Abstract
Monocolonization of germ-free (GF) mice enables the study of specific bacterial species in vivo. Lactobacillus acidophilus NCFMTM (NCFM) is a probiotic strain; however, many of the mechanisms behind its health-promoting effect remain unknown. Here, we studied the effects of NCFM on the metabolome of jejunum, cecum, and colon of NCFM monocolonized (MC) and GF mice using liquid chromatography coupled to mass-spectrometry (LC-MS). The study adds to existing evidence that NCFM in vivo affects the bile acid signature of mice, in particular by deconjugation. Furthermore, we confirmed that carbohydrate metabolism is affected by NCFM in the mouse intestine as especially the digestion of oligosaccharides (penta- and tetrasaccharides) was increased in MC mice. Additionally, levels of α-tocopherol acetate (vitamin E acetate) were higher in the intestine of GF mice than in MC mice, suggesting that NCFM affects the vitamin E acetate metabolism. NCFM did not digest vitamin E acetate in vitro, suggesting that direct bacterial metabolism was not the cause of the altered metabolome in vivo. Taken together, our results suggest that NCFM affects intestinal carbohydrate metabolism, bile acid metabolism and vitamin E metabolism, although it remains to be investigated whether this effect is unique to NCFM.
Introduction
To understand how the gut microbiota interacts with the host, previous studies have compared the metabolome of germ-free (GF) and conventionalized animals and found the gut microbiota to affect the metabolome in multiple compartments.Citation1,Citation2 Among other things, the gut microbiota has been shown to affect fat storage,Citation3 lipid metabolism,Citation4 and bile acid metabolism.Citation2,Citation5,Citation6 However, introducing a complex microbiota into GF animals does not allow for identification of separate effects of each member of the microbiota on the metabolome. Another option is monocolonization of GF animals, which enables the study of the interaction of specific bacterial species with the host.Citation7-Citation9 One bacterial species of significant interest is Lactobacillus acidophilus NCFMTM (NCFM), a probiotic bacterium that has been used commercially for more than 35 years.Citation10 Human intervention studies with consumption of NCFM have suggested a number of beneficial effects including improving mucosal integrity and motility, as well as modulating host metabolism,Citation11,Citation12 while other studies found no effects.Citation13,Citation14 It has been shown that NCFM can survive in the gastrointestinal tract,Citation10 adhere to human epithelial cells in vitro,Citation15 induce production of the anti-inflammatory cytokine IL-10 in dendritic cells, and regulate T cell functions.Citation16 Analysis of the genome of NCFM has revealed that NCFM encodes a large variety of genes related to carbohydrate metabolism enabling it to utilize a broad range of carbohydrates that escape digestion in the upper gastrointestinal tract including oligosaccharides.Citation17
Bile acids are cholesterol-derived molecules, which are synthesized in the liver and conjugated with either glycine or taurine before they are secreted into the bile and small intestine. In the small intestine, bile acids are modified by bacterial enzymes promoting secondary bile acids through deconjugation, dehydrogenation, and dehydroxylation reactions.Citation18 The genome of NCFM contains two bile salt hydrolase genes,Citation17 which in vitro have been shown to deconjugate glycine- and taurine-conjugated bile salts in a substrate specific manner.Citation19 As most studies on NCFM have been performed in vitro,Citation15,Citation16,Citation19-Citation22 our objective was to study the in vivo effect of NCFM on the metabolome of jejunum, cecum, and colon by comparing NCFM monocolonized (MC) mice with GF mice applying a non-targeted metabolomics approach using liquid chromatography coupled to mass-spectrometry (LC-MS).
Results
Effect of L. acidophilus NCFM on the intestinal metabolome
All samples were confirmed for absence of contamination, and NCFM stably colonized the MC mice at approximately 109 CFU/g of feces throughout the experiment. Luminal samples differed in number of NCFM, gradually increasing from the jejunum to colon with values of log10 (colony forming unit ± standard deviation) of 7.7 ± 0.7 in jejunum, 8.6 ± 0.2 in cecum, and 8.8 ± 0.1 in colon. We analyzed the metabolome of luminal content of jejunum, cecum, and colon by LC-MS in both negative and positive ionization mode. Replicates were closely associated in a principal component analysis (PCA) indicating a robust method (data not shown). By PCA of the whole metabolome of jejunum, cecum, colon we were able to separate the GF and MC mice (Fig. S1). This was evident in both positive and negative mode. From the multivariate analyses, it was evident that presence of NCFM affected the metabolism throughout the gut. Searching accurate masses of features of interest against the METLIN,Citation23 HMDB,Citation24 and LIPIDMAPSCitation25 databases suggested that the differing metabolites between GF and MC in the gut compartments included bile acids, carbohydrates and vitamin E.
Effect of L. acidophilus NCFM on bile acid signatures
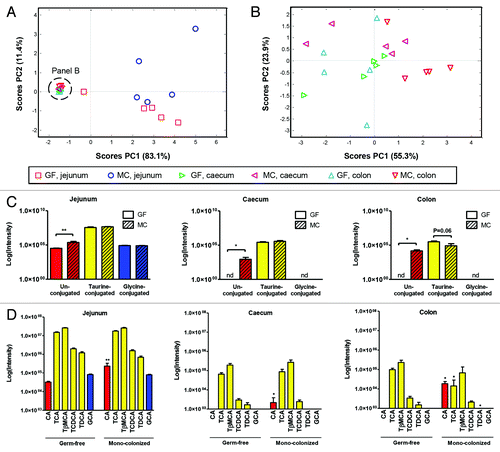
We detected a number of bile acids in jejunum, cecum and colon including unconjugated and conjugated (glycine and taurine) bile acids of GF and MC mice (). A PCA score plot constructed based on the bile acid data revealed that the bile acid composition was site-specific, and affected by microbial colonization as the GF and MC mice clustered separately, particularly in jejunum (), while less evident in cecum and colon (). The levels of taurine- and glycine-conjugated bile acids were similar in GF and MC mice (), while the MC mice had significantly higher abundance of unconjugated bile acids in jejunum (P < 0.01), cecum and colon (P < 0.05) compared with GF. In addition, a tendency for reduced abundance of taurine conjugates in MC compared with GF was observed in colon (P = 0.06). The bile acid signatures of all samples were dominated by taurine conjugates with taurocholic acid (TCA) and tauro-β-muricholic acid (TβMCA) as the most abundant taurine conjugates, while taurochenodeoxycholic acid (TCDCA) and taurodeoxycholic (TDCA) acid was found in less abundance (). The bile acid levels were generally higher in jejunum than in cecum and colon. We detected mostly primary bile acids in GF as well as MC mice. Only one secondary bile acid, TDCA, was measured in the mice. Comparison of the presence of individual bile acids in GF and MC () revealed that the level of the conjugated primary bile acid, TβMCA, was similar in jejunum, cecum, and colon, while TCA was more abundant in colon of GF compared with MC counterparts, suggesting increased absorption or metabolism of this bile acid in the colon of MC mice. Oppositely, the abundances of TCDCA were similar in GF and MC in all three gut compartments. The level of unconjugated CA was also increased in MC compared with GF in all three gut compartments indicating increased deconjugation in the gut in the presence of NCFM. The secondary bile acid, TDCA, was less abundant in colon of MC compared with GF counterparts. Glycocholic acid (GCA) was only measured in jejunum of both MC and GF indicating that this bile acid species was absorbed in jejunum. It should be noted that some of the minor bile acid species present in the gut luminal samples may not have been detected, as the analytical non-targeted LC-MS approach was optimized to detect as many metabolites as possible and not targeted specifically toward bile acids.
Figure 1. Bile acid signatures in jejunum, cecum, and colon of germ-free (GF) and monocolonized (MC) mice. (A) PCA score plot of bile acid signatures in jejunum, cecum and colon of GF and MC mice. (B) PCA score plot of bile acid signatures in cecum and colon of GF and MC mice. (C) Total relative intensity measurements of bile acids based on conjugation state in jejunum, cecum, and colon of GF and MC mice. (D) Intensity values for the measured bile acids in jejunum, cecum, and colon of GF and MC mice. The color of the bar refers to the conjugation state of the bile acid; unconjugated bile acids (red), taurine-conjugated bile acids (yellow), and glycine-conjugated bile acids (blue). Values are mean intensities ± SEM measured using LC-MS in negative ionization mode. Mann-Whitney t test vs. GF, * (P < 0.05), ** (P < 0.01). nd; not detected, CA, cholic acid; TCA, taurocholic acid; TβMCA, tauro-β-muricholic acid; TCDCA, taurochenodeoxycholic acid; TDCA, taurodeoxycholic acid; GCA, glycocholic acid. See also Table S1.
Effect of L. acidophilus NCFM on plant polysaccharide metabolism
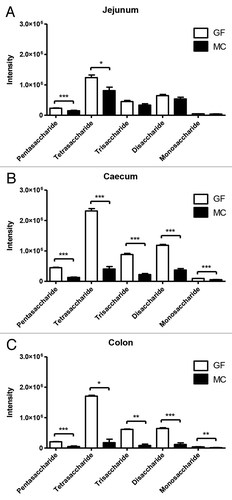
The metabolome data of the luminal samples in positive ionization mode revealed five different metabolites with low retention times that differed significantly between GF and MC. These metabolites were classified as carbohydrates, as all potential candidates from the METLIN database search were carbohydrates (Δppm < 1). The five metabolites were suggested to be a pentasaccharide, a tetrasaccharide, a trisaccharide, a disaccharide, and a monosaccharide (Table S1). The abundance of the metabolites differed between MC and GF groups along the intestinal tract, with the most pronounced differences measured in cecum (). Levels of pentasaccharides and tetrasaccharides were significantly lower in MC compared with GF in all three gut compartments, indicating that these metabolites were metabolized by NCFM. Additionally, levels of all five carbohydrate metabolites differed significantly between GF and MC in cecum and colon, where higher abundances were observed in GF than in MC.
Figure 2. Intensity values for five measured saccharides in germ-free (GF) and monocolonized (MC) mice in (A) jejunum, (B) cecum, and (C) colon. Values are mean intensities ± SEM measured using LC-MS in positive ionization mode. Stars indicate significant differences; * (P < 0.05), ** (P < 0.01), *** (P < 0.001).
Effect of L. acidophilus NCFM on vitamin E absorption and/or metabolism
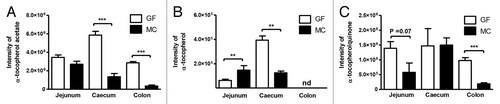
Interestingly, the abundance (intensity) of α-tocopherol acetate (vitamin E acetate) differed significantly between GF and MC in cecum and colon (P < 0.001), while in jejunum, this difference was not statistically significant (P = 0.11) (). α-tocopherol (vitamin E) was found in significantly higher levels in jejunum of MC than in GF (P < 0.01), whereas in cecum the levels of α-tocopherol were significantly lower in MC compared with GF (P < 0.01). α-tocopherol was not detected in colon. In addition, the level of α-tocopherolquinone (oxidized form of vitamin E) differed significantly in colon with higher levels observed in GF than in MC (P < 0.001), and a similar trend was observed in jejunum (P = 0.07). The observed differences suggest increased metabolism and/or absorption of α-tocopherol acetate and α-tocopherol throughout the gut in the presence of NCFM. To investigate whether NCFM degraded vitamin E acetate, we performed an in vitro digestion experiment where NCFM was grown 24 h in medium containing vitamin E acetate. Measurements of the level of vitamin E acetate and vitamin E by LC-MS before (0 h) and after (24 h) fermentation revealed that NCFM did not hydrolyze vitamin E acetate (Fig. S2).
Figure 3. Intensity values for the measured (A) α-tocopherol acetate, (B) α-tocopherol, and (C) α-tocopherolquinone in jejunum, cecum, and colon of germ-free (GF) and monocolonized (MC) mice. Values are mean intensities ± SEM measured by LC-MS in positive ionization mode. Stars indicate significant differences; * P < 0.05, ** P < 0.01, *** P < 0.001. nd, not detected.
Discussion
Here, we showed that the NCFM successfully colonized germfree mice in numbers gradually increasing from the jejunum to colon, thereby confirming the ability of NCFM to survive and proliferate in the gut of rodents.Citation26 The observed levels of NCFM in the intestine are similar to the levels found in other studies with monocolonized mice.Citation8,Citation27 A non-targeted metabolomics approach revealed indisputably that the presence of NCFM affected the gut metabolome (Fig. S1). This was in accordance with previous studies comparing GF rodents with conventionalized rodents, showing that the presence of a gut microbiota affects the metabolome in the gut.Citation1,Citation2,Citation28 Comparing the metabolome of the intestinal samples from GF and MC mice by PCA, we found that the differing metabolites were mainly bile acids, carbohydrates, and vitamin E.
We identified a selection of bile acids, which we compared between intestinal sites and between the GF and MC mice. Generally, the bile acid signature was site-specific with higher bile acid amounts measured in jejunum than in cecum and colon (), which is in accordance with the fact that most of the bile acids (95% in humans) are reabsorbed by active transport in the distal ileum.Citation29 A similar site-specific bile acid signature has been observed in other studies.Citation2,Citation5 In all intestinal samples we found taurine conjugated bile acids to be most abundant, which is in agreement with previous reports.Citation2,Citation5 Also, the predominance of TCA and TβMCA in jejunum of mice observed in the present study is confirmed by other studies.Citation5,Citation6 Comparing the bile acid signature of GF and MC mice revealed similar levels of taurine and glycine conjugated bile acids, but significantly higher amounts of unconjugated bile acids in MC than in GF mice in all three gut compartments. The increased abundance of unconjugated CA observed in MC mice supports that NCFM has the ability to deconjugate bile acids, as previously shown in vitro.Citation19 It has been suggested that probiotic bacteria reduce secondary bile acid formation in the human colon by sequestering bile acids,Citation18 and here we observed a difference in the colonic level of the secondary bile acid, TDCA, when comparing GF and MC. However, it cannot be excluded that the changed bile acid signature in MC was caused by a general response to the colonization and not induced by species-specific bile salt metabolism in the host. The monocolonization with NCFM did not induce the same drastic changes in the bile acid composition as observed after introduction of a complex microbiota.Citation2,Citation5 Nevertheless, we found that presence of a single bacterial species does indeed affect the abundance of specific bile acids, which might in turn affect the metabolism of the host, as bile acids lately have been recognized as important signal molecules regulating biosynthetic and metabolic pathways through activation of nuclear receptors, such as farnesoid X receptor (FXR), the G protein-coupled receptor TGR5, and the pregnane X receptor (PXR) in intestine and liver.Citation30,Citation31
The carbohydrate metabolism was found to be affected by the presence of NCFM (), which is consistent with the fact that NCFM is known to encode a large variety of genes related to carbohydrate metabolism.Citation17 The largest differences between GF and MC mice in terms of carbohydrate metabolism were observed in cecum and colon, which is in agreement with the fact that mice are well-known to be cecal fermenters.Citation32 Notably, the levels of penta- and tetrasaccharides differed significantly in all gut compartments between GF and MC mice, suggesting that these metabolites represented carbohydrates that were indigestible by the host enzymes, and therefore accumulated in the cecum and colon of GF mice. NCFM is capable of utilizing a variety of complex carbohydrates that escape digestion such as raffinose and fructooligosaccharides.Citation17,Citation21,Citation33
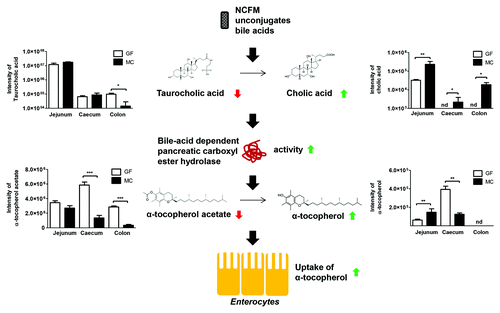
Increased metabolism and/or absorption of α-tocopherol acetate (vitamin E acetate) was observed in MC mice as compared with GF mice (). This has to our knowledge not previously been reported. α-tocopherol acetate is a chemically stable form of α-tocopherol used as a nutritional vitamin E supplement, and known to be present in the fortified chow fed to mice in this study. Before uptake into intestinal epithelial cells, dietary α-tocopherol is directly solubilized into mixed micelles by bile acids and lipids, whereas the α-tocopherol acetate requires hydrolysis prior to micellar solubilization and enterocyte uptake.Citation34 Vitamin E uptake in humans is characterized by a large inter-individual difference,Citation35 and the molecular mechanism behind the absorption of vitamin E remains unresolved.Citation36 The large inter-individual difference in vitamin E absorption has previously been suggested to be affected by gut permeability,Citation37 availability of dietary fat,Citation38 as well as by a pancreatic bile acid dependent carboxyl ester lipase (CEL) responsible for hydroxylation of tocopheryl esters.Citation39 Given that the gut microbiota affects gut permeability,Citation40 lipid metabolism,Citation4 and bile acid composition,Citation2,Citation5,Citation6 we suggest that the gut microbiota plays an important role for vitamin E absorption. Consistently, a previous report showed that the uptake of α-tocopherol was increased upon antibiotic administration to broilers leading to a changed gut microbiota and changed bile acid signature.Citation41 Here, we showed that NCFM did not digest vitamin E acetate (Fig. S2), which is in line with other studies showing that ruminal microorganisms are not able to degrade vitamin E.Citation42,Citation43 We therefore propose that NCFM influenced the vitamin E absorption in MC mice by affecting lipid emulsification and/or activation of lipolytic enzymes by changing the lipid and bile acid profiles. MC animals displayed an increased level of CA, which in vitro has been shown to be a very effective activator of CEL responsible for hydrolysis of α-tocopherol acetate prior to absorption.Citation44 This lipase is also found in mice and has previously been suggested to facilitate intestinal absorption of fat-soluble vitamins.Citation45 This suggests that the increased deconjugation of bile acids by NCFM could lead to increased activity of CEL, thereby increasing the conversion of α-tocopherol acetate to α-tocopherol leading to increased uptake of vitamin E in the MC mice (). However, it cannot be excluded that the mere presence of bacteria may alter host expression of pancreatic enzymes thereby changing the metabolism. A study in rats found that most of the absorption of α-tocopherol took place in the medial portion of the small bowel, assumed to be the ileum,Citation46 which might explain why we observed a lower amount of α-tocopherol in cecum of MC mice compared with GF mice. The decreased level of α-tocopherolquinone, which is a major product of oxidation of α-tocopherol,Citation47 observed in MC mice compared with GF mice could either be due to increased uptake of α-tocopherol in MC mice, or to a more anaerobic environment in the gut resulting from colonization of NCFM.
Figure 4. Proposed mechanism for Lactobacillus acidophilus NCFM (NCFM) mediated absorption of vitamin E (α-tocopherol). NCFM unconjugates taurocholic acid to cholic acid. Because cholic acid is an effective activator of the bile acid dependent pancreatic carboxyl ester hydrolase, this increases the activity of this enzyme, which results in an increased conversion of α-tocopherol acetate into α-tocopherol, which is the form of vitamin E that can be taken up by enterocytes through micelles. GF, germ-free; MC, monocolonized; nd, not detected.
Taken together, our data show that colonization of GF mice with NCFM alters the intestinal metabolome by modifying the bile acid signature, affecting the carbohydrate and affecting the vitamin E absorption and/or metabolism. This confirms that intestinal bacteria affect the bile acid signature and further suggests that the gut microbiota may be an important factor in vitamin E absorption, as the observed effect may not be unique to NCFM. The possible link between the gut microbiota, bile acids, and vitamin E absorption should be further explored in other studies, as the perspectives for human and animal health are of significance.Citation48,Citation49
Materials and Methods
Lactobacillus acidophilus NCFM inoculation preparation
Lactobacillus acidophilus NCFMTM was kindly provided by Danisco A/S (DuPont). The strain was grown anaerobically at 37 °C for 24 h in de Man, Rogosa, and Sharpe (MRS) broth (Oxoid Ltd). After centrifugation at 3000 g for 15 min, pellets were washed and resuspended in a sterile saline supplement with 0.1% peptone. Final concentration of the NCFM cells in the inoculum was around 6.3 × 109 CFU/ml.
Animal handling and sample collection
The animal studies were conducted according to the Federation of European Laboratory Animal Science Associations (FELASA) and Danish legislation. Germfree Swiss Webster mice, bred at the National Food Institute (Technical University of Denmark), were originally obtained from Taconic (Lille Skensved, Denmark) and kept in sterile isolators under a strict 12 h light cycle. Animals were maintained on γ-ray-irradiated Altromin-1320 fortified chow (Altromin Spezialfutter GmbH and Co) ad libitum. Absence of colonizing bacteria in GF mice was controlled by cultivation of fecal samples twice a week throughout the whole study. One group of 5 male mice at age of 5 wk was kept germfree (GF) and one group of 5 male mice at age of 5 wk was monocolonized (MC) with a single dose of 200 μL NCFM inoculum, prepared as described above, resulting in around 109 viable bacteria per dosage. After colonization, fecal samples obtained from the MC mice were cultivated twice a week as described below in order to evaluate the efficiency of the NCFM colonization and stabilization in the gastrointestinal tract. All mice were anesthetized by fentanyl and/or fluniasone (Hypnorm; Janssen Animal Health) and midazolam (Dormicum; Roche) and euthanized by decapitation at 8 wk of age. After termination, internal content (lumen) of the jejunum, cecum and colon were collected, snap-frozen in liquid nitrogen and kept at -80 °C until further use.
Enumeration of bacteria
Fecal samples from both groups of mice and luminal samples from jejunum, cecum and colon of the MC mice were suspended in sterile saline supplemented with 0.1% of peptone. NCFM of all fecal and luminal samples were counted on MRS agar (Oxoid Ltd) after anaerobic incubation at 37 °C for 48 h. Additionally, fecal samples collected twice a week and all luminal samples were screened for absence of contamination by plating on Luria-Bertani (LB; Oxoid Ltd) agar incubated at 37 °C for 48 h under aerobic and anaerobic atmosphere.
Preparation of extracts
All samples were defrosted on wet ice and kept cold throughout all procedures. Intestinal lumen samples were weighed and homogenized by grinding in glass tubes while kept on ice. 500 μL of cold 80% methanol (Fluka, Sigma-Aldrich) was added to the samples, which were then vortexed for 1 min and centrifuged at 4000 g for 5 min at 4 °C. Methanol and/or water extracts were removed. This procedure was performed three times and extracts were combined in one tube and stored at -20 °C until further use. Subsequently, methanol and/or water extracts were lyophilized in 12L Labconco Freeze Dryer (Labconco Corporation). Prior to freeze-drying, the concentration of methanol was lowered below 15% by adding cold MQ water. Dried extracts were kept at -20 °C until LC-MS analysis.
Metabolomics profiling with LC-MS
The extracts were resolubilized in 5% acetonitrile (Fluka, Sigma-Aldrich) adjusting for original weight, ultrasonicated for 10 min at 4 °C and centrifuged at 10 000 g for 7 min at 4 °C. The supernatants were removed and analyzed by LC-MS using a Dionex Ultimate 3000 RS liquid chromatograph (Dionex) coupled to a Bruker maXis time of flight mass spectrometer equipped with an electrospray interphase (Bruker Daltonics). Analytes were separated on a Kinetex pentafluorophenyl column 100 × 2.10 mm, 2.6 μm, 100 Å (Phenomenex), using the solvent system: A (5 mM ammonium formate with 0.1% formic acid; both from Fluka, Sigma-Aldrich), and B (acetonitrile, Fluka, Sigma-Aldrich, with 0.1% formic acid). Solvent programming was isocratic 0% B for 2 min followed by 5% at 5 min, then linear gradient up to 100% B at 10 min and 100% B isocratic until 12 min. The solvent composition was returned to initial conditions at 12.1 min and recalibrated to 14 min. Flow rate was 0.3 ml/min. The oven temperature was 40 °C. Injection volumes were 3 μL. The following electrospray interphase settings were used: Nebulizer pressure 2 bar, drying gas 10 L/min, 200 °C, capillary voltage 4500 V. Scan range was from 100–1000 mass/charge (m/z). Samples were analyzed in both positive and negative mode. External and internal calibration was done using sodium formate clusters (Sigma-Aldrich). Lock-mass calibration (hexakis(1H,1H,2H-perfluoroetoxy)phosphazene, Apollo Scientific) was applied in order to lower the measurement error.
In vitro digestion of vitamin E acetate
Minimal medium was prepared as previously described,Citation22 adjusted to pH 7 with sterile solutions of HCl and NaOH, and 0.5% (vol./vol.) 2 µM α-tocopherol acetate (vitamin E acetate) was added to the medium. Fermentation tests were performed as previously described.Citation22 Shortly, medium containing α-tocopherol acetate, obtained from Sigma-Aldrich, was inoculated with 1% (vol./vol.) of an overnight culture of NCFM grown in minimal medium under anaerobic conditions at 37 °C. As a negative control, sterile minimal medium was added (1% vol./vol.) to the fermentation medium instead of the overnight culture. As positive control, cholesterol ester hydrolase (CEH, specific activity 35 U/mg) from porcine pancreas obtained from Sigma-Aldrich was diluted in 0.1M phosphate buffer and added (1% vol./vol., CEH concentration of 2 U/mL) to the fermentation medium, as CEH is known to hydrolyze α-tocopherol acetate into α-tocopherol.Citation50 Inoculation with NCFM, minimal medium (negative control), and CEH (positive control) was performed in triplicates. After 0 h and 24 h of fermentation, 100 µl of sample was quenched with 100 µl cold 50% methanol (Fluka, Sigma-Aldrich), incubated for 10 min at 4 °C, and spun down at 13 000 g for 5 min at 4 °C. Subsequently, 180 µl of the supernatant was transferred to another tube and stored at -80 °C until LC-MS analysis. The abundances of α-tocopherol acetate and α-tocopherol were measured in all fermentation samples by MS in positive mode using the same system as described above with the following adjustments: A SB-C8 column 100 × 2.10 mm, 2.7 μm, 120 Å (Agilent Technologies) was used to allow better separation of vitamin E acetate and minimal medium ingredients. Solvent programming started at 50% B with a linear gradient up to 95% B at 10 min, and 95% B isocratic until 12 min. The solvent composition was returned to initial conditions at 12.1 min and recalibrated to 15 min. Flow rate was 0.4 ml/min. The oven temperature was 50 °C. Injection volumes were 2 μL.
Data analysis and statistics
The raw LC-MS data were converted to mzXML files using CompassXport software (Bruker Daltonics) and were then pre-processed through noise filtering, peak detection, and alignment using R package XCMS (v.1.36.0)Citation51 before multivariate statistical analysis. Data tables were generated comprising m/z, retention time, and intensity (peak area) for each variable in every sample, which were imported into LatentiX (version 2.11) (Latent5) for further multivariate analysis. Principal component analysis (PCA) was performed on all mean-centered data. Intensity values are expressed as mean ± SEM. Significant differences between two groups were evaluated with a two-tailed, unpaired t test or Mann-Whitney U test if variances were significantly different. Accurate masses of features of interest were searched against the METLIN,Citation23 HMDB,Citation24 and LIPID MAPSCitation25 databases. The identity of bile acids, α-tocopherol, α-tocopherol acetate, and α-tocopherolquinone was confirmed by LC-MS/MS in negative and positive ionization mode of authentic compounds giving the retention time and m/z of these metabolites, which was manually compared with the initial analysis. Authentic compounds (cholic acid, taurocholic acid, taurochenodeoxycholic acid, taurodeoxycholic acid, glycocholic acid, α-tocopherol, α-tocopherol acetate, and α-tocopherolquinone) were obtained from Sigma-Aldrich. Retention times and masses of identified metabolites are presented in Table S1.
| Abbreviations: | ||
| CA | = | Cholic acid |
| CEH | = | Cholesterol ester hydrolase |
| CEL | = | Carboxyl ester lipase |
| GCA | = | Glycocholic acid |
| FXR | = | Farnesoid X receptor |
| GF | = | Germ Free |
| MC | = | Monocolonized with NCFM |
| LC-MS | = | Liquid chromatography coupled to mass-spectrometry |
| NCFM | = | Lactobacillus acidophilus NCFM |
| PCA | = | Principal Component Analysis |
| PXR | = | Pregnane X receptor |
| TCA | = | taurocholic acid |
| TβMCA | = | tauro-β-muricholic acid |
| TCDCA | = | taurochenodeoxycholic acid |
| TDCA | = | taurodeoxycholic acid |
Additional material
Download Zip (251.8 KB)Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
Authors would like to thank Bodil Madsen, Kate Vibefeldt and Dang-Dung Nguyen for excellent technical support. Øresund Food Network (Grant no.09198-02; Bacterial Impact on the Gut Metabolome) and the Danish Council for Strategic Research (Grant no. 11-116163; Center for Gut, Grain, and Greens) funded the work.
References
- Claus SP, Tsang TM, Wang Y, Cloarec O, Skordi E, Martin FP, Rezzi S, Ross A, Kochhar S, Holmes E, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol 2008; 4:219; http://dx.doi.org/10.1038/msb.2008.56; PMID: 18854818
- Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A 2011; 108:Suppl 1 4523 - 30; http://dx.doi.org/10.1073/pnas.1006734107; PMID: 20837534
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 2004; 101:15718 - 23; http://dx.doi.org/10.1073/pnas.0407076101; PMID: 15505215
- Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, Felin J, Perkins R, Borén J, Oresic M, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res 2010; 51:1101 - 12; http://dx.doi.org/10.1194/jlr.M002774; PMID: 20040631
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 2013; 17:225 - 35; http://dx.doi.org/10.1016/j.cmet.2013.01.003; PMID: 23395169
- Martin FP, Dumas ME, Wang Y, Legido-Quigley C, Yap IK, Tang H, Zirah S, Murphy GM, Cloarec O, Lindon JC, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol 2007; 3:112; http://dx.doi.org/10.1038/msb4100153; PMID: 17515922
- Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A 2009; 106:5859 - 64; http://dx.doi.org/10.1073/pnas.0901529106; PMID: 19321416
- Derrien M, Van Baarlen P, Hooiveld G, Norin E, Müller M, de Vos WM. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader akkermansia muciniphila. Front Microbiol 2011; 2:166; http://dx.doi.org/10.3389/fmicb.2011.00166; PMID: 21904534
- Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A 2002; 99:15451 - 5; http://dx.doi.org/10.1073/pnas.202604299; PMID: 12432102
- Sanders ME, Klaenhammer TR. Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J Dairy Sci 2001; 84:319 - 31; http://dx.doi.org/10.3168/jds.S0022-0302(01)74481-5; PMID: 11233016
- Dunn SR, Simenhoff ML, Ahmed KE, Gaughan WJ, Eltayeb BO, Fitzpatrick MD, et al. Effect of oral administration of freeze-dried lactobacillus acidophilus on small bowel bacterial overgrowth in patients with end stage kidney disease: Reducing uremic toxins and improving nutrition. Int Dairy J 1998; 8:545; http://dx.doi.org/10.1016/S0958-6946(98)00081-8
- Ouwehand AC, Tiihonen K, Saarinen M, Putaala H, Rautonen N. Influence of a combination of Lactobacillus acidophilus NCFM and lactitol on healthy elderly: intestinal and immune parameters. Br J Nutr 2009; 101:367 - 75; http://dx.doi.org/10.1017/S0007114508003097; PMID: 18634707
- Larsen N, Vogensen FK, Gøbel R, Michaelsen KF, Abu Al-Soud W, Sørensen SJ, Hansen LH, Jakobsen M. Predominant genera of fecal microbiota in children with atopic dermatitis are not altered by intake of probiotic bacteria Lactobacillus acidophilus NCFM and Bifidobacterium animalis subsp. lactis Bi-07. FEMS Microbiol Ecol 2011; 75:482 - 96; http://dx.doi.org/10.1111/j.1574-6941.2010.01024.x; PMID: 21204871
- Andreasen AS, Larsen N, Pedersen-Skovsgaard T, Berg RM, Møller K, Svendsen KD, Jakobsen M, Pedersen BK. Effects of Lactobacillus acidophilus NCFM on insulin sensitivity and the systemic inflammatory response in human subjects. Br J Nutr 2010; 104:1831 - 8; http://dx.doi.org/10.1017/S0007114510002874; PMID: 20815975
- Greene JD, Klaenhammer TR. Factors involved in adherence of lactobacilli to human Caco-2 cells. Appl Environ Microbiol 1994; 60:4487 - 94; PMID: 7811085
- Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, Molle D, Lortal S, Altermann E, Klaenhammer TR, et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci U S A 2008; 105:19474 - 9; http://dx.doi.org/10.1073/pnas.0810305105; PMID: 19047644
- Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M, et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci U S A 2005; 102:3906 - 12; http://dx.doi.org/10.1073/pnas.0409188102; PMID: 15671160
- Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 2006; 47:241 - 59; http://dx.doi.org/10.1194/jlr.R500013-JLR200; PMID: 16299351
- McAuliffe O, Cano RJ, Klaenhammer TR. Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM. Appl Environ Microbiol 2005; 71:4925 - 9; http://dx.doi.org/10.1128/AEM.71.8.4925-4929.2005; PMID: 16085898
- Pfeiler EA, Azcarate-Peril MA, Klaenhammer TR. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J Bacteriol 2007; 189:4624 - 34; http://dx.doi.org/10.1128/JB.00337-07; PMID: 17449631
- Andersen JM, Barrangou R, Abou Hachem M, Lahtinen S, Goh YJ, Svensson B, Klaenhammer TR. Transcriptional and functional analysis of galactooligosaccharide uptake by lacS in Lactobacillus acidophilus. Proc Natl Acad Sci U S A 2011; 108:17785 - 90; http://dx.doi.org/10.1073/pnas.1114152108; PMID: 22006318
- Sulek K, Frandsen HL, Smedsgaard J, Skov TH, Wilcks A, Licht TR. Metabolic footprint of lactobacillus acidophilus NCFM at different pH. Metabolomics 2012; 8:244 - 52; http://dx.doi.org/10.1007/s11306-011-0305-4
- Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit 2005; 27:747 - 51; http://dx.doi.org/10.1097/01.ftd.0000179845.53213.39; PMID: 16404815
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res 2013; 41:D801 - 7; http://dx.doi.org/10.1093/nar/gks1065; PMID: 23161693
- Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res 2007; 35:W606-12; http://dx.doi.org/10.1093/nar/gkm324; PMID: 17584797
- Kaplan CW, Astaire JC, Sanders ME, Reddy BS, Kitts CL. 16S ribosomal DNA terminal restriction fragment pattern analysis of bacterial communities in feces of rats fed Lactobacillus acidophilus NCFM. Appl Environ Microbiol 2001; 67:1935 - 9; http://dx.doi.org/10.1128/AEM.67.4.1935-1939.2001; PMID: 11282651
- Martin FP, Wang Y, Sprenger N, Holmes E, Lindon JC, Kochhar S, Nicholson JK. Effects of probiotic Lactobacillus paracasei treatment on the host gut tissue metabolic profiles probed via magic-angle-spinning NMR spectroscopy. J Proteome Res 2007; 6:1471 - 81; http://dx.doi.org/10.1021/pr060596a; PMID: 17316039
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 2009; 106:3698 - 703; http://dx.doi.org/10.1073/pnas.0812874106; PMID: 19234110
- Chiang JY. Bile acids: regulation of synthesis. J Lipid Res 2009; 50:1955 - 66; http://dx.doi.org/10.1194/jlr.R900010-JLR200; PMID: 19346330
- Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res 2009; 50:1509 - 20; http://dx.doi.org/10.1194/jlr.R900007-JLR200; PMID: 19346331
- Matsubara T, Li F, Gonzalez FJ. FXR signaling in the enterohepatic system. Mol Cell Endocrinol 2013; 368:17 - 29; http://dx.doi.org/10.1016/j.mce.2012.05.004; PMID: 22609541
- Sakaguchi E. Digestive strategies of small hindgut fermenters. Anim Sci J 2003; 74:327 - 37; http://dx.doi.org/10.1046/j.1344-3941.2003.00124.x
- Barrangou R, Altermann E, Hutkins R, Cano R, Klaenhammer TR. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc Natl Acad Sci U S A 2003; 100:8957 - 62; http://dx.doi.org/10.1073/pnas.1332765100; PMID: 12847288
- Rigotti A. Absorption, transport, and tissue delivery of vitamin E. Mol Aspects Med 2007; 28:423 - 36; http://dx.doi.org/10.1016/j.mam.2007.01.002; PMID: 17320165
- Jeanes YM, Hall WL, Lodge JK. Comparative (2)H-labelled alpha-tocopherol biokinetics in plasma, lipoproteins, erythrocytes, platelets and lymphocytes in normolipidaemic males. Br J Nutr 2005; 94:92 - 9; http://dx.doi.org/10.1079/BJN20051434; PMID: 16115338
- Borel P, Preveraud D, Desmarchelier C. Bioavailability of vitamin E in humans: an update. Nutr Rev 2013; 71:319 - 31; http://dx.doi.org/10.1111/nure.12026; PMID: 23731443
- Abuasal BS, Qosa H, Sylvester PW, Kaddoumi A. Comparison of the intestinal absorption and bioavailability of γ-tocotrienol and α-tocopherol: in vitro, in situ and in vivo studies. Biopharm Drug Dispos 2012; 33:246 - 56; http://dx.doi.org/10.1002/bdd.1790; PMID: 22528033
- Bruno RS, Leonard SW, Park SI, Zhao Y, Traber MG. Human vitamin E requirements assessed with the use of apples fortified with deuterium-labeled alpha-tocopheryl acetate. Am J Clin Nutr 2006; 83:299 - 304; PMID: 16469987
- Lombardo D, Guy O. Studies on the substrate specificity of a carboxyl ester hydrolase from human pancreatic juice. II. Action on cholesterol esters and lipid-soluble vitamin esters. Biochim Biophys Acta 1980; 611:147 - 55; http://dx.doi.org/10.1016/0005-2744(80)90050-9; PMID: 7350913
- Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature 2012; 489:242 - 9; http://dx.doi.org/10.1038/nature11552; PMID: 22972297
- Knarreborg A, Lauridsen C, Engberg RM, Jensen SK. Dietary antibiotic growth promoters enhance the bioavailability of alpha-tocopheryl acetate in broilers by altering lipid absorption. J Nutr 2004; 134:1487 - 92; PMID: 15173416
- Leedle RA, Leedle JA, Butine MD. Vitamin E is not degraded by ruminal microorganisms: assessment with ruminal contents from a steer fed a high-concentrate diet. J Anim Sci 1993; 71:3442 - 50; PMID: 8294298
- Chikunya S, Demirel G, Enser M, Wood JD, Wilkinson RG, Sinclair LA. Biohydrogenation of dietary n-3 PUFA and stability of ingested vitamin E in the rumen, and their effects on microbial activity in sheep. Br J Nutr 2004; 91:539 - 50; http://dx.doi.org/10.1079/BJN20031078; PMID: 15035681
- Lauridsen C, Hedemann MS, Jensen SK. Hydrolysis of tocopheryl and retinyl esters by porcine carboxyl ester hydrolase is affected by their carboxylate moiety and bile acids. J Nutr Biochem 2001; 12:219 - 24; http://dx.doi.org/10.1016/S0955-2863(00)00156-X; PMID: 11287217
- Hui DY, Howles PN. Carboxyl ester lipase: structure-function relationship and physiological role in lipoprotein metabolism and atherosclerosis. J Lipid Res 2002; 43:2017 - 30; http://dx.doi.org/10.1194/jlr.R200013-JLR200; PMID: 12454261
- Hollander D, Rim E, Muralidhara KS. Mechanism and site of small intestinal absorption of alpha-tocopherol in the rat. Gastroenterology 1975; 68:1492 - 9; PMID: 1132630
- Wu JH, Croft KD. Vitamin E metabolism. Mol Aspects Med 2007; 28:437 - 52; http://dx.doi.org/10.1016/j.mam.2006.12.007; PMID: 17306359
- Vagni S, Saccone F, Pinotti L, Baldi A.. Vitamin E bioavailability: Past and present insights. Food and nutrition sciences 2011; 2:1088; http://dx.doi.org/10.4236/fns.2011.210146
- Maras JE, Bermudez OI, Qiao N, Bakun PJ, Boody-Alter EL, Tucker KL. Intake of alpha-tocopherol is limited among US adults. J Am Diet Assoc 2004; 104:567 - 75; http://dx.doi.org/10.1016/j.jada.2004.01.004; PMID: 15054342
- Desmarchelier C, Tourniaire F, Prévéraud DP, Samson-Kremser C, Crenon I, Rosilio V, Borel P. The distribution and relative hydrolysis of tocopheryl acetate in the different matrices coexisting in the lumen of the small intestine during digestion could explain its low bioavailability. Mol Nutr Food Res 2013; 57:1237 - 45; http://dx.doi.org/10.1002/mnfr.201200720; PMID: 23520193
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 2006; 78:779 - 87; http://dx.doi.org/10.1021/ac051437y; PMID: 16448051
