Abstract
The human gastrointestinal tract hosts a large number of microbial cells which exceed their mammalian counterparts by approximately 3-fold. The genes expressed by these microorganisms constitute the gut microbiome and may participate in diverse functions that are essential to the host, including digestion, regulation of energy metabolism, and modulation of inflammation and immunity. The gut microbiome can be modulated by dietary changes, antibiotic use, or disease. Different ailments have distinct associated microbiomes in which certain species or genes are present in different relative quantities. Thus, identifying specific disease-associated signatures in the microbiome as well as the factors that alter microbial populations and gene expression will lead to the development of new products such as prebiotics, probiotics, antimicrobials, live biotherapeutic products, or more traditional drugs to treat these disorders. Gained knowledge on the microbiome may result in molecular lab tests that may serve as personalized tools to guide the use of the aforementioned products and monitor interventional progress.
The Human Body is a Community of Microbes
Only one of 4 cells of the human body is actually human in origin. The remaining 75% of cells are composed of the microorganisms that colonize the internal and external surfaces of our bodies. Although it is often stated that the number of microbial cells is approximately 10 times the number of human cells, recent studies have estimated we harbor 37 trillion human cellsCitation1as well as at least 100 trillion microbial cells and a quadrillion viruses in and on our bodies.Citation2 Given these calculations, the ratio microbial-to-human cell is around 3-fold. The microbiota is defined as the full complement of microbes, including bacteria, viruses, fungi, and protozoa that reside in and on our bodies.Citation3-Citation5 The microbiome has been defined as “the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space.”Citation6 The two may be distinguished as the microbiome includes the collective genomes.Citation3-Citation5 In 1985 researchers demonstrated that bacteria which can be cultivated in the laboratory represent only a small fraction of the microbes present in the environment, and dubbed this phenomenon the “great plate count anomaly.”Citation7 Recently, culture-independent high-throughput sequencing has greatly expanded our repertoire of known microbes.Citation8 This has allowed for measurement of the structure and dynamics of microbial communities, the relationships between their members, what substances are produced and consumed, the interaction with the host, and differences between healthy hosts and those with disease. It is hypothesized that the composition of the microbiome may influence the host’s health by contributing to its metabolic and immune functions. In this review, we summarize the emerging work on the relationship of the gut microbiome with human health, its development and establishment, and the factors including diet, antibiotic use, and disease, that shape it.
The Microbiome and Human Health
An increasing level of evidence reveals that the human microbiome plays a major role in health. For this reason it is often referred to as the “forgotten organ.”Citation9 All surfaces of the human body that are exposed to the environment including skin, respiratory system, urogenital tract and gastrointestinal (GI) tract, are colonized with microorganisms. The majority of microbes composing the human microbiome are in the GI tractCitation3 and thus this review will focus on the human gut microbiome. The gut microbiome provides metabolic functions such as energy harvest and storage, fermentation and absorption of indigestible carbohydrates,Citation10,Citation11 synthetic functions such as the production of vitamins KCitation12,Citation13 and B,Citation14 and immune roles such as the modulation of innate immunity, the promotion, maturation and development of cell-mediated immunity, and the maintenance of appropriate immune response to pathogens. In addition, the microbiome plays an important part in the preservation of the intestinal barrier function.Citation15-Citation17
The gut microbiome expresses the enzymatic machinery to process otherwise non-digestible carbohydrates such as fructo-oligosaccharides (FOS), galacto-oligosaccharides (GOS), and inulin, and thus release monosaccharides that could be utilized by the host for metabolic purposes.Citation18-Citation20 Some bacteria express colonization factors, which consist of polysaccharide utilization loci containing the machinery to degrade mucosal polysaccharides, and survive in epithelial crypts.Citation21 The breakdown of mucosal polysaccharides results in the release of smaller sugars that can be consumed by other GI microorganisms. Additionally, fermentation of FOS and other dietary fibers by some of the bacterial symbionts results in the production of short chain fatty acids (SCFAs), specifically in the colon, which promote functions that are beneficial to the human body and provide an energy source for colonocytes. Microorganisms with such capabilities include bacteria from the Bacteroides and Bifidobacterium genera. For example, increases in SCFAs and glucagon-like peptide-1 (GLP-1) levels, were seen in humans after fiber intake.Citation22 Conversely, GLP-1 levels inversely correlated with SCFA levels and the presence of a microbiome in a mouse study. This result was attributed to a GLP-1-dependent delay of gastric transit to improve energy harvest in germ free mice.Citation23 SCFA signaling has also been implicated in a number of immune and anti-inflammatory functions such as activation of regulatory T cells, downregulation of proinflammatory cytokines by peripheral blood mononuclear cells, promotion of chemotaxis and phagocytosis by neutrophils, and the maintenance of intestinal epithelial barrier integrity.Citation24-Citation28
The gut microbiome also influences the host health status through enzymatic transformation of bile acids (BAs), natural detergents with novel signaling functions including regulation of cholesterol synthesis and absorption, modulation of inflammatory responses, and energy homeostasis.Citation29 Some microorganisms in the gut express enzymes that hydrolyse taurine- or glycine- conjugated BAs to change their physical and chemical properties. Other bacterial enzymes dehydroxylate or sulfate the resulting deconjugated primary BAs.Citation30 The composition of the gut microbiome determines the diversity and size of the BA pool in different host compartments.Citation31,Citation32 As a result, transformed BAs can be re-absorbed into mesenteric circulation and exert signaling functions in different tissues through nuclear farnesoid X receptor (FXR) or G-protein coupled receptor (TGR-5) among others.Citation31 It has been shown that BA signaling through FXR results in the reduction of triglycerides, cholesterol, and fatty acids,Citation33,Citation34 improvement of glucose metabolism,Citation35 and hepatoprotection.Citation36 In contrast, BA signaling through TGR-5 plays a role in immune system modulation,Citation37 energy metabolism,Citation38,Citation39 glucose metabolism,Citation39-Citation41 intestinal motility, and gall bladder filling and fluid secretion.Citation42-Citation44 These examples illustrate how the gut microbiome can trigger diverse mechanisms to maintain homeostasis, either by fermentation of food that is otherwise non-digestible by the host, or by transformation of metabolites in the gut.
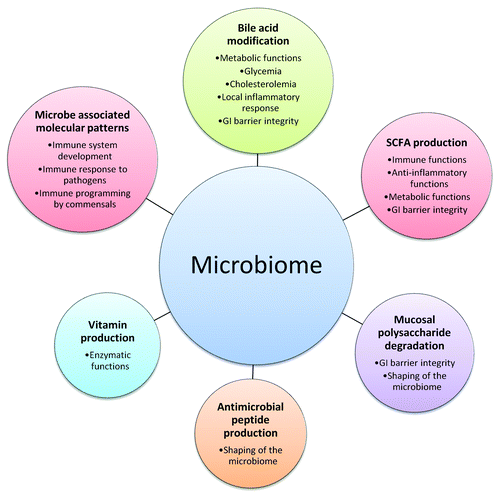
Further, the effects of the microbiome in human health have been attributed to numerous mechanistic pathways. Specific functions, as outlined in , include (1) degradation of dietary and mucosal polysaccharides with subsequent modulation of the microbial ecology and release of metabolites, and preservation of mucosal barrier integrity (2) regulation of host metabolic, inflammatory and immune responses through the release of SCFAs, (3) regulation of the host metabolism and inflammatory responses through modification of BAs, (4) enhancement of the host enzymatic functions by release of bacteria-derived co-factors such as vitamins, (5) development and modulation of the immune system by expression of extracellular molecular patterns, and (6) release of antimicrobial peptides with further shaping of the microbiome. Although evidence from in vitro studies as well as gene deficient, germ-free or gnotobiotic mouse models sustains these hypotheses, proof in animals with a natural microbiota is more difficult to obtain. Simplified models present an excellent opportunity to explain punctual mechanisms but fail to account for changes in bacterial populations caused by the release of metabolites as observed in wild type animals. Further, these studies are restricted to rodents, which differ from humans in both host genome and microbiome, and involve only a small set of genes within the gut microbiome, leaving most of the metagenome unexplored.
Changes to the Microbiome over Time
Understanding the stability of the microbiota within an individual over time is an important step in facilitating prediction of disease and the development of therapies to correct imbalance. It is known that several factors such as host genetics, diet, environment, antibiotic use, and age influence the development and composition of the human gut microbiome considerably.Citation45-Citation47 While significant interindividual variability exists, the microbiota of most individuals can be categorized into one of three distinct variants of bacterial communities dominated by Bacteroides, Prevotella, or Ruminococcus, regardless of the geographic origin of the sample.Citation48 Interestingly, the abundance of each of these three dominant genera correlate strongly, either positively or negatively, with those of other genera, meaning different microorganisms co-occur or avoid each other. The enterotypes are thus determined by favored community compositions.Citation48 Overall, the microbiome is considered robust at both the phylogenetic and functional level. A 5-year-long study indicated that strains recovered from fecal samples may have been present in the hosts for decades.Citation49 Despite transient environmental perturbations caused by dietary changes, antibiotic use, or disease may result in significant variations in microbial populations, several studies report that the microbiome could be restored and is stable over time.Citation50-Citation52
Development of the microbiome
The human microbiota starts developing shortly after birth, evolves and establishes itself through infancy and adapts to metabolic changes of the body even during old age.Citation45 Traditionally, babies have been considered sterile in utero and thus open to colonization by microbiota at each generation. However, the baby’s first intestinal discharge, the meconium, is not sterile and may contain a low diversity microbiome.Citation53,Citation54 It is hypothesized that mothers may transfer their microorganisms in utero and no global differences were observed when comparing the meconia from babies delivered either via caesarean section or vaginally.Citation55 In contrast, other studies show newborn babies are exposed to microbes from different environments immediately upon birth and are rapidly colonized through vertical acquisition of the microbes depending on delivery mode.Citation56 Infants born vaginally have communities resembling those found in the vaginal microbiota of their mothersCitation56 while infants delivered by caesarean section harbor a microbiota characteristic of skin, which are dominated by Staphylococcus and Propionibacterium.Citation56 Delivery mode has also been hypothesized to influence immunological functions during the first year of life via gut microbiota development, with babies delivered by caesarean section having lower bacterial cell counts in fecal samples and a higher number of antibody secreting cells.Citation57 Diversity of bacteria in the infant gut is initially low with the early colonizers being aero-tolerant as the gut contains oxygen but are replaced by anaerobes typical of the adult gut microbiota.Citation58 Detailed time series show that the phylogenetic diversity increases gradually over time and is punctuated by major shifts in taxonomic compositions of the microbiota associated with events such as introduction of solid foods and antibiotic use.Citation52 Interestingly, metagenomic analysis shows that the infant microbiome is enriched in genes to facilitate lactate utilization in milk and that the functional capacity to utilize plant derived glycans is present before introduction of solid foods.Citation52 Once the microbiota reaches maturity it remains relatively constant until old age.Citation52,Citation59 Finally, the elderly microbiota is generally less diverse and presents increased levels of the Bacteroidetes phylum as well as the Clostridia genus.Citation59
Establishment of the microbiome
The formation of a stable microbiome during the first years of life, followed by maintenance of a mature microbiome in adulthood, raises questions such as how the adult microbiome is established, what factors influence colonization and which organisms are involved. The most numerous organisms of the microbiome are members of the Firmicutes and Bacteriodetes phyla, which are appropriate candidates for studying the factors affecting colonization. Germ-free mice mono-associated with single Bacteroides species have been found resistant to colonization by the same, but not different, species.Citation60 In vivo genetic screening of a B. fragilis strain representative of Bacteroides identified a unique polysaccharide utilization loci, commensal colonization factor (CCF), which when deleted results in colonization deficit and reduced horizontal transmission.Citation60 This strain of B. fragilis penetrates the colonic mucus and resides within crypt channels. The CCF system of genes enables this microorganism to use crypt mucus as a nutrient source and was found to be required for colonization following microbiome disruption by infection or oral antibiotics.Citation60 Specifically, the crypts represent a preferred environmental niche and reservoir for B. fragilis allowing for durable and long-term colonization.Citation60 This is an example of how commensal bacteria have evolved molecular mechanisms to promote their own health in a symbiotic relationship with their human hosts. In agreement with these observations, it was shown that glycosylation patterns in the colonic mucus, determined by host genetics can modulate the composition of the microbiome, favoring species that utilize specific mucus terminal sugars as a source of energy. For example, in the absence of terminal fucose in the mucus as a result of the host’s deficiency in fucosyl transferase enzymatic activity, there is a significant increase in the populations of Bacteroides and a decrease in Clostridiales, as shown in a mouse study.Citation47 Additionally, fucosyl transferase mutations decrease the susceptibility to infections of Campylobacter jejuni, Helicobacter pylori, and Norwalk virus in humans.Citation61-Citation63
Effects of host genetics, gender and environment on the microbiome have been analyzed in mice with different genetic backgrounds that have been housed with mice of the same strain, or co-housed with mice of different strains or gender. The study showed a highly significant contribution of genetic background and co-housing to the microbiome composition. Interestingly, strain genetic distances correlated positively with the microbiome distances.Citation64 The effects of host genetics on the microbiome were also described in co-twins, who share gut metagenomes with a core of redundant functionalities. Nonetheless, these metagenomic studies show monozygotic twins share less than 50% of species-level bacterial taxa, and only 17% of the gene clusters in the microbiome.Citation65 Despite equal genomes, many co-twin pairs have discordant health conditions which are associated with differences in the microbiome. Transplantation of cultured or uncultured microbiota from obese co-twins into germ free mice resulted in greater increases in the recipient’s weight and body fat as compared with the changes seen in mice receiving microbiota components from lean co-twins.Citation66 These series of elegant and well controlled studies show that the establishment of the microbiome is not only influenced by genes of the microorganisms that populate the gut and their mutual interactions, but also by host genetics. In another respect, the use of knockout mice to study the effects of host genetics on the microbiome addresses only the effect of one particular gene at a time in an inbred genetic background. Considering the more diverse genetic background of humans, it is difficult to confirm whether mutations in one gene are causing changes in the human microbiome; however, it does provide evidence of the potential for cross-talk. As mentioned above, emerging studies show other environmental effects, such as diet and antibiotic use, affect the microbiome as well, and are discussed in the following sections.
Diet affects the microbiome
Throughout life, diet plays a significant role in determining the gut microbial community. Dietary patterns associate with predominant variants, including Bacteroides, Prevotella, and Ruminococcus.Citation48 Bacteroides-dominated enterotypes are associated with a diet high in animal fat and derive energy primarily from carbohydrates and proteins through saccharolytic fermentation and proteolysis.Citation48 The Prevotella-dominated enterotype correlates with a diet rich in carbohydratesCitation67 and contains many species capable of desulphating and degrading mucin glycoproteins present in the mucosal layer of the gut.Citation48 The Ruminococcus enterotype is dominated by species which efficiently bind and hydrolyze mucins, as well as uptake the resulting simple sugars.Citation48 While the basis of enterotype clustering is unknown, it has been shown to be independent of nationality, sex, age, or body mass index (BMI). Adults who shifted from a high-fat/low fiber diet, generally a Bacteroides-dominated enterotype, to a low-fat and/or high-fiber diet display notable changes in the gut microbiota within 24 h.Citation67 Research combining cross-sectional and interventional studies show that long-term diet is correlated with enterotype clustering. However, while changes seen during dietary intervention are significant and rapid, the magnitude was shown to be insufficient to switch individuals between enterotypes clusters within 10 d.Citation67 Additionally, this study found that several factors such as BMI, red wine, and aspartame intake were significantly correlated with microbiome composition but not necessarily with enterotype, indicating that enterotype may not be predicative of BMI or dietary intake of individual foods.
The effect of diet on the microbiota composition was studied in gnotobiotic mice with well-defined diets composed of only four ingredients. Dietary perturbations evoked changes in microbial abundances and a statistical model developed from the experimental data could predict up to 60% of the experimental results.Citation68 Diets rich in plant polysaccharides stimulate the growth of bacteria within the Bacteroidetes phylum. The latter have richer genomes including glycan degrading capabilities to release monosaccharides from these carbohydrates. In the presence of polysaccharide-degrading Bacteroidetes, bacteria of the Firmicute phylum upregulate their amino acid and sugar transporter genes to perform glycolysis, and utilize acetate released by the former bacteria for energy harvesting. As a result, Firmicutes release butyrate, which is beneficial for human metabolism.Citation69 Moreover, in the presence of fructan rich diets, B. tetraiotaomicron produces acetate and formate, wherein the latter is consumed by the archaebacteria Methanobrevibacter smithii to release methane.Citation70
Recent studies have shown that diet-induced changes in the microbiome can occur very rapidly, notably after ingestion of an animal-based diet in comparison to a plant-based diet.Citation71 Switching to a diet richer in fat and protein derived from animal products may increase populations of Bilophila, Alistipes, and Bacteroides genera in the gut microbiota. As a consequence, these microorganisms release SCFAs such as isobutyrate, isovalerate and valerate. In contrast, when the diet is changed to plant-based, the gut microbiota presents increases in the Roseburia, Eubacterium, Faecalibacterium, and Bifidobacterium genera, and their derived SCFAs such as acetate, butyrate and propionate.Citation71 Whereas an animal-based diet was shown to trigger microbial expression of genes associated with vitamin synthesis, polycyclic aromatic hydrocarbons, or b-lactamase genes, a plant-based diet induced microbial expression of saccharolytic genes. It is believed that changes in gene expression in the microbiome are the combined result of regulatory and selective pressure.Citation71
Consumption of inulin or FOS was shown to increase populations of Bifidobacteria by approximately 10-fold,Citation72 which warranted studies on the effect of prebiotic intake on the microbiome of healthy human subjects.Citation73-Citation77 However, in individuals suffering from gastrointestinal disorders, evidence of prebiotic intervention affecting the microbiome and/or clinical outcomes is mixed. In such cases, dose and type of prebiotic is critical to the clinical outcome where high doses may result in worsening of symptoms.Citation78 A review of dietary interventions, in the form of prebiotics, and their effect on the microbiome is shown in . Taken together, it is clear that long-term dietary habits and host genetic backgroundCitation47 determine the types of bacterial communities that makeup the gut microbiome and that short-term dietary changes can have a significant and immediate impact, but may not be enough to change the composition and function of the microbiome in the short-term.
Table 1. Prebiotics, their effects on the microbiome and associated observed clinical benefit
Antibiotics affect the microbiome
Observational and interventional studies suggest there is relative stability of the microbiome throughout adult lifeCitation59 and that long-term stability is maintained by geneticCitation47 and environmental forcesCitation79 and governed by the laws of microbial ecology.Citation60 Oral antibiotic use results in immediate and significant alterations in the gut microbiota with the affected taxa varying between individuals.Citation46,Citation80 There is at least partial recovery of taxonomic composition over time; however, some taxa do not recover even months after treatment and generally there is a long-term decrease in bacterial diversity.Citation46 After oral antibiotic use there is an immediate reduction in the stability of the microbial community, a collapse in diversity, oscillatory population dynamics, and colonization by opportunistic organisms followed by repopulation by the commensal flora.Citation46,Citation80 As the microbiota recovers, there is a reduced resistance to colonization allowing foreign microbes to outgrow commensal organisms; however, most patients report normal GI function suggesting functional redundancy of the intestinal mirobiota.Citation46 It has been suggested that repeated use of oral antibiotics increases the reservoir of antibiotic resistant genes and overuse may be associated with an increase in antibiotic-resistant pathogens.Citation81 Certainly, dysbiosis of the microbiome has been reported with both clinical and subclinical antibiotic use and is the cause of alterations in immune and metabolic functions. A healthy and rich microbiome protects the GI tract from bacterial or viral infections and antibiotic use often triggers the emergence of certain pathogens such as Salmonella enterica serovar Typhimurium or Clostridium difficile. These infections are facilitated by the fluctuations in microbial populations that result in transient increases of certain bacteria, such as B. tetaiotaomicron,Citation82 which releases sialic acid from the intestinal mucus via sialidase activity. Free sialic acid is then utilized by the pathogen for propagation. Gnotobiotic mice colonized with a sialidase-deficient B. tetaiotaomicron demonstrated impaired expansion of the pathogen. This effect was reverted by oral administration of free sialic acid.Citation82 Overall, antibiotic use disrupts the integrity and richness of the microbiome, the latter of which is not only beneficial for the host metabolism but also protects it from pathogenic infections.
Microbiome and the Immune System
Humans and their constituent microbiota have coevolved over millions of years. Approximately 70% of the human immune system is found in the GI tract.Citation83 The gut associated lymph tissue (GALT) is comprised of several types of lymphoid tissue that store immune cells, such as T and B lymphocytes, that carry out attacks and defend against pathogens.Citation17 Innate and adaptive immune systems have evolved to require microbial interactions during their development. It has been shown that germ-free mice have reduced gut secretory IgA, defects in gut-associated lymphoid tissue development, smaller Peyer’s patches, and mesenteric lymph nodes.Citation17 As well, it is believed that the progressive loss of microbial diversity, due to reduced transmission, may result in an adaptive immune system which is not adequately educated and in turn an increase in the incidence of autoimmune disease.Citation84 The innate immune system recognizes microbe-associated molecular patterns (MAMPs) that are present across diverse lineages of bacteria such as lipopolysaccharides (LPS), peptidoglycan, and flagellin.Citation15 Toll-like receptors (TLRs) are one of several proteins that recognize MAMP antigens, and when not expressed the gut, immune systems do not form normally.Citation85 TLRs play an important function in host defense by either promoting appropriate inflammatory signals to pathogens via inflammatory cytokines such as interleukin-1β (IL-1β), IL-6, and tumor necrosis factor-α (TNF-α), or promoting immune tolerance through the production of anti-inflammatory cytokines such as IL-10.Citation86 The adaptive immune system is also programmed by the commensal microbiota and microbes have been shown to impact the differentiation of T cell populations.Citation16 Pathogenic bacteria are capable of inducing the proliferation of effector T cells, whereas commensal bacteria induce the proliferation of T regulatory cell population, which can be educated by the commensal microbiota.Citation16 Finally, microbes participate in a symbiotic relationship with their hosts, modulating the host immune system to promote their own fitness.Citation16 Thus, it is clear that a primary function of the gut microbiome is to educate and maintain host immune function at multiple levels including maintenance of anatomical barrier function, development and maintenance of innate immunity as well as to temper the adaptive cell mediated immune response. The roles and relationships of the microbiome within the immune system have been reviewed extensively elsewhere.Citation24,Citation87-Citation90
Microbiome in Health and Disease
In the past, research on microbial interactions and disease has been related to single pathogenic microbes. While microbial communities benefit the host by providing functions such as digestion of nutrients or protection against infection,Citation91 studies on the communities of non-pathogenic microbes were less prevalent as their influence on human health was deemed to pale in comparison with that of pathogenic microbes.Citation91 Recent analysis has revealed that complex phenotypes are associated more appropriately with a dysbiotic gut microflora rather than the presence of a single pathogenic microbe.Citation45 Dysbiosis is a pathological imbalance in a microbial community and is becoming increasingly appreciated as a “central environmental factor” that is affected by host genetics, diet and antibiotic use. Furthermore, dysbiosis likely results from or is a cause of numerous disease states including atopy, autoimmune disease, inflammatory bowel disease (IBD), obesity, diabetes, non-alcoholic fatty liver disease (NAFLD), and hyperlipidemia.Citation92 Emerging clinical studies comparing the gut microbiomes of healthy and disease populations using 16s DNA or metagenomic sequencing analyses have identified associations between certain bacterial families and disease, as presented in . Several studies show Firmicutes from diverse families, namely Clostridiales, Erysipelotrichaceae, Ruminococcaceae, Eubacteriaceae, and Lachnospiraceae have been associated with healthy populations.Citation66,Citation93-Citation96 In particular, bacteria from the Faecalibacterium and Roseburia genera, among the most abundant organisms in the colonic microbiome, are present in significantly lower numbers in diabetic, obese, and atherosclerotic populations. These genera express active bile salt hydrolases with significant activity in the hydrolysis of both glycine and taurine conjugated BAsCitation97 and may exert metabolic benefits to the host by modifying BA pool size and composition. In addition, Roseburia and Faecalibacterium are known SCFA producers, and deficiencies in these genera were observed in patients with ulcerative colitis.Citation98 These studies establish associations between disease and microbiota compositions, with microbial genes identified in some cases. However, there is little evidence of whether the composition of microbial ecology is causative of disease or a result of the host’s metabolism.
Table 2. Association of microbial species or families in health and disease
Researchers believe that reduced microbial diversity, selective disappearance of the microbiota and coincident increases in metabolic and autoimmune disease may be the result of a loss of horizontal and vertically transmitted microorganisms.Citation84 One hypothesis helping to describe the finding of hereditary dysbiosis, is that there is a loss of maternal transmission of microbes to their offspring (i.e., vertical transmission) as well as transmission of microbes through fecally contaminated drinking and bathing water (i.e., horizontal transmission).Citation84 Part of this modern “hygiene hypothesis” is that a lack of early childhood exposure to infectious agents, symbiotic microorganisms, and parasites increases susceptibility to allergic diseases by suppressing natural development of the immune system.Citation84,Citation99 Other recent changes in human ecology include cleaner water, an increase in caesarean sections, increased used of pre-term antibiotics, reduced breastfeeding, smaller family size, and widespread antibiotic use.Citation84 Diminished horizontal transmission resulting from changes in human ecology makes it more difficult to overcome losses in vertical transmission, and this then manifests as a birth cohort phenomenon. This is supported by the finding that over the past 50 years there has been a progressive decline in infectious disease coinciding with increased incidence of immune disorders. Furthermore, over this period there has also been a significant increase in the incidence of metabolic disease due to the widespread adoption of a more sedentary lifestyle and Western diet.Citation84,Citation99 These changes are increasingly thought to be associated with the development of a dysbiotic microflora, and conversely a dysbiotic microflora has been shown to lead to changes in metabolism.Citation48,Citation67,Citation100
Dysbiosis in the form of reduced microbial diversity at the gene and species level does define a subset of individuals who are at increased risk of obesity related metabolic disorders.Citation96 Comparison of gene number across obese and non-obese individuals shows a bimodal distribution of bacterial genes in DanishCitation96 and FrenchCitation100 individuals. It has been suggested that the obesity-associated signal in the human gut microbiome may be much more pronounced than that presently known in the human genome.Citation96 Based on microbiome gene richness there are two groups of individuals, including a group of high gene count (HGC) individuals and a group with 40% fewer genes called low gene count (LGC) individuals.Citation96 While dietary intervention with a high protein energy restricted diet has been shown to correct low gene richness as well as improve metabolic endpoints, it appears less effective in changing markers of inflammation.Citation96 Interestingly, low diversity of gut microbiota has been reported in a wide range of cohorts, including patients with IBD,Citation101-Citation103 elderly patients with inflammation,Citation59 obese individualsCitation96 and individuals after oral antibiotic treatment.Citation104
Individuals with low microbiome gene richness are characterized by more marked adiposity, insulin resistance and dyslipidemia as well as a more pronounced inflammatory phenotype when compared with individuals with high bacterial richness.Citation96 An increase in the Firmicutes phylum and a decrease in Bacteroidetes has been shown to be associated with obese individuals in several studies, with others reporting no association. As well, increased Actinobacteria has been shown in obese individuals. Further, LGC individuals appear to have a phylogenetic shift in favor of Proteobacteria and Bacteroidetes as compared with HGC indivudals, who show increased Verrucomicrobia, Actinobacteria, and Euryarchaeota. Beyond metabolic dysfunctions, low-grade inflammation is associated with a plethora of chronic diseases. Whether a low gut bacterial richness is common in all cases could be revealed by exploring gut microbiota at a deep metagenomic level in a broad variety of these afflictions. Going forward, it may be possible to develop stratified approaches, based on gene richness, for treatment and prevention of chronic disorders.Citation96 In addition, knowledge of the microbiome in determined disease states supports the use of probiotics, as summarized in , either alone or in combination with prebiotics, to potentially affect phenotypes to which the microbiota is linked. The use of probiotics in the management of disease has been reviewed extensively elsewhere.Citation105-Citation107
Table 3. Probiotics, their effects on the microbiome and associated clinical benefit
Conclusions and Future Prospects
Understanding the interactions between host genetics, diet, and the microorganisms that live in the gut provides insight on environmental perturbations and how the microbiome changes over time. The use of advanced mathematical models of ecology and machine learning in combination with metatranscriptomics, metabolomics, and proteomics data will allow scientists to identify defined signatures or patterns associated with disease states. As described, a great body of evidence originates from animal models, sometimes using human fecal transplantation. Although these studies are well controlled, the use of murine models introduces concerns when results are extended to humans due to a number of reasons. For example, rats do not have gall bladders. Also, the BA composition of mice and rats differ from humans which is problematic when many of the beneficial mechanisms of the microbiome in health rely on BA metabolism and signaling through its receptors. Further, animal diets and metabolisms differ from those of the human, which may account for differences in microbial ecologies and in turn make results difficult to interpret. Nevertheless, these investigations provide compelling evidence that may warrant further human studies. It is also plausible that given the availability of knockout mice encompassing families of genes involved in metabolic, inflammatory, immune, structural, and developmental activities, future studies will include these resources to study the microbiome.
Another research question that should be better addressed is whether the dysbiotic microbiome is causative or a consequence of disease. The available proof of causation is based on a few animal studies where germ-free mice were transplanted with microbiomes from lean or obese individuals, as described above.Citation66,Citation108 In order to answer this question, there is a need for more transplantation studies in animals as well as longitudinal studies in humans where the microbiome and host health are followed from the early onset of disease.
A better understanding of the microbiome should also result in the discovery of tools to modulate the microbiome composition toward a “healthy state” for the treatment of a determined set of metabolic or inflammatory ailments. In particular, knowledge on the effect of prebiotics on the gut microbiome can lead to the development of therapeutics (e.g., non-digestible fibers) that will favor the increase of certain commensal species with beneficial effects on the host that may be otherwise depleted in a disease state. In the same line of thought, specific microorganisms or a cocktail of select microorganisms including bacteria and yeast could be delivered to either enrich a dysbiotic microbiota, or supply metabolites or enzymatic activities that may be present at low levels in disease states. On the other hand, compositions of bacterial populations are regulated by antimicrobial agents as well as quorum sensing molecules released by the same native microorganisms. Thus, identifying genes that participate in the release of these molecules in healthy and disease populations may serve to introduce new therapies to control the growth of pathogenic species while favoring beneficial ones. This approach could, in certain cases, act as a useful alternative to broad range antibiotics that have detrimental effects on both microbiome and host health, as described.
In addition, identification of genetic markers within the microbiome that are associated with disease will lead to the development of cost-effective lab tests to evaluate the suitability of therapeutics in a personalized manner. Currently, evaluating the microbiome is performed through metagenomic sequencing, which is time-consuming and expensive, despite a significant cost reduction in recent years. For instance, the development of digital PCR-based assays for the quantification of specific microbial gene copy number or gene expression will reduce test times and make the analysis more accessible for patients and health care practitioners.
As an example, dysbiosis of the gut microbiome and impaired microbiota enzymatic activity has been observed in patients with IBDCitation109 and IBS.Citation110 BA dysmetabolism, often observed in patients with inflammatory disorders, is commonly associated with a deficiency in bile salt hydrolase (BSH) gene derived from the Firmicute phylum,Citation111 Therefore, supplementing diet with a probiotic delivering high BSH activity may be a suitable solution for patients suffering from these maladies. As well, lab tests that are able to accurately detect GI tract BSH activity, gene expression or gene copy number may prove useful to determine who may benefit most from such a treatment. Further research in the field is well supported based on the available evidence.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, et al. An estimation of the number of cells in the human body. Ann Hum Biol 2013; 40:463 - 71; http://dx.doi.org/10.3109/03014460.2013.807878; PMID: 23829164
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 1998; 95:6578 - 83; http://dx.doi.org/10.1073/pnas.95.12.6578; PMID: 9618454
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science 2005; 307:1915 - 20; http://dx.doi.org/10.1126/science.1104816; PMID: 15790844
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006; 124:837 - 48; http://dx.doi.org/10.1016/j.cell.2006.02.017; PMID: 16497592
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature 2007; 449:804 - 10; http://dx.doi.org/10.1038/nature06244; PMID: 17943116
- Lederberg J. 'Ome Sweet 'Omics–A Genealogical Treasury of Words. The Scientist 2001; 15
- Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol 1985; 39:321 - 46; http://dx.doi.org/10.1146/annurev.mi.39.100185.001541; PMID: 3904603
- Tringe SG, Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol 2008; 11:442 - 6; http://dx.doi.org/10.1016/j.mib.2008.09.011; PMID: 18817891
- O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep 2006; 7:688 - 93; http://dx.doi.org/10.1038/sj.embor.7400731; PMID: 16819463
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 2008; 6:121 - 31; http://dx.doi.org/10.1038/nrmicro1817; PMID: 18180751
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 2005; 307:1955 - 9; http://dx.doi.org/10.1126/science.1109051; PMID: 15790854
- LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol 2013; 24:160 - 8; http://dx.doi.org/10.1016/j.copbio.2012.08.005; PMID: 22940212
- Resta SC. Effects of probiotics and commensals on intestinal epithelial physiology: implications for nutrient handling. J Physiol 2009; 587:4169 - 74; http://dx.doi.org/10.1113/jphysiol.2009.176370; PMID: 19596893
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 2009; 6:279 - 89; http://dx.doi.org/10.1016/j.chom.2009.08.003; PMID: 19748469
- Beg AA. ComPPARtmentalizing NF-kappaB in the gut. Nat Immunol 2004; 5:14 - 6; http://dx.doi.org/10.1038/ni0104-14; PMID: 14699401
- Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 2011; 10:311 - 23; http://dx.doi.org/10.1016/j.chom.2011.10.004; PMID: 22018232
- Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol 2012; 20:467 - 76; http://dx.doi.org/10.1016/j.tim.2012.07.002; PMID: 22902802
- Hardy H, Harris J, Lyon E, Beal J, Foey AD. Probiotics, prebiotics and immunomodulation of gut mucosal defences: homeostasis and immunopathology. Nutrients 2013; 5:1869 - 912; http://dx.doi.org/10.3390/nu5061869; PMID: 23760057
- Mitsuoka T, Hidaka H, Eida T. Effect of fructo-oligosaccharides on intestinal microflora. Nahrung 1987; 31:427 - 36; http://dx.doi.org/10.1002/food.19870310528; PMID: 3657917
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 2003; 299:2074 - 6; http://dx.doi.org/10.1126/science.1080029; PMID: 12663928
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 2013; 501:426 - 9; http://dx.doi.org/10.1038/nature12447; PMID: 23955152
- Freeland KR, Wilson C, Wolever TM. Adaptation of colonic fermentation and glucagon-like peptide-1 secretion with increased wheat fibre intake for 1 year in hyperinsulinaemic human subjects. Br J Nutr 2010; 103:82 - 90; http://dx.doi.org/10.1017/S0007114509991462; PMID: 19664300
- Wichmann A, Allahyar A, Greiner TU, Plovier H, Lundén GO, Larsson T, Drucker DJ, Delzenne NM, Cani PD, Bäckhed F. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe 2013; 14:582 - 90; http://dx.doi.org/10.1016/j.chom.2013.09.012; PMID: 24237703
- Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol 2013; 14:676 - 84; http://dx.doi.org/10.1038/ni.2640; PMID: 23778795
- Cox MA, Jackson J, Stanton M, Rojas-Triana A, Bober L, Laverty M, Yang X, Zhu F, Liu J, Wang S, et al. Short-chain fatty acids act as antiinflammatory mediators by regulating prostaglandin E(2) and cytokines. World J Gastroenterol 2009; 15:5549 - 57; http://dx.doi.org/10.3748/wjg.15.5549; PMID: 19938193
- Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, Brezillon S, Dupriez V, Vassart G, Van Damme J, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem 2003; 278:25481 - 9; http://dx.doi.org/10.1074/jbc.M301403200; PMID: 12711604
- Patel KK, Stappenbeck TS. Autophagy and intestinal homeostasis. Annu Rev Physiol 2013; 75:241 - 62; http://dx.doi.org/10.1146/annurev-physiol-030212-183658; PMID: 23216414
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341:569 - 73; http://dx.doi.org/10.1126/science.1241165; PMID: 23828891
- Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev 2009; 89:147 - 91; http://dx.doi.org/10.1152/physrev.00010.2008; PMID: 19126757
- Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev 2005; 29:625 - 51; http://dx.doi.org/10.1016/j.femsre.2004.09.003; PMID: 16102595
- de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab 2013; 17:657 - 69; http://dx.doi.org/10.1016/j.cmet.2013.03.013; PMID: 23602448
- Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A 2011; 108:Suppl 1 4523 - 30; http://dx.doi.org/10.1073/pnas.1006734107; PMID: 20837534
- Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, Auwerx J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest 2004; 113:1408 - 18; http://dx.doi.org/10.1172/JCI21025; PMID: 15146238
- Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A 2006; 103:1006 - 11; http://dx.doi.org/10.1073/pnas.0506982103; PMID: 16410358
- Schaap FG. Role of fibroblast growth factor 19 in the control of glucose homeostasis. Curr Opin Clin Nutr Metab Care 2012; 15:386 - 91; http://dx.doi.org/10.1097/MCO.0b013e3283547171; PMID: 22617565
- Matsubara T, Li F, Gonzalez FJ. FXR signaling in the enterohepatic system. Mol Cell Endocrinol 2013; 368:17 - 29; http://dx.doi.org/10.1016/j.mce.2012.05.004; PMID: 22609541
- Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor κ light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology 2011; 54:1421 - 32; http://dx.doi.org/10.1002/hep.24525; PMID: 21735468
- Maruyama T, Tanaka K, Suzuki J, Miyoshi H, Harada N, Nakamura T, Miyamoto Y, Kanatani A, Tamai Y. Targeted disruption of G protein-coupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J Endocrinol 2006; 191:197 - 205; http://dx.doi.org/10.1677/joe.1.06546; PMID: 17065403
- Sato H, Genet C, Strehle A, Thomas C, Lobstein A, Wagner A, Mioskowski C, Auwerx J, Saladin R. Anti-hyperglycemic activity of a TGR5 agonist isolated from Olea europaea. Biochem Biophys Res Commun 2007; 362:793 - 8; http://dx.doi.org/10.1016/j.bbrc.2007.06.130; PMID: 17825251
- Katsuma S, Hirasawa A, Tsujimoto G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun 2005; 329:386 - 90; http://dx.doi.org/10.1016/j.bbrc.2005.01.139; PMID: 15721318
- Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab 2009; 10:167 - 77; http://dx.doi.org/10.1016/j.cmet.2009.08.001; PMID: 19723493
- Lavoie B, Balemba OB, Godfrey C, Watson CA, Vassileva G, Corvera CU, Nelson MT, Mawe GM. Hydrophobic bile salts inhibit gallbladder smooth muscle function via stimulation of GPBAR1 receptors and activation of KATP channels. J Physiol 2010; 588:3295 - 305; http://dx.doi.org/10.1113/jphysiol.2010.192146; PMID: 20624794
- Li T, Holmstrom SR, Kir S, Umetani M, Schmidt DR, Kliewer SA, Mangelsdorf DJ. The G protein-coupled bile acid receptor, TGR5, stimulates gallbladder filling. Mol Endocrinol 2011; 25:1066 - 71; http://dx.doi.org/10.1210/me.2010-0460; PMID: 21454404
- Vassileva G, Golovko A, Markowitz L, Abbondanzo SJ, Zeng M, Yang S, Hoos L, Tetzloff G, Levitan D, Murgolo NJ, et al. Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation. Biochem J 2006; 398:423 - 30; http://dx.doi.org/10.1042/BJ20060537; PMID: 16724960
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell 2012; 148:1258 - 70; http://dx.doi.org/10.1016/j.cell.2012.01.035; PMID: 22424233
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 2008; 6:e280; http://dx.doi.org/10.1371/journal.pbio.0060280; PMID: 19018661
- Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A 2013; 110:17059 - 64; http://dx.doi.org/10.1073/pnas.1306070110; PMID: 24062455
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al, MetaHIT Consortium. Enterotypes of the human gut microbiome. Nature 2011; 473:174 - 80; http://dx.doi.org/10.1038/nature09944; PMID: 21508958
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science 2013; 341:1237439; http://dx.doi.org/10.1126/science.1237439; PMID: 23828941
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science 2009; 326:1694 - 7; http://dx.doi.org/10.1126/science.1177486; PMID: 19892944
- Vanhoutte T, Huys G, Brandt E, Swings J. Temporal stability analysis of the microbiota in human feces by denaturing gradient gel electrophoresis using universal and group-specific 16S rRNA gene primers. FEMS Microbiol Ecol 2004; 48:437 - 46; http://dx.doi.org/10.1016/j.femsec.2004.03.001; PMID: 19712312
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 2011; 108:Suppl 1 4578 - 85; http://dx.doi.org/10.1073/pnas.1000081107; PMID: 20668239
- Gosalbes MJ, Llop S, Vallès Y, Moya A, Ballester F, Francino MP. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy 2013; 43:198 - 211; http://dx.doi.org/10.1111/cea.12063; PMID: 23331561
- Moles L, Gómez M, Heilig H, Bustos G, Fuentes S, de Vos W, Fernández L, Rodríguez JM, Jiménez E. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One 2013; 8:e66986; http://dx.doi.org/10.1371/journal.pone.0066986; PMID: 23840569
- Hu J, Nomura Y, Bashir A, Fernandez-Hernandez H, Itzkowitz S, Pei Z, Stone J, Loudon H, Peter I. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS One 2013; 8:e78257; http://dx.doi.org/10.1371/journal.pone.0078257; PMID: 24223144
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 2010; 107:11971 - 5; http://dx.doi.org/10.1073/pnas.1002601107; PMID: 20566857
- Huurre A, Kalliomäki M, Rautava S, Rinne M, Salminen S, Isolauri E. Mode of delivery - effects on gut microbiota and humoral immunity. Neonatology 2008; 93:236 - 40; http://dx.doi.org/10.1159/000111102; PMID: 18025796
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol 2007; 5:e177; http://dx.doi.org/10.1371/journal.pbio.0050177; PMID: 17594176
- Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A 2011; 108:Suppl 1 4586 - 91; http://dx.doi.org/10.1073/pnas.1000097107; PMID: 20571116
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 2013; 501:426 - 9; http://dx.doi.org/10.1038/nature12447; PMID: 23955152
- Ikehara Y, Nishihara S, Yasutomi H, Kitamura T, Matsuo K, Shimizu N, Inada K, Kodera Y, Yamamura Y, Narimatsu H, et al. Polymorphisms of two fucosyltransferase genes (Lewis and Secretor genes) involving type I Lewis antigens are associated with the presence of anti-Helicobacter pylori IgG antibody. Cancer Epidemiol Biomarkers Prev 2001; 10:971 - 7; PMID: 11535550
- Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Lindblad L, Stewart P, LePendu J, Baric R. Human susceptibility and resistance to Norwalk virus infection. Nat Med 2003; 9:548 - 53; http://dx.doi.org/10.1038/nm860; PMID: 12692541
- Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem 2003; 278:14112 - 20; http://dx.doi.org/10.1074/jbc.M207744200; PMID: 12562767
- Hildebrand F, Nguyen TL, Brinkman B, Yunta RG, Cauwe B, Vandenabeele P, Liston A, Raes J. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol 2013; 14:R4; http://dx.doi.org/10.1186/gb-2013-14-1-r4; PMID: 23347395
- Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A 2010; 107:7503 - 8; http://dx.doi.org/10.1073/pnas.1002355107; PMID: 20363958
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013; 341:1241214; http://dx.doi.org/10.1126/science.1241214; PMID: 24009397
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 2006; 444:1022 - 3; http://dx.doi.org/10.1038/4441022a; PMID: 17183309
- Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 2011; 333:101 - 4; http://dx.doi.org/10.1126/science.1206025; PMID: 21596954
- Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A 2009; 106:5859 - 64; http://dx.doi.org/10.1073/pnas.0901529106; PMID: 19321416
- Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A 2006; 103:10011 - 6; http://dx.doi.org/10.1073/pnas.0602187103; PMID: 16782812
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505:559 - 63; http://dx.doi.org/10.1038/nature12820; PMID: 24336217
- Gibson GR, Beatty ER, Wang X, Cummings JH. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 1995; 108:975 - 82; http://dx.doi.org/10.1016/0016-5085(95)90192-2; PMID: 7698613
- Calame W, Weseler AR, Viebke C, Flynn C, Siemensma AD. Gum arabic establishes prebiotic functionality in healthy human volunteers in a dose-dependent manner. Br J Nutr 2008; 100:1269 - 75; http://dx.doi.org/10.1017/S0007114508981447; PMID: 18466655
- Costabile A, Kolida S, Klinder A, Gietl E, Bäuerlein M, Frohberg C, Landschütze V, Gibson GR. A double-blind, placebo-controlled, cross-over study to establish the bifidogenic effect of a very-long-chain inulin extracted from globe artichoke (Cynara scolymus) in healthy human subjects. Br J Nutr 2010; 104:1007 - 17; http://dx.doi.org/10.1017/S0007114510001571; PMID: 20591206
- Elli M, Cattivelli D, Soldi S, Bonatti M, Morelli L. Evaluation of prebiotic potential of refined psyllium (Plantago ovata) fiber in healthy women. J Clin Gastroenterol 2008; 42:Suppl 3 Pt 2 S174 - 6; http://dx.doi.org/10.1097/MCG.0b013e31817f183a; PMID: 18685505
- Lefranc-Millot C, Guérin-Deremaux L, Wils D, Neut C, Miller LE, Saniez-Degrave MH. Impact of a resistant dextrin on intestinal ecology: how altering the digestive ecosystem with NUTRIOSE®, a soluble fibre with prebiotic properties, may be beneficial for health. J Int Med Res 2012; 40:211 - 24; http://dx.doi.org/10.1177/147323001204000122; PMID: 22429361
- Ramnani P, Gaudier E, Bingham M, van Bruggen P, Tuohy KM, Gibson GR. Prebiotic effect of fruit and vegetable shots containing Jerusalem artichoke inulin: a human intervention study. Br J Nutr 2010; 104:233 - 40; http://dx.doi.org/10.1017/S000711451000036X; PMID: 20187995
- Whelan K. Mechanisms and effectiveness of prebiotics in modifying the gastrointestinal microbiota for the management of digestive disorders. Proc Nutr Soc 2013; 72:288 - 98; http://dx.doi.org/10.1017/S0029665113001262; PMID: 23680358
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011; 334:105 - 8; http://dx.doi.org/10.1126/science.1208344; PMID: 21885731
- Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, Eismann K, Otto W, Rojo D, Bargiela R, von Bergen M, et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 2013; 62:1591 - 601; http://dx.doi.org/10.1136/gutjnl-2012-303184; PMID: 23236009
- Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N, Pan Y, Li J, Zhu L, Wang X, et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun 2013; 4:2151; http://dx.doi.org/10.1038/ncomms3151; PMID: 23877117
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013; 502:96 - 9; http://dx.doi.org/10.1038/nature12503; PMID: 23995682
- Weiner HL, da Cunha AP, Quintana F, Wu H. Oral tolerance. Immunol Rev 2011; 241:241 - 59; http://dx.doi.org/10.1111/j.1600-065X.2011.01017.x; PMID: 21488901
- Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota?. Nat Rev Microbiol 2009; 7:887 - 94; http://dx.doi.org/10.1038/nrmicro2245; PMID: 19898491
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010; 328:228 - 31; http://dx.doi.org/10.1126/science.1179721; PMID: 20203013
- Frazão JB, Errante PR, Condino-Neto A. Toll-like receptors’ pathway disturbances are associated with increased susceptibility to infections in humans. Arch Immunol Ther Exp (Warsz) 2013; 61:427 - 43; http://dx.doi.org/10.1007/s00005-013-0243-0; PMID: 24057516
- Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol 2013; 14:646 - 53; http://dx.doi.org/10.1038/ni.2604; PMID: 23778791
- Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol 2013; 14:660 - 7; http://dx.doi.org/10.1038/ni.2611; PMID: 23778793
- Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol 2013; 14:668 - 75; http://dx.doi.org/10.1038/ni.2635; PMID: 23778794
- Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 2013; 14:685 - 90; http://dx.doi.org/10.1038/ni.2608; PMID: 23778796
- Weinstock GM. Genomic approaches to studying the human microbiota. Nature 2012; 489:250 - 6; http://dx.doi.org/10.1038/nature11553; PMID: 22972298
- Brown K, DeCoffe D, Molcan E, Gibson DL. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients 2012; 4:1095 - 119; http://dx.doi.org/10.3390/nu4081095; PMID: 23016134
- Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Bäckhed F, Nielsen J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun 2012; 3:1245; http://dx.doi.org/10.1038/ncomms2266; PMID: 23212374
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012; 490:55 - 60; http://dx.doi.org/10.1038/nature11450; PMID: 23023125
- Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 2010; 5:e9085; http://dx.doi.org/10.1371/journal.pone.0009085; PMID: 20140211
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al, MetaHIT consortium. Richness of human gut microbiome correlates with metabolic markers. Nature 2013; 500:541 - 6; http://dx.doi.org/10.1038/nature12506; PMID: 23985870
- Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A 2008; 105:13580 - 5; http://dx.doi.org/10.1073/pnas.0804437105; PMID: 18757757
- Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014; 63:1275 - 83; http://dx.doi.org/10.1136/gutjnl-2013-304833; PMID: 24021287
- Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med 2002; 347:911 - 20; http://dx.doi.org/10.1056/NEJMra020100; PMID: 12239261
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, et al, ANR MicroObes consortium. Dietary intervention impact on gut microbial gene richness. Nature 2013; 500:585 - 8; http://dx.doi.org/10.1038/nature12480; PMID: 23985875
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al, MetaHIT Consortium. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464:59 - 65; http://dx.doi.org/10.1038/nature08821; PMID: 20203603
- Lepage P, Häsler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A, Ott S, Kupcinskas L, Doré J, Raedler A, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 2011; 141:227 - 36; http://dx.doi.org/10.1053/j.gastro.2011.04.011; PMID: 21621540
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006; 55:205 - 11; http://dx.doi.org/10.1136/gut.2005.073817; PMID: 16188921
- Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 2012; 488:621 - 6; http://dx.doi.org/10.1038/nature11400; PMID: 22914093
- Behnsen J, Deriu E, Sassone-Corsi M, Raffatellu M. Probiotics: properties, examples, and specific applications. Cold Spring Harb Perspect Med 2013; 3:a010074; http://dx.doi.org/10.1101/cshperspect.a010074; PMID: 23457295
- Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB, Sherman PM, Mayer EA. An update on the use and investigation of probiotics in health and disease. Gut 2013; 62:787 - 96; http://dx.doi.org/10.1136/gutjnl-2012-302504; PMID: 23474420
- Whelan K, Quigley EM. Probiotics in the management of irritable bowel syndrome and inflammatory bowel disease. Curr Opin Gastroenterol 2013; 29:184 - 9; http://dx.doi.org/10.1097/MOG.0b013e32835d7bba; PMID: 23286925
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 2004; 101:15718 - 23; http://dx.doi.org/10.1073/pnas.0407076101; PMID: 15505215
- Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, Thomas G, Barbu V, Humbert L, Despras G, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013; 62:531 - 9; http://dx.doi.org/10.1136/gutjnl-2012-302578; PMID: 22993202
- Duboc H, Rainteau D, Rajca S, Humbert L, Farabos D, Maubert M, Grondin V, Jouet P, Bouhassira D, Seksik P, et al. Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil 2012; 24:513 - 20, e246-7; http://dx.doi.org/10.1111/j.1365-2982.2012.01893.x; PMID: 22356587
- Ogilvie LA, Jones BV. Dysbiosis modulates capacity for bile acid modification in the gut microbiomes of patients with inflammatory bowel disease: a mechanism and marker of disease?. Gut 2012; 61:1642 - 3; http://dx.doi.org/10.1136/gutjnl-2012-302137; PMID: 22490526
- Welters CF, Heineman E, Thunnissen FB, van den Bogaard AE, Soeters PB, Baeten CG. Effect of dietary inulin supplementation on inflammation of pouch mucosa in patients with an ileal pouch-anal anastomosis. Dis Colon Rectum 2002; 45:621 - 7; http://dx.doi.org/10.1007/s10350-004-6257-2; PMID: 12004211
- Wong JM, Kendall CW, de Souza R, Emam A, Marchie A, Vidgen E, Holmes C, Jenkins DJ. The effect on the blood lipid profile of soy foods combined with a prebiotic: a randomized controlled trial. Metabolism 2010; 59:1331 - 40; http://dx.doi.org/10.1016/j.metabol.2009.12.017; PMID: 20096897
- Drakoularakou A, Tzortzis G, Rastall RA, Gibson GR. A double-blind, placebo-controlled, randomized human study assessing the capacity of a novel galacto-oligosaccharide mixture in reducing travellers’ diarrhoea. Eur J Clin Nutr 2010; 64:146 - 52; http://dx.doi.org/10.1038/ejcn.2009.120; PMID: 19756029
- Silk DB, Davis A, Vulevic J, Tzortzis G, Gibson GR. Clinical trial: the effects of a trans-galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment Pharmacol Ther 2009; 29:508 - 18; http://dx.doi.org/10.1111/j.1365-2036.2008.03911.x; PMID: 19053980
- Parisi GC, Zilli M, Miani MP, Carrara M, Bottona E, Verdianelli G, Battaglia G, Desideri S, Faedo A, Marzolino C, et al. High-fiber diet supplementation in patients with irritable bowel syndrome (IBS): a multicenter, randomized, open trial comparison between wheat bran diet and partially hydrolyzed guar gum (PHGG). Dig Dis Sci 2002; 47:1697 - 704; http://dx.doi.org/10.1023/A:1016419906546; PMID: 12184518
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013; 155:1451 - 63; http://dx.doi.org/10.1016/j.cell.2013.11.024; PMID: 24315484
- Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013; 2:e01202; http://dx.doi.org/10.7554/eLife.01202; PMID: 24192039
- Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, McGilvray ID, Allard JP. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 2013; 58:120 - 7; http://dx.doi.org/10.1002/hep.26319; PMID: 23401313
- Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol 2013; 11:868 - 75, e1-3; http://dx.doi.org/10.1016/j.cgh.2013.02.015; PMID: 23454028
- Whorwell PJ, Altringer L, Morel J, Bond Y, Charbonneau D, O’Mahony L, Kiely B, Shanahan F, Quigley EM. Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Am J Gastroenterol 2006; 101:1581 - 90; http://dx.doi.org/10.1111/j.1572-0241.2006.00734.x; PMID: 16863564
- Weizman Z, Asli G, Alsheikh A. Effect of a probiotic infant formula on infections in child care centers: comparison of two probiotic agents. Pediatrics 2005; 115:5 - 9; PMID: 15629974
- Cleusix V, Lacroix C, Vollenweider S, Le Blay G. Glycerol induces reuterin production and decreases Escherichia coli population in an in vitro model of colonic fermentation with immobilized human feces. FEMS Microbiol Ecol 2008; 63:56 - 64; http://dx.doi.org/10.1111/j.1574-6941.2007.00412.x; PMID: 18028400
- Jones ML, Martoni CJ, Prakash S. Cholesterol lowering and inhibition of sterol absorption by Lactobacillus reuteri NCIMB 30242: a randomized controlled trial. Eur J Clin Nutr 2012; 66:1234 - 41; http://dx.doi.org/10.1038/ejcn.2012.126; PMID: 22990854
- Jones ML, Martoni CJ, Parent M, Prakash S. Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults. Br J Nutr 2012; 107:1505 - 13; http://dx.doi.org/10.1017/S0007114511004703; PMID: 22067612
- Jones ML, Martoni CJ, Ganopolsky JG, Sulemankhil I, Ghali P, Prakash S. Improvement of gastrointestinal health status in subjects consuming Lactobacillus reuteri NCIMB 30242 capsules: a post-hoc analysis of a randomized controlled trial. Expert Opin Biol Ther 2013; 13:1643 - 51; http://dx.doi.org/10.1517/14712598.2013.833601; PMID: 24074303
- Jones ML, Martoni CJ, Prakash S. Letter to the editor regarding the report of Duboc et al: Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel disease. Gut 2013; 62:654 - 5; http://dx.doi.org/10.1136/gutjnl-2012-303867; PMID: 23148122
- Jones ML, Martoni CJ, Prakash S. Oral supplementation with probiotic L. reuteri NCIMB 30242 increases mean circulating 25-hydroxyvitamin D: a post hoc analysis of a randomized controlled trial. J Clin Endocrinol Metab 2013; 98:2944 - 51; http://dx.doi.org/10.1210/jc.2012-4262; PMID: 23609838
- Szajewska H, Ruszczyński M, Radzikowski A. Probiotics in the prevention of antibiotic-associated diarrhea in children: a meta-analysis of randomized controlled trials. J Pediatr 2006; 149:367 - 72; http://dx.doi.org/10.1016/j.jpeds.2006.04.053; PMID: 16939749
- McFarland LV. Meta-analysis of probiotics for the prevention of antibiotic associated diarrhea and the treatment of Clostridium difficile disease. Am J Gastroenterol 2006; 101:812 - 22; http://dx.doi.org/10.1111/j.1572-0241.2006.00465.x; PMID: 16635227
- Szymański H, Pejcz J, Jawień M, Chmielarczyk A, Strus M, Heczko PB. Treatment of acute infectious diarrhoea in infants and children with a mixture of three Lactobacillus rhamnosus strains--a randomized, double-blind, placebo-controlled trial. Aliment Pharmacol Ther 2006; 23:247 - 53; http://dx.doi.org/10.1111/j.1365-2036.2006.02740.x; PMID: 16393304
- Hickson M, D’Souza AL, Muthu N, Rogers TR, Want S, Rajkumar C, Bulpitt CJ. Use of probiotic Lactobacillus preparation to prevent diarrhoea associated with antibiotics: randomised double blind placebo controlled trial. BMJ 2007; 335:80; http://dx.doi.org/10.1136/bmj.39231.599815.55; PMID: 17604300
- Guandalini S. Probiotics for prevention and treatment of diarrhea. J Clin Gastroenterol 2011; 45:Suppl S149 - 53; http://dx.doi.org/10.1097/MCG.0b013e3182257e98; PMID: 21992955
- Allen SJ, Martinez EG, Gregorio GV, Dans LF. Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst Rev 2010; CD003048; PMID: 21069673
- Swidsinski A, Loening-Baucke V, Verstraelen H, Osowska S, Doerffel Y. Biostructure of fecal microbiota in healthy subjects and patients with chronic idiopathic diarrhea. Gastroenterology 2008; 135:568 - 79; http://dx.doi.org/10.1053/j.gastro.2008.04.017; PMID: 18570896
- Kruis W, Fric P, Pokrotnieks J, Lukás M, Fixa B, Kascák M, Kamm MA, Weismueller J, Beglinger C, Stolte M, et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 2004; 53:1617 - 23; http://dx.doi.org/10.1136/gut.2003.037747; PMID: 15479682
- Tursi A, Brandimarte G, Papa A, Giglio A, Elisei W, Giorgetti GM, Forti G, Morini S, Hassan C, Pistoia MA, et al. Treatment of relapsing mild-to-moderate ulcerative colitis with the probiotic VSL#3 as adjunctive to a standard pharmaceutical treatment: a double-blind, randomized, placebo-controlled study. Am J Gastroenterol 2010; 105:2218 - 27; http://dx.doi.org/10.1038/ajg.2010.218; PMID: 20517305
- Sood A, Midha V, Makharia GK, Ahuja V, Singal D, Goswami P, Tandon RK. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin Gastroenterol Hepatol 2009; 7:1202 - 9, e1; http://dx.doi.org/10.1016/j.cgh.2009.07.016; PMID: 19631292
- Gionchetti P, Rizzello F, Helwig U, Venturi A, Lammers KM, Brigidi P, Vitali B, Poggioli G, Miglioli M, Campieri M. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology 2003; 124:1202 - 9; http://dx.doi.org/10.1016/S0016-5085(03)00171-9; PMID: 12730861
- Arthur JC, Gharaibeh RZ, Uronis JM, Perez-Chanona E, Sha W, Tomkovich S, Mühlbauer M, Fodor AA, Jobin C. VSL#3 probiotic modifies mucosal microbial composition but does not reduce colitis-associated colorectal cancer. Sci Rep 2013; 3:2868; http://dx.doi.org/10.1038/srep02868; PMID: 24100376