Abstract
Background
Interactions between the microbial flora of the intestine and the human host play a critical role in maintaining intestinal health and in the pathophysiology of a wide variety of disorders such as antibiotic associated diarrhea, Clostridium difficile infection, and inflammatory bowel disease. Prebiotics can confer health benefits by beneficial effects on the intestinal microbiome, whereas antibiotics can disrupt the microbiome leading to diarrhea and other side effects.
Aim
To compare the effects of the prebiotic, polysaccharopeptide from Trametes versicolor, to those of the antibiotic, amoxicillin, on the human gut microbiome
Methods
Twenty-four healthy volunteers were randomized to receive PSP, amoxicillin, or no treatment (control). Stool specimens were analyzed using bTEFAP microbial ecology methods on seven occasions over 8 weeks from each participant in the active treatment groups and on three occasions for the controls.
Results
Twenty-two of 24 participants completed the protocol. PSP led to clear and consistent microbiome changes consistent with its activity as a prebiotic. Despite the diversity of the human microbiome we noted strong microbiome clustering among subjects. Baseline microbiomes tended to remain stable and to overshadow the treatment effects. Amoxicillin treatment caused substantial microbiome changes most notably an increase in Escherichia/Shigella. Antibiotic associated changes persisted to the end of the study, 42 days after antibiotic therapy ended.
Conclusions
The microbiomes of healthy individuals show substantial diversity but remain stable over time. The antibiotic amoxicillin alters the microbiome and recovery from this disruption can take several weeks. PSP from T. versicolor acts as a prebiotic to modulate human intestinal microbiome composition.
Introduction
Microbes found in the human intestine represent a diverse ecosystem of multiple co-existing species.Citation1,Citation2 There are trillions of microbes in the normal human gut of which bacteria are most abundant forming almost 60% of fecal mass and numbering >1012 per gram of stool.Citation1-Citation3 Recent research has established many vital functions for gut microbes including: metabolic activities that lead to the salvage of energy and absorbable nutrients, trophic effects on the intestinal epithelium, crucial maturation and development effects on immune structure and function, and protection of the host against colonization by pathogenic microbes.Citation1,Citation2 Interactions between the microbial flora of the intestine and the host appear to play a critical role in maintaining intestinal health and in the pathophysiology of a variety of intestinal diseases such as inflammatory bowel disease (IBD), antibiotic associated diarrhea and Clostridium difficile infection (CDI).Citation1,Citation2 With new data emerging about the importance of gut microbes in intestinal health, the use of prebiotic and probiotic agents are increasingly being investigated.Citation1,Citation2
Prebiotics are a category of edible agents that are selectively fermented and induce changes in the composition and/or activity of the gastrointestinal microbiome that in turn can confer benefits upon host well-being and health.Citation1,Citation2,Citation4-Citation7 Trametes versicolor (also known as Coriolus versicolor or turkey tail) is one of the most common mushrooms in North American woods, found virtually anywhere that there are decomposing hardwood logs, tree stumps, or, less commonly, conifer wood.Citation8,Citation9 Polysaccharopeptide (PSP) is a protein-bound polysaccharide extracted from the mycelia of T. versicolor. It has been well known in Chinese literature for centuries and is used frequently with the goal of regulating the immune system.Citation8,Citation9 PSP from T. versicolor culture extract has been used in Japan, China, and Russia as an anti-tumor agent.Citation8-Citation11 It is generally considered to be safe and is distributed as an over the counter nutritional supplement with no known significant toxicity in humans.Citation8-Citation11 More recently it was shown that T. versicolor PSP modifies human fecal microbiome composition in vitro, indicating that it may have prebiotic properties.Citation12
Conversely, antimicrobial drugs disrupt the normal gut microbiome, usually as an inadvertent side effect, during their therapeutic use for systemic infections.Citation13-Citation15 This disruption frequently results in candida overgrowth, simple AAD, and, occasionally, in the more serious condition of CDI.Citation14-Citation17 In the case of CDI, disruption of the intestinal microbiome during antibiotic treatment weakens the colonization resistance conferred by a healthy gut microbiome and allows for opportunistic CDI.Citation14,Citation15
Thus, understanding how the intestinal microbiome are altered by prebiotic and by antibiotic treatments is of central importance to understanding the pathophysiology of AAD and CDI.Citation15,Citation16 It is also relevant to disease mechanisms in many other gastrointestinal disorders such as IBD and IBS.Citation18,Citation19 Here we present the results of an open label, randomized clinical trial to compare and contrast the effects of a prebiotic, PSP from T. versicolor, and an antibiotic, amoxicillin, on the intestinal microbiome of healthy adults. Our study findings indicate persisting effects of antibiotic therapy on the human intestinal microbiome as well as a role for PSP as a prebiotic agent in humans.
Results
Clinical study results
Eight subjects were recruited for each study group (PSP, Amoxicillin, and Control). Two subjects, both in the PSP group, did not comply with the study protocol and their samples were excluded from the analysis. compares the characteristics of study subjects in the three groups. The participants ranged in age from 21 to 51 years (mean 30.6) with 15 (68.2%) being females. No serious adverse events were recorded. Minor adverse events or symptoms were reported on 11 occasions by eight patients. Of the 11 events, 3 were felt to be associated with the study agent and one with the dietary restrictions. Two patients receiving amoxicillin reported a softer (but still formed) consistency of stool. One participant in the PSP arm reported increased gas and burping. Symptoms in all three participants were limited to the duration of active study agent consumption. A fourth participant became constipated after entering the study but resumed regular bowel habit upon resuming consumption of kimchi after the study. One participant in the PSP arm reported improved bowel regularity while taking the supplement. Four control subjects failed to comply completely with the dietary restriction (ingested 1 cup of miso soup [day 24], 1 cup of yogurt [day 40], 1/4 cup Parmesan cheese [day 46], 1 cup of yogurt [day 46]).
Table 3. Comparison of different study populations
Evaluation of overall microbial diversity
A total of 1 821 806 sequences were derived among the 121 samples from the study. After stringent quality sequence curation a total of 1 127 082 sequences identified within the bacterial kingdom were utilized for final microbiome analyses with an average of 9238 sequences per sample. Rarefaction and Operational Taxonomic Unit (OTU) analyses were performed on each of the samples. There were no significant differences between the samples.
The most clear and consistent finding from the analyses in this study is the heterogeneity of the human microbiome. Healthy humans tend to have diverse, even unique, microbial assemblages due to highly interactive factors that include varying diets, environments, stress, sleep cycles, and genetics among many other potential impacts. The diversity of the human microbiome notwithstanding, in the current study, we noted a strong clustering of β diversity among subjects prior to treatment. These baseline microbiomes tended to remain stable over time and showed a strong tendency to overshadow differentiation due to treatment effects. Note in that when utilizing clustering of the microbial populations, at the genus level—based upon Ward’s Minimum Variance and Manhattan distance—that the individual subject’s samples cluster together over time. Note also that this individual clustering is more consistent and more dominant than treatment-related effects. Put differently, the samples from an individual subject tended to show a consistent microbiome over time despite the different treatments being administered.
Figure 2. Dual Hierarchal dendrogram with subjects clustered on the X-axis and coded by subject number, treatment group and sample day. AMX = amoxicillin, CON = control and PSP = PolySaccharoPeptide. Thus, the first sample on the far left “16 AMX minus 7” denotes subject 16, amoxicillin treatment group, day minus 7 (screening visit) stool sample. Subjects with more similar microbial populations are closer together. Each sample time point is represented for each of the 9 subjects shown. This figure illustrates that each subject maintains a relatively stable microbial population during the course of the study and that these populations are identifiable to the individual. The heatmap represents the relative percentages of each bacterial genus. The predominant genera are represented along the right Y-axis. The legend for the heatmap is provided in the upper left corner representing the relative percentages of each bacterial genera within each sample. We can see that the genera that are most abundant are Faecalibacterium, Bacteroides, Rosburia, Clostridium, and Ruminococcus.
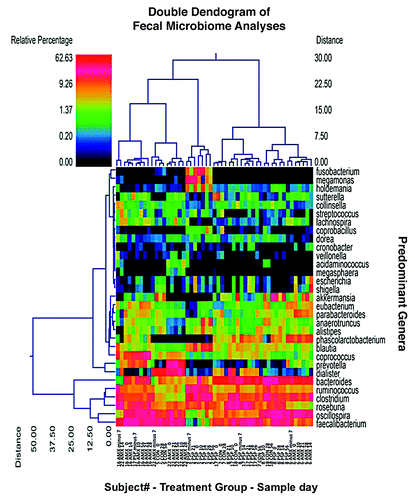
A heatmap is also shown in and represents the relative percentages of each bacterial genus identified. The predominant genera are listed along the right Y-axis of the figure. The genera which are most abundant are Faecalibacterium, Bacteroides, Rosburia, Clostridium, and Ruminococcus.
Prevalence of bacterial genera
To evaluate the baseline microbiome for each of the groups we identified the most prevalent genera within each group using the sampling points prior to study interventions. There were no statistically significant differences in baseline predominant genera between groups. However, there were notable differences in the prevalence of Faecalibacterium between the groups at baseline. This exemplifies the way in which random group assignments can introduce inherent population biases into human microbial ecology studies.
When evaluating the predominant bacterial genera based upon age groups (by rounded to nearest decade) and gender we found that several genera were significantly different after accounting for repeated measurements. In relation to age groups Bacteroides and Sutterella were significantly higher in the age 30 group (with males accounting for this significant difference). On the other hand Coprococcus was higher in the 40 and 50 y age-groups with females in the 50 y age group having significantly higher abundance. Prevotella was also significantly higher in abundance in the 40 and 50 y age-groups.
Differences in genera over time and between study groups
There were no significant differences in the predominant genera over time within the control group. This reflects the relative stability of an individual’s intestinal microbiome.
provides a summary of the top 16 bacterial genera during the study periods and across groups. Several variations in the prevalence of bacterial genera were evident. However, the most substantial shift was that Escherichia/Shigella was significantly higher during antibiotic treatment (as determined using repeated measures ANOVA).
Table 4. The most prevalent bacterial genera within the study population (the top 16 are listed)
To evaluate the effects of treatment on the microbiome we evaluated the total microbial assemblages within each treatment group. A redundancy plot analysis is illustrated in with each point representing a treatment group at a single time point. The region surrounding the “core microbiomes” (solid circle) includes all pretreatment and control samples (a total of 52 samples from 22 participants). Alterations in the microbiome during or after treatment can be seen as movement away from the core. Once again the diversity of the microbiome between individuals yet its consistency within individuals over time is reflected by the evident clustering of the Control, PSP and amoxicillin groups data points over time.
Figure 3. Biplot of Redundancy Analysis: Each point represents a treatment group at a single time point. The region within the circle encompasses all of the control and pre-treatment samples creating a baseline or “core microbiome.” Effects of treatment can be seen as movement away from or outside the core. Maximal divergence of the antibiotic group from the core is seen at Day 14 (end of therapy, arrow 1.). A return toward the core is evident after antibiotic treatment ended but the group’s microbiome still appears outside the core on Day 56 (42 d after stopping amoxicillin, arrow 2.). Divergence of the PSP group from the core is far less dramatic but the group lies outside the core on Day 56 after treatment has ended (arrow 3.).
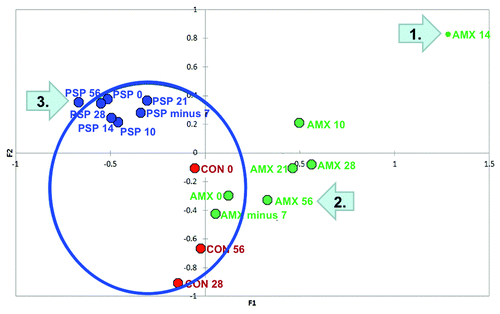
This clustering notwithstanding the antibiotic treatment group, particularly at the end of therapy on day 14, showed a notable population shift away from the core microbiome. After treatment with amoxicillin ended the microbiome of this group began to recover and returned closer to its baseline position within the core. However, even at day 56 (42 d after completing amoxicillin) the antibiotic-treated subjects manifest microbiomes that were still outside the baseline or core parameters.
Divergence of the PSP group’s microbiomes is less dramatic than for the antibiotic treatment group. It is maximal on Day 56, after treatment has ended, when the microbiome parameters lie just outside the core (arrow 3.).
Several predominant bacterial genera were significantly changed during the treatment period when evaluating each treatment group as a whole (). The most marked change was that Escherichia/Shigella was significantly increased in the antibiotic group during treatment returning to elevated, but not significantly so, levels after treatment. Fusobacterium was increased in the PSP group during treatment returning to non-significantly different levels after treatment. These apparent changes in Fusobacterium prevalence arose largely from a single subject (see , subject 2, PSP) with unusually high pretreatment Fusobacterium levels. This subject was a 51 y/o Asian male with mild HTN (controlled by lifestyle modification) and history of gout (inactive at enrollment and not on chronic treatment). It is possible that the higher levels of Fusobacterium in this individual may be due to pre-enrollment consumption of two over the counter health supplements; “Long Jack,” (a proprietary blend of herbs and natural supplements) and “shou wu” (also known as “Polygonum Multiforum” or Chinese knotweed).
To further evaluate which treatments created notable population shifts we evaluated the microbial assemblage using discriminant analysis and biplots. The analysis for the antibiotic group () shows the control group clustered together with the pre-treatment antibiotic group samples (as expected). However, both during and after antibiotic treatment there are marked microbiome shifts. The group’s microbiomes during antibiotic treatment cluster closely together as do the group’s microbiomes after amoxicillin treatment has ended. However, the pre- and post-treatment microbiomes also differ leading to three distinct groupings: baseline, antibiotic treatment, and recovery after antibiotic treatment. It is notable that the post-antibiotic treatment microbiome compositions differ from baseline out to the end of the study (to study day 56, which is 42 d after antibiotic therapy ended).
Figure 4. (A) Discriminant analysis biplots: Biplot A shows data for the Control and Amoxicillin groups. Three distinct groupings are evident: (1) Baseline (bottom left), controls cluster with the antibiotic group prior to therapy to form one grouping; (2) During antibiotic treatment (top left); and (3) Incomplete recovery after antibiotic treatment (bottom right). (B) Discriminant analysis biplots: Biplot B shows data for the Control and PSP groups (before, during and after treatment). Three distinct groupings are evident: (1) The PSP group (bottom left) clusters together both pre- and post-treatment but is separated clearly from (2) the Control group (top left), with no overlap, and (3) the PSP group during treatment (bottom right).
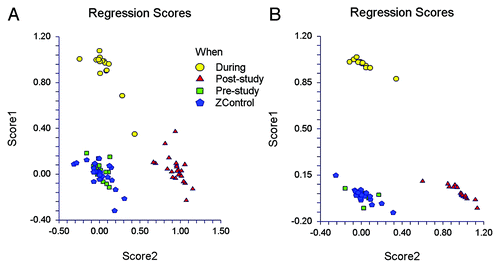
The PSP group () clusters together both pre- and post-study, but is separated clearly from the Control group, with no overlap. This again reflects the consistency of an individual’s microbiome and also a, presumably random, separation of the PSP group from the control and antibiotic treatment groups at baseline. The PSP group’s microbiomes clearly shifted during treatment to form a separate but closely related cluster. This cluster does not overlap with the pre- or post-treatment microbiomes. This demonstrates that PSP ingestion has the ability to elicit distinctive changes in the human microbiome consistent with its activity as a prebiotic.
Discussion
In this study we analyzed the fecal microbiome of healthy volunteers over a two month time period and examined the nature and the duration of effects associated with antibiotic (Amoxicillin) use and ingestion of polysaccharopeptide from the mushroom T. versicolor. Our findings demonstrate the diversity of the human microbiome, the relative stability of an individual’s microbiome over time and during the study interventions, the clear and persisting effects of antibiotic treatment as well as effects of PSP consistent with its role as a prebiotic agent.
Our sequencing studies revealed almost 2 million sequences within the 121 study samples averaging 15,056 sequences per sample, which after quality curation yielded 9238 sequences per sample for our final analyses. The genera, which are most abundant in our subjects, are Faecalibacterium, Bacteroides, Rosburia, Clostridium, and Ruminococcus. Although the breadth of microbiome diversity was similar across study subjects microbiome composition varied substantially between individuals with each subject showing a distinctive and relatively stable pattern. These individual patterns clustered to varying extents leading to microbiome groupings as recently described by other investigators.Citation20 Whether, and under which circumstances, microbiome constituents can influence host health or disease outcomes remains to be determined and is a burgeoning field of current research.
We also identified several age and gender associated patterns in microbiome composition. Men aged 25 to 34 y had significantly higher counts of certain genera (Bacteroides and Sutterella). Women aged 45 to 54 had significantly higher counts of Coprococcus while Prevotella was significantly more abundant in those aged 35 to 54 from both genders. These findings illustrate the complex interactions between individual physiology (or pathophysiology) and the make-up of the gut microbiome. It is possible that these patterns may contribute to age or gender associated disease susceptibility or resistance attributes.
Not surprisingly perhaps, the largest shifts in microbiome composition were seen in study subjects who received the antibiotic amoxicillin. The microbiome population shift was most notable at the end of antibiotic administration (day 14). Thereafter, recovery toward baseline was clearly evident. However, significant alterations in the microbiome were still evident 42 d after the final antibiotic dose. This finding is consistent with the clinical finding of susceptibility to Clostridium difficile infection that is at its greatest during and shortly after antibiotic therapy but can persist for as long as 2 to 3 mo after completing antibiotic treatment. Our study examined healthy subjects; recovery of the colonic microbiome after antibiotic therapy may well be delayed further in elderly, frail patients, on multiple medications and often unable to tolerate their normal diets.
In this human study, amoxicillin therapy was associated with changes in the prevalence of several bacterial genera the most notable being an increase in Escherichia/Shigella. This is in keeping with previous reports of increases in E. coli following antibiotic therapy in pigs.Citation21,Citation22 Although the Escherichia/Shigella group is considered commensal, notable increases in its populations are not typically considered healthy. Increases in E. coli population have been reported during periods of non-specific stresses, such as weaning in livestock and are associated with diarrheal disease-related deaths in weaned pigs.Citation23,Citation24 Moreover, it has also been shown that the severity and duration of disease can be mitigated by decreasing the E. coli population through dietary interventions.Citation25 Increases in the relative abundance of intestinal Enterobacteriaceae, particularly Escherichia coli have also been reported in human IBD and in murine colitis caused by chemicals, pathogens or immune alterations.Citation26-Citation33 Hence the increased prevalence of Escherichia/Shigella, as seen in this study of human antibiotic recipients, may be associated with an increased predisposition to intestinal dysfunction or disease. On the other hand, certain host factors such as obesity may be associated with microbiomes that are inherently resistant to perturbation secondary to administration of antibiotics such as amoxicillin.Citation34
Analysis of microbial population shifts by discriminant analysis and biplots confirmed marked microbiome changes during antibiotic treatment that persisted after the end of the antibiotic exposure as discussed above. Using similar analyses we found that the PSP group differed from the Control and Antibiotic groups at baseline, presumably by random chance. These differences notwithstanding the microbiome composition of the PSP group showed a clear and consistent change during treatment consistent with a prebiotic effect. However, the divergence from the baseline parameters of the PSP group is quite small. Hence, we cannot be certain that similar observations would be made if the baseline microbiome of this group was clustered with the control and amoxicillin groups.
Many carbohydrates act as prebiotics and extensive studies have shown that carbohydrates, primary oligosaccharides and polysaccharides from plants, bacteria or milk, could stimulate the growth of certain probiotic populations and suppress pathogenic species.Citation35-Citation39 The polysaccharides derived from higher fungi are reported to elicit diverse beneficial effects in numerous studies and while more recent research suggests that the beneficial health effects of fungal polysaccharides may result from their role as prebiotics.Citation40-Citation42
The carbohydrate moiety of PSP is a heteropolysaccharide with a primary structure of β-1,3-glucan with β-1,6 branches. Its polysaccharide also contains other 6 monosaccharides and has α-1,4 glucosidic bonds.Citation8,Citation9,Citation12 These molecular characteristics render PSP resistant to human digestive enzymes. To the present, very little is known about whether PSP can alter the human intestinal microbiome and thus render a prebiotic effect. Yu et al. reported an in vitro study finding that PSP modified human fecal microbiome composition by increasing populations of Bifidobacterium spp and Lactobacillus spp while reducing Clostridium spp population, and decreasing culture pH.Citation12 To our knowledge, our study is the first in vivo study to demonstrate a regulatory or prebiotic activity of PSP on the human intestinal microbiome. Conversely given it’s reported efficacy as an immunomodulator, it is possible that PSP shaped microbiome composition not by acting directly on the microbes, but instead by eliciting host responses that, in turn, reshaped the microbiome.
Our study findings suggest several opportunities for additional study. One avenue is to expand fecal analyses to incorporate an examination of metabolic profiles and pathways (metabolome) as well as study of other microorganisms (virome, fungome). Regarding the observed antibiotic effects on the microbiome it is of interest to determine whether these vary, either in extent or in duration of recovery, in the young and in the elderly. It is also of interest to examine the effects of feeding (or inability to take a normal diet) both on the baseline microbiome and on antibiotic effects and recovery therefrom. It also appears likely that medications other than antimicrobial agents, such as immunomodulators or chemotherapeutic agents, may exert substantial influences on the intestinal microbiome. Similarly, the effects of PSP in these subject populations is of interest to determine whether it’s prebiotic effects can modulate microbial composition in the elderly, during and after antibiotic therapy, in immunosuppression or during periods of impaired oral nutrition.
Materials and Methods
Clinical study design
This is a single-center, open-label, randomized controlled trial in healthy volunteers. All Study visits were held at the Harvard Catalyst, Clinical and Translational Science Center. The study protocols and procedures were approved by the BIDMC ethics committee (Committee on Clinical Investigation).
Interested subjects attended a screening visit (day -28 to -7) when the study protocol including risks and benefits was explained and written informed consent was obtained. Study inclusion and exclusion criteria are described in and included age 18 to 65 y (male or female), good general health and absence of any known immunocompromise or allergy to mushrooms or penicillin. Subjects were enrolled in this study between 08/29/2011 and 12/16/2011. Consented subjects were randomized, in blocks of four, using a computer generated randomization sequence to receive either Amoxicillin 250 mg 3 times daily at least 1 h before meals during days 8 to 14 (Amox Group), PSP from T. versicolor (“I'm- Yunity ®” supplied by Integrated Chinese Medicine Holdings Ltd.) 1200 mg, 3 times daily on an empty stomach during days 1 to 14 (PSP Group), Saccharomyces boulardii 250 mg 3 times during days 1 to 14 or no intervention (Control group). Studies on S. boulardii were subsequently expanded to an additional 12 subjects and their results will be reported separately when the data from this larger group become available. PSP is an approved category II drug in china and the dosage used in this study was based on standard medication instructions (360 mg capsules, 3 capsules orally, three times daily).
Table 1. Inclusion and exclusion criteria
Participants were provided with a list of foods to avoid strictly between screening and the end of the study (day 56) consisting of foods that are believed to alter the gut microflora such as yogurt with active cultures, aged cheeses, or naturally fermented vegetables (). Participants were instructed to avoid making any other changes to their usual diet. Compliance with taking the study agent and with dietary restrictions were assessed and adverse events recorded at each study visit (days 0, 14, and 56). provides a summary of the study design, visits and sample collections.
Table 2. Foods to Avoid
Figure 1. Summary illustration of the study design and stool sample collection timelines. PSP, PolySaccharoPeptide; AMX, Amoxicillin.
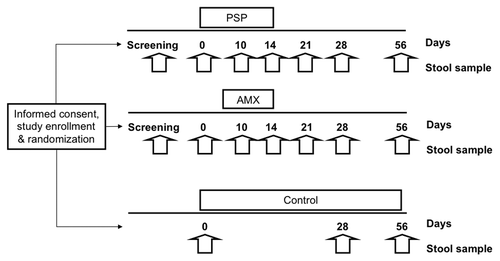
Participants randomized to the control group provided three stool samples (days 0, 28, and 56). Participants receiving PSP or amoxicillin provided seven stool samples prior to (screening and day 0), during (days 10 and 14) and after treatment (days 21, 28, and 56). Stool was frozen immediately (at -20 °C if passed away from BIDMC) and transferred to -80 °C as soon as feasible but always within 48 h.
Microbiome analyses
DNA was extracted from swabs as described previously.Citation43-Citation47 Amplicon pyrosequencing (bTEFAP®) was originally described by Dowd et al. and has been utilized in describing a wide range of environmental and health related microbiomes including the intestinal populations of a variety of sample types including human, cattle, mouse and rat, as well a wide array of environmental samples.Citation43-Citation47 The 16S universal Eubacterial primers (27Fmod 5′ AGRGTTTGAT CMTGGCTCAG and 519Rmodbio 5′GTNTTACNGC GGCKGCTG) were utilized to evaluate the microbial ecology of fecal samples based upon the bTEFAP® process. A single-step 30 cycle PCR using HotStarTaq Plus Master Mix Kit (Qiagen) was used under the following conditions: 94 °C for 3 min, followed by 28 cycles of 94 °C for 30 s; 53 °C for 40 s, and 72 °C for 1 min; after which a final elongation step at 72 °C for 5 min was performed. Following PCR, all amplicon products from different samples were mixed in equal concentrations and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation). Samples were sequenced utilizing Roche 454 FLX titanium instruments and reagents following manufacturer’s guidelines.
The Q25 sequence data was processed using a proprietary analysis pipeline (www.mrdnalab.com, MR DNA). Sequences are depleted of barcodes and primers and then short sequences <200 bp are removed, sequences with ambiguous base calls removed, and sequences with homopolymer runs exceeding 6bp removed. Sequences are then de-noised and chimeras removed. Operational taxonomic units (OTUs) were defined after removal of singleton sequences, clustering at 3% divergence (97% similarity).Citation43-Citation47 OTUs were then taxonomically classified using BLASTn against a curated GreenGenes/RDP/NCBI derived database and compiled into each taxonomic level into both “counts” and “percentage” files.Citation48 Counts files contain the actual number of sequences while the percent files contain the relative (proportion) percentage of sequences within each sample that map to the designated taxonomic classification.
Statistical analysis was performed using a variety of computer packages including XLstat, NCSS 2007, “R” and NCSS 2010. Alpha and β diversity analysis was conducted as described previously.Citation43-Citation47 Alpha diversity (number of different bacterial species) was determined by the number of operational taxonomic units (OTU) identified in a sample. Beta diversity (an analysis of bacterial community structure) was evaluated using phylogenetic trees, without regard for taxonomy. “Core microbiome” is defined as the grouping of microbial community structures that encompasses the control subjects as well as the treated subjects prior to any treatment and/or intervention. A principal coordinate analysis was then performed to allow for visualization of 10 separate jackknife iterative comparisons. The multidimensional space was then described within the 3 primary vectors. Data were evaluated in a multivariate manner to determine the effects of individuals, age, and gender. Significance reported for any analysis is defined as P < 0.05.
| Abbreviations: | ||
| PSP | = | Polysaccharopeptide |
| CDI | = | Clostridium difficile infection |
| IBD | = | inflammatory bowel disease |
| AAD | = | antibiotic associated diarrhea |
| IBS | = | irritable bowel syndrome |
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
This work was conducted with support from: Harvard Catalyst, The Harvard Clinical and Translational Science Center, NIH Award 8UL1TR000170-05, Integrated Chinese Medicine Holdings Ltd, and Hong Kong Association for Health Care Ltd.
References
- Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology 2009; 136:65 - 80; http://dx.doi.org/10.1053/j.gastro.2008.10.080; PMID: 19026645
- Guarner F, Malagelada JR. Gut flora in health and disease. Lancet 2003; 361:512 - 9; http://dx.doi.org/10.1016/S0140-6736(03)12489-0; PMID: 12583961
- Stephen AM, Cummings JH. The microbial contribution to human faecal mass. J Med Microbiol 1980; 13:45 - 56; http://dx.doi.org/10.1099/00222615-13-1-45; PMID: 7359576
- Jirillo E, Jirillo F, Magrone T. Healthy effects exerted by prebiotics, probiotics, and symbiotics with special reference to their impact on the immune system. Int J Vitam Nutr Res 2012; 82:200 - 8; http://dx.doi.org/10.1024/0300-9831/a000112; PMID: 23258401
- Silk DB, Davis A, Vulevic J, Tzortzis G, Gibson GR. Clinical trial: the effects of a trans-galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment Pharmacol Ther 2009; 29:508 - 18; http://dx.doi.org/10.1111/j.1365-2036.2008.03911.x; PMID: 19053980
- Bodera P. Influence of prebiotics on the human immune system (GALT). Recent Pat Inflamm Allergy Drug Discov 2008; 2:149 - 53; http://dx.doi.org/10.2174/187221308784543656; PMID: 19076004
- Lenoir-Wijnkoop I, Sanders ME, Cabana MD, Caglar E, Corthier G, Rayes N, Sherman PM, Timmerman HM, Vaneechoutte M, Van Loo J, et al. Probiotic and prebiotic influence beyond the intestinal tract. Nutr Rev 2007; 65:469 - 89; http://dx.doi.org/10.1111/j.1753-4887.2007.tb00272.x; PMID: 18038940
- Ng TB. A review of research on the protein-bound polysaccharide (polysaccharopeptide, PSP) from the mushroom Coriolus versicolor (Basidiomycetes: Polyporaceae). Gen Pharmacol 1998; 30:1 - 4; http://dx.doi.org/10.1016/S0306-3623(97)00076-1; PMID: 9457474
- Wasser SP, Weis AL. Therapeutic effects of substances occurring in higher Basidiomycetes mushrooms: a modern perspective. Crit Rev Immunol 1999; 19:65 - 96; PMID: 9987601
- Kidd PM. The use of mushroom glucans and proteoglycans in cancer treatment. Altern Med Rev 2000; 5:4 - 27; PMID: 10696116
- Fisher M, Yang LX. Anticancer effects and mechanisms of polysaccharide-K (PSK): implications of cancer immunotherapy. Anticancer Res 2002; 22:1737 - 54; PMID: 12168863
- Yu ZT, Liu B, Mukherjee P, Newburg DS. Trametes versicolor extract modifies human fecal microbiota composition in vitro. Plant Foods Hum Nutr 2013; 68:107 - 12; http://dx.doi.org/10.1007/s11130-013-0342-4; PMID: 23435630
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 2008; 6:e280; http://dx.doi.org/10.1371/journal.pbio.0060280; PMID: 19018661
- Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology 2010; 156:3216 - 23; http://dx.doi.org/10.1099/mic.0.040618-0; PMID: 20705661
- De La Cochetière MF, Durand T, Lalande V, Petit JC, Potel G, Beaugerie L. Effect of antibiotic therapy on human fecal microbiota and the relation to the development of Clostridium difficile. Microb Ecol 2008; 56:395 - 402; http://dx.doi.org/10.1007/s00248-007-9356-5; PMID: 18209965
- Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol 2004; 42:1203 - 6; http://dx.doi.org/10.1128/JCM.42.3.1203-1206.2004; PMID: 15004076
- Sullivan A, Edlund C, Nord CE. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect Dis 2001; 1:101 - 14; http://dx.doi.org/10.1016/S1473-3099(01)00066-4; PMID: 11871461
- Swidsinski A, Loening-Baucke V, Kirsch S, Doerffel Y. [Functional biostructure of colonic microbiota (central fermenting area, germinal stock area and separating mucus layer) in healthy subjects and patients with diarrhea treated with Saccharomyces boulardii]. Gastroenterol Clin Biol 2010; 34:Suppl 1 S79 - 92; http://dx.doi.org/10.1016/S0399-8320(10)70025-7; PMID: 20889010
- Marteau P, Lepage P, Mangin I, Suau A, Doré J, Pochart P, Seksik P. Review article: gut flora and inflammatory bowel disease. Aliment Pharmacol Ther 2004; 20:Suppl 4 18 - 23; http://dx.doi.org/10.1111/j.1365-2036.2004.02062.x; PMID: 15352889
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al, MetaHIT Consortium. Enterotypes of the human gut microbiome. Nature 2011; 473:174 - 80; http://dx.doi.org/10.1038/nature09944; PMID: 21508958
- Allen HK, Looft T, Bayles DO, Humphrey S, Levine UY, Alt D, Stanton TB. Antibiotics in feed induce prophages in swine fecal microbiomes. MBio 2011; 2; http://dx.doi.org/10.1128/mBio.00260-11; PMID: 22128350
- Janczyk P, Pieper R, Souffrant WB, Bimczok D, Rothkötter HJ, Smidt H. Parenteral long-acting amoxicillin reduces intestinal bacterial community diversity in piglets even 5 weeks after the administration. ISME J 2007; 1:180 - 3; http://dx.doi.org/10.1038/ismej.2007.29; PMID: 18043627
- Looft T, Allen HK. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 2012; 3:463 - 7; http://dx.doi.org/10.4161/gmic.21288; PMID: 22825498
- Wu XY, Chapman T, Trott DJ, Bettelheim K, Do TN, Driesen S, Walker MJ, Chin J. Comparative analysis of virulence genes, genetic diversity, and phylogeny of commensal and enterotoxigenic Escherichia coli isolates from weaned pigs. Appl Environ Microbiol 2007; 73:83 - 91; http://dx.doi.org/10.1128/AEM.00990-06; PMID: 17056683
- Fairbrother JM, Nadeau E, Gyles CL. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim Health Res Rev 2005; 6:17 - 39; http://dx.doi.org/10.1079/AHR2005105; PMID: 16164007
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007; 104:13780 - 5; http://dx.doi.org/10.1073/pnas.0706625104; PMID: 17699621
- Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, Jian R, Doré J. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut 2003; 52:237 - 42; http://dx.doi.org/10.1136/gut.52.2.237; PMID: 12524406
- Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 2004; 127:80 - 93; http://dx.doi.org/10.1053/j.gastro.2004.03.054; PMID: 15236175
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007; 2:204; http://dx.doi.org/10.1016/j.chom.2007.08.002; PMID: 18030708
- Heimesaat MM, Fischer A, Siegmund B, Kupz A, Niebergall J, Fuchs D, Jahn HK, Freudenberg M, Loddenkemper C, Batra A, et al. Shift towards pro-inflammatory intestinal bacteria aggravates acute murine colitis via Toll-like receptors 2 and 4. PLoS One 2007; 2:e662; http://dx.doi.org/10.1371/journal.pone.0000662; PMID: 17653282
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun 2008; 76:907 - 15; http://dx.doi.org/10.1128/IAI.01432-07; PMID: 18160481
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 2010; 8:292 - 300; http://dx.doi.org/10.1016/j.chom.2010.08.004; PMID: 20833380
- Wohlgemuth S, Haller D, Blaut M, Loh G. Reduced microbial diversity and high numbers of one single Escherichia coli strain in the intestine of colitic mice. Environ Microbiol 2009; 11:1562 - 71; http://dx.doi.org/10.1111/j.1462-2920.2009.01883.x; PMID: 19245530
- Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, van Nood E, Holleman F, Knaapen M, Romijn JA, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol 2014; 60:824 - 31; http://dx.doi.org/10.1016/j.jhep.2013.11.034; PMID: 24316517
- Badia R, Lizardo R, Martinez P, Brufau J. Oligosaccharide structure determines prebiotic role of β-galactomannan against Salmonella enterica ser. Typhimurium in vitro. Gut Microbes 2013; 4:72 - 5; http://dx.doi.org/10.4161/gmic.22728; PMID: 23137964
- Van den Abbeele P, Verstraete W, El Aidy S, Geirnaert A, Van de Wiele T. Prebiotics, faecal transplants and microbial network units to stimulate biodiversity of the human gut microbiome. Microb Biotechnol 2013; 6:335 - 40; http://dx.doi.org/10.1111/1751-7915.12049; PMID: 23594389
- Kondepudi KK, Ambalam P, Nilsson I, Wadström T, Ljungh A. Prebiotic-non-digestible oligosaccharides preference of probiotic bifidobacteria and antimicrobial activity against Clostridium difficile. Anaerobe 2012; 18:489 - 97; http://dx.doi.org/10.1016/j.anaerobe.2012.08.005; PMID: 22940065
- Polari L, Ojansivu P, Mäkelä S, Eckerman C, Holmbom B, Salminen S. Galactoglucomannan extracted from spruce (Picea abies) as a carbohydrate source for probiotic bacteria. J Agric Food Chem 2012; 60:11037 - 43; http://dx.doi.org/10.1021/jf303741h; PMID: 23067113
- Oozeer R, van Limpt K, Ludwig T, Ben Amor K, Martin R, Wind RD, Boehm G, Knol J. Intestinal microbiology in early life: specific prebiotics can have similar functionalities as human-milk oligosaccharides. Am J Clin Nutr 2013; 98:561S - 71S; http://dx.doi.org/10.3945/ajcn.112.038893; PMID: 23824728
- Rop O, Mlcek J, Jurikova T. Beta-glucans in higher fungi and their health effects. Nutr Rev 2009; 67:624 - 31; http://dx.doi.org/10.1111/j.1753-4887.2009.00230.x; PMID: 19906249
- Chang ST, Wasser SP. The role of culinary-medicinal mushrooms on human welfare with a pyramid model for human health. International journal of medicinal mushrooms 2012; 14(2): 95-134.
- Chou WT, Sheih IC, Fang TJ. The applications of polysaccharides from various mushroom wastes as prebiotics in different systems. J Food Sci 2013; 78:M1041 - 8; http://dx.doi.org/10.1111/1750-3841.12160; PMID: 23701736
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26:2460 - 1; http://dx.doi.org/10.1093/bioinformatics/btq461; PMID: 20709691
- Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 2008; 8:125; http://dx.doi.org/10.1186/1471-2180-8-125; PMID: 18652685
- Eren AM, Zozaya M, Taylor CM, Dowd SE, Martin DH, Ferris MJ. Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PLoS One 2011; 6:e26732; http://dx.doi.org/10.1371/journal.pone.0026732; PMID: 22046340
- Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog Dis 2008; 5:459 - 72; http://dx.doi.org/10.1089/fpd.2008.0107; PMID: 18713063
- Swanson KS, Dowd SE, Suchodolski JS, Middelbos IS, Vester BM, Barry KA, Nelson KE, Torralba M, Henrissat B, Coutinho PM, et al. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J 2011; 5:639 - 49; http://dx.doi.org/10.1038/ismej.2010.162; PMID: 20962874
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069 - 72; http://dx.doi.org/10.1128/AEM.03006-05; PMID: 16820507
