillin G and ampicillin alone, and strains 1, 3, 8, and 18 showed low level resistance to vancomycin with MIC values of 4 to 8 μg/ml (). Since C. ramosum and C. innocuum are reported to have intrinsic resistance to glycopeptides and lipopeptides (vancomycin),Citation9 and genes identical to VanB2 ligase of Enterococcus spp. can be found in C. hathewayi, C. boltae, and C. inocuum-like bacteria,Citation11 it is not surprising that strains 1, 3, 8 and 18, which have 16S sequence similarities with those species, showed low level resistance to vancomycin. Overall, none of the 17 strains have high-level resistance to antibiotics tested; rather they show a benign safety profile. Further careful characterization of the strains and pre-clinical studies will be required before clinical translation.
Mechanisms of Treg Accumulation by the 17 Strains
The precise mechanism underlying how the 17 strains of Clostridia stimulate the induction of colonic Treg cells remains to be further elucidated. One suggested mechanism is the production of short chain fatty acids (SCFAs), which have multiple metabolic and immune functions.Citation12 In the context of Treg induction, SCFAs can elicit a TGF-β1 response in epithelial cells, which can contribute to de novo induction of peripheral Treg cells (pTreg).Citation5 SCFAs, particularly butyrate, can suppress dendritic cell activation through suppression of expression of the NFκB component RelB.Citation13 It has also been shown that butyrate activates signaling pathways through GPR109a to induce anti-inflammatory genes in dendritic cells.Citation14 In addition to its effects on dendritic cells, butyrate can directly stimulate thymic Treg cell (tTreg) proliferation through activation of GPR43Citation15 and the differentiation of naïve CD4+ T cells into pTreg cells through histone H3 acetylation of the Foxp3 gene intronic enhancer by inhibition of histone deacetylase (HDAC).Citation13,Citation16
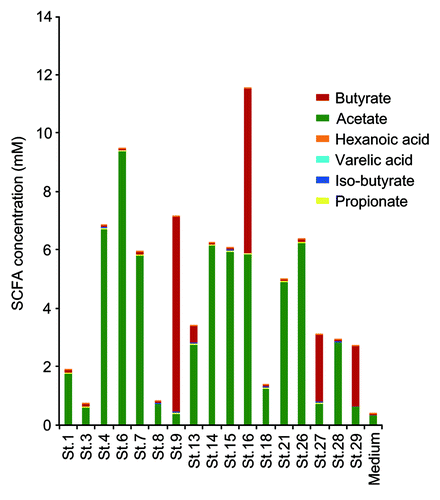
The genomes of the 17 strains contain abundant genes predicted to be involved in the biosynthesis of acetate and butyrate (). SCFA production by each of the 17 strains when cultured in vitro in glucose-supplemented medium was analyzed by a validated liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) systemCitation17 (). Among SCFAs, acetate and butyrate were detected at high concentrations in most of the culture supernatants except those of strains 1, 3, 8, and 18. Notably, strains 9, 13, 16, 27, and 29 showed very high butyrate production (). These findings are consistent with previous reports showing that bacterial strains belonging to Clostridia cluster IV and XIVa were positive for butyryl-CoA:acetate CoA transferase, an enzyme responsible for butyrate production.Citation18 Importantly, mono-colonization of GF mice with one of each of the 17 strains was insufficient to induce Treg cells in vivoCitation5; however, the mixture of 17 strains is effective, suggesting synergistic effects in a microbial community-dependent manner.
Figure 2. SCFA biosynthesis pathways and corresponding gene copy numbers in the 17 strains. Potential metabolic pathways leading to the production of acetate and butyrate from acetyl-CoA are shown. The thickness of the arrow and the number on the left of the arrow indicates the copy number of genes identified in the genomes of the 17 strains.
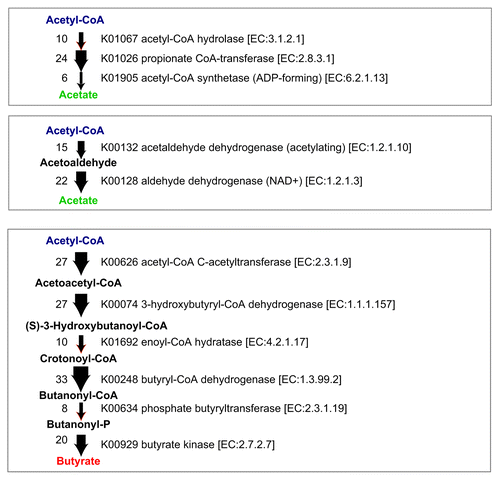
Figure 3. SCFA production by each of the 17 strains. Liquid chromatography-electrospray ionization-tandem mass spectrometry (LC–ESI-MS/MS) analysis of short chain fatty acid metabolites was performed on 48 h cultures of peptone-yeast extract-Fildes solution supplemented with glucose at a concentration of 1% (w/v). Fatty acids in the culture supernatant were derivatized with 2-nitrophenylhydrazine and purified by liquid/liquid extraction.Citation28 Short chain fatty acid peaks were identified by both their specific MS/MS ion-transitions and comparison of the retention times with those of known short chain fatty acids in a standard solution.
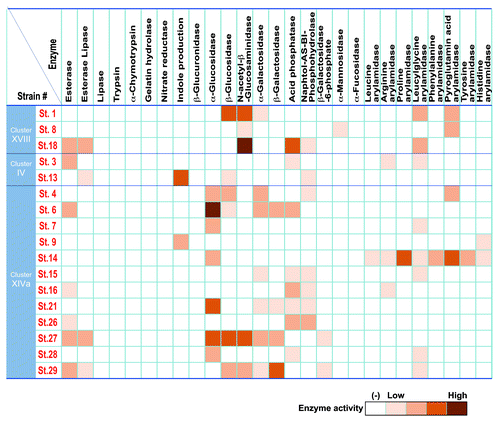
We also examined enzymatic properties of the 17 strains using API ZYM, Rapid ID 32A, and API 20A systems in vitro (). Enzyme activities related to virulence such as trypsin, α-chymotrypsin, and gelatin hydrolase, which have been implicated in infective endocarditis, and β-glucuronidase, which may release toxic substances by cleaving glucuronic acid-conjugation in the intestine, were very low or absent in the 17 strains. Of note, 17 strains showed variety of enzyme activities. Some strains were positive for the activity of principal mucin degrading enzymes, such as β-galactosidase and N-acetyl-β-glucosaminidase, and many strains exhibited strong activity of α-glucosidase, which is an intestinal enzyme that breaks down a variety of carbohydrates. Strain 9 and 13 could convert tryptophan into indole, which is reported to enhance epithelial barrier function and contribute to suppression of colitis.Citation19 Strain 14 was positive for proline arylamidase and pyroglutamic acid arylamidase activities. Together with SCFA production profiles, the 17 strains are diverse and heterogeneous in terms of enzymatic and metabolic properties. Considering that the dysbiotic microbiota associated with inflammatory bowel disease (IBD) is frequently accompanied by significantly decreased diversity,Citation20 the use of a functionally diverse mixture like the 17 strains of Clostridia described here may be reasonable for clinical applications to correct dysbiosis.
Figure 4. Enzymatic activity of the 17 strains. APIZYM and Rapid API 32A tests (Sysmex BioMérieux) were used for determination of enzymatic activities of each of the 17 strains. Cells from cultures grown on Eggerth-Gagnon blood agar plates for 48 h at 37 °C in an anaerobic chamber were suspended in saline and the turbidity was adjusted to 5–6 in the McFarland scale (approx. 1.5–1.9 × 109 CFU/ml). Aliquots of 65 μl of the suspensions were inoculated into cupules in the strip. In the case of Rapid API 32A, the turbidity was matched to 4 in the McFarland scale and aliquots of 55 μl were applied to the cupules. The strips were incubated at 37 °C for 4 h and the reactions were determined according to the manufacturer’s instructions.
Future Directions
Imbalance in the gut microbiota, termed dysbiosis, significantly contributes to various human diseases. Several studies have reported decreased levels of Clostridia and Bacteroidetes in fecal samples from IBD patients.Citation21 Marked decreases in the prevalence of Clostridia have been associated with increased risk of airway hypersensitivity and atopic dermatitis.Citation22,Citation23 Animal studies have also shown that bacterial dysbiosis helps perpetuate the cycle of chronic inflammation characteristic of IBD.Citation24 Therefore, manipulation of the gut microbiota as a therapeutic strategy holds great promise for immuno-inflammatory diseases including IBD and allergy.
Alteration of the gut microbiome via fecal microbiota transplantation (FMT), which involves placing stool from a healthy donor via duodenal tubing, colonoscopy, or enema, has been shown effective in patients with pseudomembranous colitis induced by Clostridium difficile infection,Citation25 and researchers and patients are interested in testing the potential of FMT for the treatment of other disease associated with disruption of the intestinal microbiota, including insulin resistance, multiple sclerosis, and IBD.Citation26 While FMT has been established as a proof of principle for the feasibility of manipulating the human microbiome as a therapeutic strategy, the development of commercial products based on fecal transplants faces a number of hurdles from manufacturing, quality assurance, pathogen contamination risk, donor selection, and patient acceptance perspectives. Therefore, treatment with a composite of well-characterized benign microorganisms is more desirable from many perspectives for medical purposes.
There are many probiotics with a history of safe use in humans for medicinal purposes. However, probiotics currently in use have generally been selected based on properties such as ease of culture and tolerance to acid and oxygen and are not among the major colonizers of the human gut.Citation27 In other words, they have not been isolated based on their ability to correct microbiome dysbiosis associated with human disease or to boost specific arms of the host immune system. Presumably as a result, the dysbiotic microbiota are refractory to treatment with currently available individual probiotic strains, and most probiotics tested to date have demonstrated, at best, mediocre effects in the clinic. Thus, there is a compelling need to identify more robust therapeutic organism compositions that are compatible and symbiotic to the host and, ideally, able to induce broader changes to the microbial ecosystem to correct dysbiosis and drive the immune system to normal homeostasis.
In this context, we believe that the 17 strains of Clostridia isolated in our study can form the basis for a future live biotherapeutic product to treat certain forms of IBD, allergy, and other immune-inflammatory diseases. Compositions based on these strains could (1) help correct microbiome imbalances; (2) be easily administered orally and not necessarily frequently, providing patients with a convenient therapeutic option; (3) be safe for human use, since they are commensal strains that are long-term colonizers of the healthy human gut, do not have prominent virulence factors, and are sensitive to antibiotics; (4) be viewed favorably by patients given that they are natural compositions; and (5) be manufactured with traditional fermentation methods and developed under the live biotherapeutics route outlined by the FDA, thus circumventing the manufacturing, regulatory, and patient acceptance hurdles that hamper commercialization of fecal transplants.
Our next step should be optimization of the therapeutic mixture by elimination of nonessential components of the 17 strains for Treg cell induction. In addition to this, we need to identify and isolate other bacterial strains from human intestine that can regulate differentiation and activation of other immune cell subsets, such as Th17 cells. Our long-term goal is to offer a diverse but minimal consortium of microbes that can colonize the human gut and restore and maintain immune homeostasis to prevent and cure immune-inflammatory diseases.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol 2012; 30:759 - 95; http://dx.doi.org/10.1146/annurev-immunol-020711-074937; PMID: 22224764
- Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe 2012; 12:496 - 508; http://dx.doi.org/10.1016/j.chom.2012.09.009; PMID: 23084918
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139:485 - 98; http://dx.doi.org/10.1016/j.cell.2009.09.033; PMID: 19836068
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011; 331:337 - 41; http://dx.doi.org/10.1126/science.1198469; PMID: 21205640
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500:232 - 6; http://dx.doi.org/10.1038/nature12331; PMID: 23842501
- Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 1994; 44:812 - 26; http://dx.doi.org/10.1099/00207713-44-4-812; PMID: 7981107
- Itoh K, Mitsuoka T. Characterization of clostridia isolated from faeces of limited flora mice and their effect on caecal size when associated with germ-free mice. Lab Anim 1985; 19:111 - 8; http://dx.doi.org/10.1258/002367785780942589; PMID: 3889493
- Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun 2012; 80:3786 - 94; http://dx.doi.org/10.1128/IAI.00647-12; PMID: 22890996
- Alexander CJ, Citron DM, Brazier JS, Goldstein EJ. Identification and antimicrobial resistance patterns of clinical isolates of Clostridium clostridioforme, Clostridium innocuum, and Clostridium ramosum compared with those of clinical isolates of Clostridium perfringens. J Clin Microbiol 1995; 33:3209 - 15; PMID: 8586704
- Nava GM, Stappenbeck TS. Diversity of the autochthonous colonic microbiota. Gut Microbes 2011; 2:99 - 104; http://dx.doi.org/10.4161/gmic.2.2.15416; PMID: 21694499
- Ballard SA, Grabsch EA, Johnson PD, Grayson ML. Comparison of three PCR primer sets for identification of vanB gene carriage in feces and correlation with carriage of vancomycin-resistant enterococci: interference by vanB-containing anaerobic bacilli. Antimicrob Agents Chemother 2005; 49:77 - 81; http://dx.doi.org/10.1128/AAC.49.1.77-81.2005; PMID: 15616278
- Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 2002; 217:133 - 9; http://dx.doi.org/10.1111/j.1574-6968.2002.tb11467.x; PMID: 12480096
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013; 504:451 - 5; http://dx.doi.org/10.1038/nature12726; PMID: 24226773
- Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014; 40:128 - 39; http://dx.doi.org/10.1016/j.immuni.2013.12.007; PMID: 24412617
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341:569 - 73; http://dx.doi.org/10.1126/science.1241165; PMID: 23828891
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504:446 - 50; http://dx.doi.org/10.1038/nature12721; PMID: 24226770
- Morikawa T, Kajimura M, Nakamura T, Hishiki T, Nakanishi T, Yukutake Y, Nagahata Y, Ishikawa M, Hattori K, Takenouchi T, et al. Hypoxic regulation of the cerebral microcirculation is mediated by a carbon monoxide-sensitive hydrogen sulfide pathway. Proc Natl Acad Sci U S A 2012; 109:1293 - 8; http://dx.doi.org/10.1073/pnas.1119658109; PMID: 22232681
- Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 2009; 294:1 - 8; http://dx.doi.org/10.1111/j.1574-6968.2009.01514.x; PMID: 19222573
- Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, Takeda K. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One 2013; 8:e80604; http://dx.doi.org/10.1371/journal.pone.0080604; PMID: 24278294
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006; 55:205 - 11; http://dx.doi.org/10.1136/gut.2005.073817; PMID: 16188921
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007; 104:13780 - 5; http://dx.doi.org/10.1073/pnas.0706625104; PMID: 17699621
- Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep 2012; 13:440 - 7; http://dx.doi.org/10.1038/embor.2012.32; PMID: 22422004
- Candela M, Rampelli S, Turroni S, Severgnini M, Consolandi C, De Bellis G, Masetti R, Ricci G, Pession A, Brigidi P. Unbalance of intestinal microbiota in atopic children. BMC Microbiol 2012; 12:95; http://dx.doi.org/10.1186/1471-2180-12-95; PMID: 22672413
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 2007; 131:33 - 45; http://dx.doi.org/10.1016/j.cell.2007.08.017; PMID: 17923086
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368:407 - 15; http://dx.doi.org/10.1056/NEJMoa1205037; PMID: 23323867
- Smits LP, Bouter KE, de Vos WM, Borody TJ, Nieuwdorp M. Therapeutic potential of fecal microbiota transplantation. Gastroenterology 2013; 145:946 - 53; http://dx.doi.org/10.1053/j.gastro.2013.08.058; PMID: 24018052
- O’Toole PW, Cooney JC. Probiotic bacteria influence the composition and function of the intestinal microbiota. Interdiscip Perspect Infect Dis 2008; 2008:175285; PMID: 19277099
- Miwa H, Yamamoto M. High-performance liquid chromatographic analysis of serum short-chain fatty acids by direct derivatization. J Chromatogr 1987; 421:33 - 41; http://dx.doi.org/10.1016/0378-4347(87)80376-6; PMID: 3429573
