Abstract
Consistent with an important role for adaptive immunity in modulating interactions between intestinal bacteria and host, dramatic alteration in the composition of gut microbes during chronic HIV infection was recently reported by ourselves and independently by four other research groups. Here we evaluate our results in the context of these other studies and delve into the effects of antiretroviral therapy (ART). Although gut microbiota of HIV-positive individuals on ART usually does not resemble that of HIV-negative individuals, the degree to which ART restores health-associated prevalence varies across bacterial taxa. Finally, we discuss potential drivers and health consequences of gut microbiota alterations. We propose that understanding the mechanism of HIV-associated gut microbiota changes will elucidate the role of adaptive immunity in shaping gut microbiota composition, and lay the foundation for therapeutics targeting the microbiota to attenuate HIV disease progression and reduce the risk of gut-linked disease in people with HIV.
HIV infection causes rapid and substantial depletion of lamina propria CD4+ T cells that are important for modulating interaction with intestinal bacteria.Citation1 In five recently published, independent research studies that used the 16S ribosomal RNA (rRNA) gene to compare gut microbiota composition between individuals with chronic HIV infection and seronegative controls,Citation2-Citation6 strong and characteristic compositional differences were observed in the relative abundance of bacteria in fecesCitation2,Citation6 as well as in mucosal samples from the ileum through the rectosigmoid colon.Citation2,Citation4,Citation5 Gut bacteria have been proposed to play a key role in HIV disease progression, since translocation of bacterial products such as lipopolysaccharide to blood may cause systemic activation of T cells, and HIV preferentially infects activated T cells.Citation7-Citation9 Furthermore, many diseases that increase in prevalence with chronic HIV infection have been linked with gut microbiota composition including metabolic and cardiovascular disease.Citation10-Citation12 Now that it has been well established that gut microbiota compositional changes occur with HIV infection, understanding why they occur and the health consequences of these alterations is of paramount importance.
There was some consistency in the types of bacteria reported to differ with chronic HIV infection across the five independent studies. For instance, the most profoundly enriched genus in feces of untreated HIV-infected individuals in our study (Prevotella) was significantly enriched with chronic HIV infection in two other studiesCitation2,Citation4 and the most profoundly depleted genus in our study (Bacteroides) was significantly depleted in three other studies.Citation2,Citation4,Citation5 These shared observations occurred despite substantial methodological differences, including (1) the types of samples evaluated (e.g., mucosal biopsies from different regions of the intestine vs. feces), (2) whether the HIV-infected individuals were receiving ART, and (3) the techniques used to characterize gut microbiota composition, including DNA extraction protocol, PCR primers, use of hybridization arrays (the PhyloChip) vs. next generation sequencing, which sequencing platform was used, and how data were bioinformatically analyzed.
Given the substantial methodological differences between the studies, it is also not surprising that there were notable ways in which the results varied across studies. For instance, various types of Proteobacteria significantly increased in relative abundance with HIV infection in all three papers that evaluated biopsiesCitation2,Citation4,Citation5 but with the exception of taxa in the Desulfovibrio genus in feces, increases in Proteobacteria were not observed in fecal and rectal sponge samples.Citation3,Citation6 Consistent with this observation, Dillon et al.Citation2 detected a significant increase in Proteobacteria in mucosal samples but not matched fecal samples. Since increased Proteobacteria has been linked with inflammatory states in the gut,Citation13 mucosal samples from HIV-infected individuals appear to have a more pro-inflammatory composition than do fecal samples. Differential detection of Proteobacteria between mucosal vs. luminal samples is consistent with the observation that Escherichia coli (one of many species within this broad phylum) was augmented in the mucosa but not the lumen of patients with inflammatory bowel disease (IBD).Citation14 This indicates that changes in some taxa may only occur in tightly host-associated niches of the gut.
It does not appear to be the case, however, that only bacteria that tightly associate with the host mucosa change with HIV infection, with strong and sometimes unique compositional changes observed in evaluations of fecal samples. For instance, the most highly depleted Operational Taxonomic Unit (OTU; cluster in which sequences have ≥97% identity over their aligned 16S rRNA genes, which approximates assignment to the same speciesCitation15) in the fecal samples in our study had a representative 16S rRNA gene sequence that was 100% identical to Bacteroides uniformis, a species previously reported to be found in higher abundance in the lumen than in the mucosa.Citation16 We did not detect a significant change in Bacteroides fragilis,Citation6 a species that is much more prevalent in mucosal samples than feces.Citation16 In contrast, by using BLAST to find the most closely affiliated species of the significantly depleted PhyloChip probes in mucosal samples from HIV-positive individuals in Vujkovic-Cvijin et al., we observed that a probe with 99% identity over aligned 16S rRNA genes to B. fragilis (probeset ID 69014) was among the most significantly depleted in HIV-infected individuals, showing a 5.6-fold depletion, whereas no significantly depleted probes were highly related to B. uniformis.Citation5 Thus with regard to species in the Bacteroides group, those with niches that are relatively luminal or mucosal were both depleted with chronic HIV infection, but change was more readily detected in the appropriate sample type.
Effect of ART on the Composition of Gut Microbiota
One conclusion drawn independently by all research groups whose cohorts included individuals undergoing ART, is that individuals on long-term successful treatment rarely had a gut microbiota composition that resembled that of HIV-negative individuals,Citation4-Citation6 suggesting that gut microbiota alterations could contribute to the pathogenesis of diseases that occur and/or persist with ART. Here we expand our analyses by doubling the size of our ART cohort from 14 to 28 individuals ().
Table 1. Patient clinical characteristics
In this augmented cohort, our original result was supported by the finding that at the community level, individuals on ART usually resemble individuals with untreated HIV infection more than HIV-negative controls. Specifically, various taxa that significantly decrease with HIV infection, such as the genera Bacteroides and Odoribacter or an OTU classified as Parabacteroides distasonis, remain at low prevalence in the majority of individuals on ART (). Similarly, some taxa that significantly increase in relative abundance with HIV infection, including the genus Prevotella, the family Paraprevotellaceae, and an OTU classified as Eubacterium biforme, which we previously have shown to induce a pro-inflammatory cytokine profile in in vitro stimulations,Citation6 have increased variance in relative abundance in individuals undergoing ART, but overall do not decrease toward levels typical of HIV-negative individuals (). In contrast, some taxa that were increased with untreated HIV infection, such as the genus Peptococcus, significantly decrease with ART (). Other taxa that had increased representation in people with untreated HIV infection, such as the Desulfovibrio and Catenibacterium genera, with ART trended back toward proportions seen in HIV seronegative individuals, although these decreases were not statistically significant after correcting for multiple comparisons (). These taxa furthermore showed intermediate levels in individuals on ART for less than one year compared with those undergoing long-term treatment (1–9 years; ). Differential recovery of various taxa with ART may reflect differences in the recovery of various components of the host immune system in gut-associated lymphoid tissue (GALT) and different underlying factors that control prevalence.
Figure 1. The degree to which ART restores health-associated prevalence to taxa that change in relative abundance varies across different bacterial taxa. The plots in this figure were selected across many significantly varying taxa to illustrate the point that taxa that change in relative abundance with untreated HIV infection differ in the degree to which they respond to ART. Panel A shows taxa with significantly decreased relative abundance with untreated HIV infection that did not generally return to levels typical of HIV negative controls with ART. Panel B shows taxa with significantly increased relative abundance with untreated HIV infection that rarely had levels typical of HIV negative controls with ART. Panel C shows taxa that significantly increase with untreated HIV infection that decrease in relative abundance progressively with short (<1 y) and long (1–9 y)-term ART. The relative abundance of each taxa in each fecal sample was estimated from 16S rRNA sequence data as described in reference Citation6 except that 14 additional individuals on ART, one additional individual with chronic untreated infection, and two additional HIV-negative controls were added to the previously described cohortCitation6 (). Briefly, the V4 region of 16S rRNA was PCR amplified and sequenced on a MiSeq sequencer (Illumina). Sequences were demultiplexed and quality filtered using QIIME 1.8Citation55 and binned into 97% ID OTUs and filtered for chimeras using usearchCitation56 and the greengenes 3_8 reference database.Citation57 OTUs were assigned to higher level taxa using the RDP classifier trained on the greengenes database and the number of sequences per sample were standardized to 9864 before performing statistical tests. The p-values listed on each plot indicates the significance of differences across HIV-negative, chronic HIV-positive untreated, and ART cohorts as calculated using a Kruskal-Wallis test. The p-values after correcting for multiple comparisons with the FDR techniqueCitation58 are also listed and were calculated with the group_significance.py script of QIIME. The bar on each plot indicates the median.
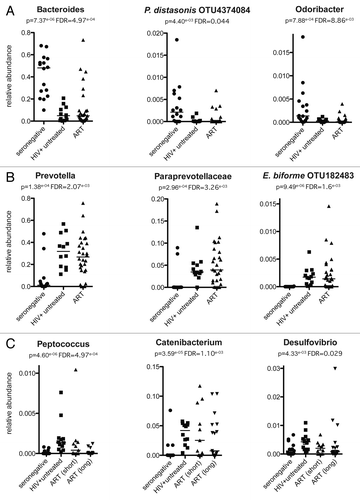
Occasionally, the microbiota composition of individuals on long-term ART resembled HIV-negative individuals more than those with untreated HIV infection. For instance a minority of individuals on ART had Bacteroides to Prevotella ratio typical of HIV-negative individuals. We showed previously that an overall “correction” of microbiota composition was not associated with the duration of ARTCitation6; however, other factors that may be involved have not been explored. These additional factors may include the duration of HIV infection and extent of CD4+ T cell depletion (i.e., CD4+ T cell nadir) before ART commenced, the specific class of antiretroviral drugs used, or the state of CD4+ T cell recovery in GALT.
Our ART-treated cohort also had lower stool microbiota diversity as measured with the Shannon diversity index compared with individuals with untreated HIV infection (P = 0.029: T test with Welch’s correction). This result is supported by Mutlu et al. who observed a significant decrease in mucosal bacterial diversity in their HIV-positive cohort who were receiving ART compared with seronegative controls.Citation4 Studies that compared diversity in mucosal samples from untreated HIV-infected individuals to seronegative controls did not see a significant difference in α diversity,Citation2,Citation5 and we actually saw a significant increase in phylogenetic diversity and the Shannon diversity index in the feces of individuals with untreated infection compared with HIV-negative controls.Citation6 A decrease in α diversity exclusively with ART may be related to direct effects of the antiretroviral drugs on the function of the gastrointestinal tract; some antiretroviral drugs induce noninfectious diarrhea as a side-effect, with up to 40% of HIV-positive individuals receiving ART experiencing moderate to severe diarrhea.Citation17,Citation18 Decreased α diversity has been described in many different inflammatory and/or disturbed states of the gut, including diarrhea.Citation19 Taken together, this suggests that the combined effects of ART and of HIV infection itself, whose effects are not entirely mitigated by ART, may induce a “double hit” to the microbiota. However, the effects of ART on microbiota composition appear to be subtle compared the effects of HIV infection itself. Further study of the effect of different types of antiretroviral drugs on the microbiota is an important future direction.
Why Does HIV-Induced Immunodeficiency Cause a Compositional Change?
Now that alteration in gut microbiota composition with chronic HIV infection has been demonstrated, important questions that remain are: (1) Why do these specific changes occur? and (2) What are the implications for health?
One possible driving factor is that the loss of effector CD4+ T cells results in the failure to mount an effective immune response to pathobionts, resulting in the outgrowth of certain bacteria. However, this view ignores the concept that adaptive immunity has the means to both promote and to temper innate immune responses programmed for microbial clearance.Citation20 Compositional change with HIV infection may also result from the loss of mutualistic bacteria that depend on interaction with the adaptive immune system for persistence (). Consistent with this notion, B. fragilis, a species that was significantly depleted in relative abundance in mucosa samples from HIV-infected subjectsCitation5 relies on a CD4+ T cell-interacting coat polysaccharide called polysaccharide A (PSA) to establish a niche in the gut; B. fragilis isolates in which the PSA operon has been knocked out have a decreased ability to colonize the mouse colon mucosa.Citation21 PSA is a member of a family of zwitterionic coat polysaccharides (ZPSs) that after processing by antigen presenting cells can activate CD4+ T cells to induce IL-10 producing anti-inflammatory FoxP3+ CD4+ T regulatory cells (Tregs).Citation22,Citation23 The dependence of B. fragilis on CD4+ T cell-interacting PSA for persistence in the mucosa is consistent with the depletion of B. fragilis in HIV-infected individuals who have reduced numbers of CD4+ T cells in GALT.
Figure 2. Certain symbiotic bacteria, such as B. fragilis, require CD4+ T cell interacting molecular factors for persistence in the gut mucosa. For instance, some bacteria produce zwitterionic coat polysaccharides (ZPS) which when processed by the innate immune system and presented to CD4+ T cells cause differentiation into Treg cells that produce the anti-inflammatory cytokine IL-10. By inducing IL-10 production, B. fragilis effectively limits the activity of both effector T cells and innate immune cells thereby enabling B. fragilis to persist in the gut mucosa with limited immune system interventionCitation21 (A). Both effector and regulatory CD4+ T cells are specifically targeted and killed by HIV and the loss of these cells may leave beneficial bacteria, such as B. fragilis, without the protection of IL-10 and may thus result in killing by innate immune cells (B).
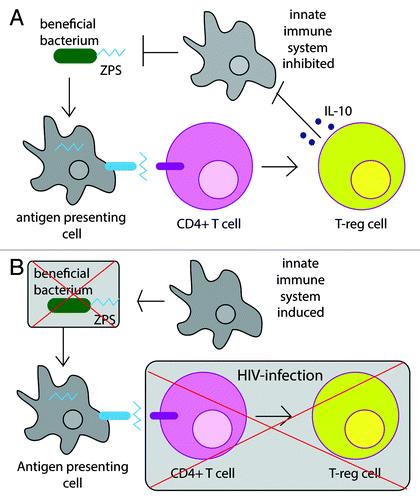
This leads to questions of whether other Treg-inducing bacteria may also depend on CD4+ T cell interaction to establish a niche in the gut. In fact, several of the phylotypes that are decreased with chronic HIV infection are known Treg inducers. These include P. distasonisCitation24 and B. uniformis, which induce Tregs through a shared T cell receptor,Citation25 as well as Bacteroides thetaiotaomicron and Bacteroides massiliensis.Citation26 It is also possible that the loss of Clostridia species with HIV infection observed by some groupsCitation2,Citation5 may indicate a loss of Treg inducers since some Clostridia have Treg-inducing properties.Citation27 However, Clostridia is a broad phylogenetic group containing taxa with very diverse properties, and it has not been established that the types of Clostridia that decrease with HIV infection induce Tregs.
Do Alterations in Gut Microbiota Mediate Chronic Inflammation in HIV-Infected Individuals?
Immune activation and chronic inflammation have been linked with HIV disease progressionCitation28-Citation30 and co-morbidities that occur in both untreated and treated chronic infection, including metabolic and cardiovascular disease.Citation31-Citation35 Determining whether gut microbiota alterations that occur with HIV infection are direct drivers of inflammation will establish whether these alterations may be a part of the etiology of these diseases. Consistent with a role for gut microbiota alterations in HIV disease progression, the only long-term non-progressor (LTNP, i.e., a person in an “elite” group of suppressors in whom HIV disease does not progress despite not undergoing ART) evaluated in any of the existing studies had a gut microbiota composition that was more similar to HIV seronegative subjects than to HIV-positive subjects.Citation5
Interestingly, gut microbiota composition correlated with markers of immune activation in both blood and gut mucosal tissues, although in all reports no effort was made to correct for multiple comparisons or potentially confounding variables such as viral load, shedding some doubt on the strength of the associations. Both Dillon et al. and Vujkovic-Cvijin et al. detected a correlation between overall microbiota composition in rectosigmoid biopsies and HLA-DR/CD38 expression on blood and mucosal CD4+ and CD8+ T cells.Citation2,Citation5 Specifically, Dillon et al. found that the relative abundance of the genus Prevotella was positively correlated with the number of activated (CD38+ HLA-DR+) CD4+ and CD8+ T cells per gram of mucosal tissue and the level of CD1c+ mDC activation based on CD40 expression.Citation2 Associations were also seen between the relative abundance of specific bacterial taxa in mucosal tissues and plasma cytokines and/or chemokines levels including IL-6, TNF-α, IL-10, and IP-10. For example, Mutlu et al. showed that plasma IL-6 levels were inversely associated with Bacteroides relative abundance,Citation4 which is consistent with anti-inflammatory properties of at least some of the species in this genus. Dillon et al. also found that systemic biomarkers of bacterial translocation in plasma, sCD14 and LPS, were correlated in an increase in Lachnospira and Roseburia, respectively, in the gut.Citation2 Although the presence of these associations is intriguing, correlations do not define cause and effect and may just be a reflection of HIV disease progression. No associations were found between changes in the composition of gut microbiota and CD4+ T cell count or viral load in blood in any of the studies.
Positive or negative correlations between specific bacteria and inflammatory markers are particularly compelling if the identified bacteria were independently shown to have pro- or anti-inflammatory properties. As already noted, several species that decrease in relative abundance with HIV infection are known to stimulate anti-inflammatory Tregs. Furthermore, a couple of these Treg inducers have been shown to protect against colitis in mouse models, at least in part by competitively inhibiting the colonization of pathogenic bacteria. B. fragilis protected against colonization of Helicobacter hepaticus and of 2,4,6-trinitrobenzenesulfonic acid (TNBS) -induced colitis in a PSA dependent manner.Citation36,Citation37 Furthermore, oral treatment of mice with P. distasonis lysate reduced the severity of dextran sodium sulfate (DSS)-induced intestinal inflammation. This protection was mediated at least in part by the adaptive immune system, since severe combined immunodeficient (SCID) mice were not protected.Citation24
As already noted, phylotypes related to Proteobacteria that can be pro-inflammatory, such as Escherichia and Campylobacter, were increased in mucosal but not fecal samples of HIV-infected individuals.Citation4,Citation5,Citation38 Heat-killed commensal E. coli exposure has previously been shown to enhance HIV-1 replication, CD4+ T cell activation and infection in in vitro experiments with lamina propria mononuclear cells (LPMCs)Citation38 although the relative abundance of Escherichia did not correlate with systemic or mucosal markers of immune activation in vivo.Citation2 The high relative abundance of the genus Prevotella in HIV-infected individuals and a positive correlation with immune activation markers in mucosal tissues has led to speculation about whether bacteria in the Prevotella genus are themselves pro-inflammatory and Prevotella-rich communities have indeed been associated with inflammation in other studies. For instance, in a recent study that correlated Prevotella copri with disease in new-onset untreated Rheumatoid Arthritis (RA) patients,Citation39 the colonization of antibiotic-treated C57BL-6 mice with P. copri by oral gavage resulted in increased inflammation and led to more severe symptoms in DSS-induced colitis.Citation39 In another study, deficiency of the NLRP6 inflammasome in mouse colonic epithelial cells resulted in a shift in the microbiota to one with a significant increase in Prevotellaceae, and transmission of this microbiota to wildtype mice via co-housing also lead to more severe symptoms in DSS-induced colitis.Citation40
Although these studies both indicate an association of Prevotella with inflammation and that Prevotella-rich communities may fail to protect against bacteria-mediated inflammation upon chemical injury, neither directly show that Prevotella itself is driving the inflammation. Consistent with the Prevotella-dominated community type in humans, an increased representation of Prevotella was associated with other complex community changes, including in both cases a community shift away from the anti-inflammatory Bacteroides genus just discussed.Citation39 Genomic analysis of P. copri revealed genes that allow for the tolerance of inflammation, such as superoxide dismutase, but not genes that promote inflammation.Citation39 Furthermore, our stimulations of human peripheral blood mononuclear cells (PBMC) with P. copri lysate showed that the cytokine production was not any more pro-inflammatory (as assessed by TNF-α/IL-10 ratio) than two different Bacteroides species testedCitation6 and stimulations of human dendritic cells with three different species in the Prevotella genus showed them to have a moderate pro-inflammatory profile that was weaker than the assayed opportunistic pathogens Hemophilus spp and Moraxella spp.Citation41 Although multiple studies have suggested that sulfatases may allow Prevotella to degrade mucins and induce inflammation by that mechanism,Citation2,Citation42 many of the Bacteroides species that decrease with HIV infection are also known to degrade mucins using sulfatases.Citation43 Taken together, these results suggest that it may be aspects of the Prevotella-rich community type, including a corresponding loss of anti-inflammatory Bacteroides or co-occurrence of directly pro-inflammatory bacteria, rather than Prevotella-richness itself that results in an inflammatory phenotype.
It is also important to note that Prevotella-richness is commonly observed in health and associated with diet. A Prevotella-rich and/or Bacteroides-poor community profile highly similar to that observed in HIV-infected patients is regularly observed in healthy individuals in agrarian cultures, including Malawi, the Amazonas states of Venezuela, and Burkina Faso.Citation6,Citation44,Citation45 Among healthy Thai subjects, vegetarians had high abundance of Prevotella and non-vegetarians of Bacteroides,Citation46 and within the United States population, Prevotella-richness has been described in healthy individuals who consume diets relatively rich in carbohydrates and poor in animal products.Citation47
Mouse studies have suggested that at least some of the differences between the microbiota that is typical of Western vs. agrarian cultures (and HIV-negative vs. -positive people in the USA) have the potential to protect against health consequences of a Western diet. As an example, B. uniformis is highly enriched in HIV-negative individuals in the USA compared with HIV-positive and compared with individuals in Malawi and Venezualean American Indians,Citation6,Citation45 and oral administration of B. uniformis reduced high fat diet (HFD)-induced inflammation and metabolic disease in mice.Citation11 This protection was mediated, at least in part, by interaction with the adaptive arm of the immune system. B. uniformis intake restored an otherwise compromised capacity of intestinal dendritic cells to induce a T cell proliferative response to LPS, and reduced the prominence of Enterobacteraceae and the overall pro-inflammatory signal from the gut (as assessed by in vitro stimulations of macrophage and dendritic cells with feces).Citation11 However, in the absence of the HFD, the mice did not have an overall pro-inflammatory signal in the gut, nor did they have as high populations of Enterobacteraceae, indicating that the presence of B. uniformis only had a health benefit in a particular dietary context.
In our work, we did not collect dietary information on study participants, and so we do not know if dietary differences between HIV-negative and HIV-infected persons were influencing observed differences in the fecal microbiome. Dillon et al. also observed an increase in Prevotella and decrease in Bacteroides with chronic HIV infection and determined that this relationship was not associated with dietary differences between their cohorts.Citation2 Interestingly, Bacteroides relative abundance positively correlated with red-meat intake in HIV-negative, but not HIV-positive, individuals.Citation2 This observation, together with the finding that certain Bacteroides species protect against metabolic disease in HFD-fed mice, suggests that dysfunction of the adaptive immune system that occurs with HIV infection compromises the ability to select for bacteria that protect against metabolic disease in individuals who are eating a Western HFD, potentially resulting in a mismatch between diet and microbiota composition that is detrimental to health (). Consistent with this notion, HIV-positive individuals who eat a diet more consistent with their microbiota, one high in fiber, have a reduced incidence of lipodystrophy, a metabolic disease manifested by lipoatrophy in the face, extremities, and buttocks with or without visceral fat accumulation,Citation48-Citation50 dyslipidemia, impaired glucose tolerance, and insulin resistance.Citation51,Citation52 Similarly among Africans in Rwanda, lipodystrophy was observed in 48.5% of ART-treated individuals living in cities but in only 17.3% of rural populations who likely consume a more plant-based diet.Citation33
Figure 3. A healthy gut microbiota is context specific. Species that are protective against a Western HFD in mice (e.g., B. uniformis) are also present in increased relative abundance in people who generally consume that diet (e.g., the US population) than those that do not (agrarian cultures). The “Western” diet-associated microbiota may thus in some respects be the result of the positive selection of bacteria that help us to optimally digest our diet. That bacteria that are beneficial in the context of a HFD increase with a HFD in HIV-negative but not in HIV-positive individuals suggests that the adaptive immune system plays a role in molding microbial composition to one that is optimal for our diet.
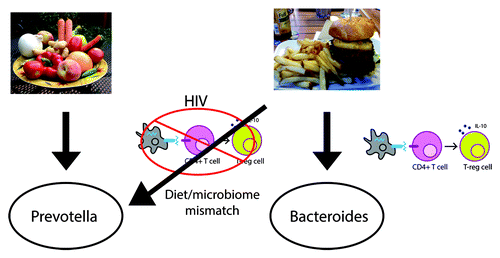
Prevotella-richness in the context of a HFD may also be linked to an increased prevalence of cardiovascular disease and/or atherosclerosis in HIV-infected individuals. A Prevotella-rich community type has been associated with atherosclerosis in HIV-negative individuals, because Prevotella-rich communities produce more trimethylamine (TMA) from L-carnitine, a nutrient in red meat.Citation12 TMA is converted by human enzymes to pro-atherosclerotic trimethylamine-N-oxide (TMAO). Although TMA-production capabilities have not been demonstrated in Prevotella itself, it has been shown in multiple species of Desulfovibrio,Citation53 which are increased in HIV-infected individualsCitation5,Citation6 (). More work is needed to determine whether a Prevotella-rich microbiota type and its co-occurring microbes such as TMA producing Desulfovibrio species are associated with heart disease in HIV-infected patients.
Finally, given that multiple species that are depleted in the gut of HIV-infected individuals protect against HFD-induced inflammation in mice, including both B. uniformis and Akkermansia mucinophila,Citation4,Citation10 it is also of interest that an atherogenic diet (AD) high in saturated fat and cholesterol has been shown to accelerate the progression of simian immunodeficiency virus (SIV) disease in macaques.Citation54 SIV-infected macaques fed an AD had higher levels of proinflammatory cytokines, specifically IL-18, which strongly correlated with viral load and is known to be elevated in obese individuals and associated with lipodystrophy in HIV-infected individuals.Citation54
Conclusions
HIV infection clearly is associated with alterations of the gut microbiota composition. New challenges are (1) to understand why the change occurs, (2) to determine whether this altered microbiota plays a role in HIV disease progression and in the high prevalence of co-morbidities such as cardiovascular and metabolic diseases, and (3) to determine the influence of ART. The pronounced loss of Bacteroides spp. such as B. uniformis, which is associated with the reduction of HFD-induced inflammation, suggests that HIV-infected individuals who consume diets high in fat and protein and low in carbohydrates and fiber may have an increase in inflammation because of the absence of beneficial bacteria that attenuate diet-driven inflammation. This notion is consistent with previous reports that diet modulation has the potential to attenuate both metabolic diseases in HIV-infected individualsCitation51,Citation52 and SIV disease progression in SIV-infected macaques.Citation54 Tailoring diet modulations aimed at promoting health to the microbiota composition of HIV-positive individuals and developing probiotic therapies that are based on a mechanistic understanding of the drivers and consequences of the compositional changes that occur with HIV infection, are promising strategies for promoting health in HIV-infected individuals.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
This work was supported by a LHMP sponsored project (UO1HL098996 to A.F. and T.C.) and by the Colorado Clinical and Translational Sciences Institute (UL1TR000005). C.L. was supported by K01DK090285. We would particularly like to thank the study participants.
References
- Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med 2004; 200:749 - 59; http://dx.doi.org/10.1084/jem.20040874; PMID: 15365096
- Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, Gianella S, Siewe B, Smith DM, Landay AL, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 2014; 7:983 - 94; http://dx.doi.org/10.1038/mi.2013.116; PMID: 24399150
- McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, Anton P, Braun J. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome 2013; 1:26; http://dx.doi.org/10.1186/2049-2618-1-26; PMID: 24451087
- Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, French A, Demarais P, Sun Y, Koenig L, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 2014; 10:e1003829; http://dx.doi.org/10.1371/journal.ppat.1003829; PMID: 24586144
- Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, Hernandez RD, Lederman MM, Huang Y, Somsouk M, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 2013; 5:93ra91; http://dx.doi.org/10.1126/scitranslmed.3006438; PMID: 23843452
- Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, Knight R, Fontenot AP, Palmer BE. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013; 14:329 - 39; http://dx.doi.org/10.1016/j.chom.2013.08.006; PMID: 24034618
- Brenchley JM, Douek DC. HIV infection and the gastrointestinal immune system. Mucosal Immunol 2008; 1:23 - 30; http://dx.doi.org/10.1038/mi.2007.1; PMID: 19079157
- Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 2006; 12:1365 - 71; http://dx.doi.org/10.1038/nm1511; PMID: 17115046
- Moriyama K, Ando C, Tashiro K, Kuhara S, Okamura S, Nakano S, Takagi Y, Miki T, Nakashima Y, Hirakawa H. Polymerase chain reaction detection of bacterial 16S rRNA gene in human blood. Microbiol Immunol 2008; 52:375 - 82; http://dx.doi.org/10.1111/j.1348-0421.2008.00048.x; PMID: 18667036
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 2013; 110:9066 - 71; http://dx.doi.org/10.1073/pnas.1219451110; PMID: 23671105
- Gauffin Cano P, Santacruz A, Moya Á, Sanz Y. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS One 2012; 7:e41079; http://dx.doi.org/10.1371/journal.pone.0041079; PMID: 22844426
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 2013; 19:576 - 85; http://dx.doi.org/10.1038/nm.3145; PMID: 23563705
- Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play?. Nat Rev Gastroenterol Hepatol 2012; 9:219 - 30; http://dx.doi.org/10.1038/nrgastro.2012.14; PMID: 22349170
- de Souza HL, de Carvalho VR, Romeiro FG, Sassaki LY, Keller R, Rodrigues J. Mucosa-associated but not luminal Escherichia coli is augmented in Crohn's disease and ulcerative colitis. Gut Pathog 2012; 4.
- Stackebrandt E, Goebal BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int Syst Bacteriol 1994; 44:846 - 9; http://dx.doi.org/10.1099/00207713-44-4-846
- Namavar F, Theunissen EB, Verweij-Van Vught AM, Peerbooms PG, Bal M, Hoitsma HF, MacLaren DM. Epidemiology of the Bacteroides fragilis group in the colonic flora in 10 patients with colonic cancer. J Med Microbiol 1989; 29:171 - 6; http://dx.doi.org/10.1099/00222615-29-3-171; PMID: 2746627
- Max B, Sherer R. Management of the adverse effects of antiretroviral therapy and medication adherence. Clin Infect Dis 2000; 30:Suppl 2 S96 - 116; http://dx.doi.org/10.1086/313859; PMID: 10860894
- Kartalija M, Sande MA. Diarrhea and AIDS in the era of highly active antiretroviral therapy. Clin Infect Dis 1999; 28:701 - 5, quiz 706-7; http://dx.doi.org/10.1086/515191; PMID: 10825021
- Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 2008; 197:435 - 8; http://dx.doi.org/10.1086/525047; PMID: 18199029
- Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012; 489:231 - 41; http://dx.doi.org/10.1038/nature11551; PMID: 22972296
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011; 332:974 - 7; http://dx.doi.org/10.1126/science.1206095; PMID: 21512004
- Tzianabos AO, Onderdonk AB, Rosner B, Cisneros RL, Kasper DL. Structural features of polysaccharides that induce intra-abdominal abscesses. Science 1993; 262:416 - 9; http://dx.doi.org/10.1126/science.8211161; PMID: 8211161
- Avci FY, Kasper DL. How bacterial carbohydrates influence the adaptive immune system. Annu Rev Immunol 2010; 28:107 - 30; http://dx.doi.org/10.1146/annurev-immunol-030409-101159; PMID: 19968562
- Kverka M, Zakostelska Z, Klimesova K, Sokol D, Hudcovic T, Hrncir T, Rossmann P, Mrazek J, Kopecny J, Verdu EF, et al. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin Exp Immunol 2011; 163:250 - 9; http://dx.doi.org/10.1111/j.1365-2249.2010.04286.x; PMID: 21087444
- Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011; 478:250 - 4; http://dx.doi.org/10.1038/nature10434; PMID: 21937990
- Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med 2014; 6:20ra11; http://dx.doi.org/10.1126/scitranslmed.3008051; PMID: 24452263
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500:232 - 6; http://dx.doi.org/10.1038/nature12331; PMID: 23842501
- Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, Shih R, Lewis J, Wiley DJ, Phair JP, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis 1999; 179:859 - 70; http://dx.doi.org/10.1086/314660; PMID: 10068581
- Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, Hsue P, Emu B, Krone M, Lampiris H, et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis 2008; 197:126 - 33; http://dx.doi.org/10.1086/524143; PMID: 18171295
- Salazar-Gonzalez JF, Martinez-Maza O, Nishanian P, Aziz N, Shen LP, Grosser S, Taylor J, Detels R, Fahey JL. Increased immune activation precedes the inflection point of CD4 T cells and the increased serum virus load in human immunodeficiency virus infection. J Infect Dis 1998; 178:423 - 30; http://dx.doi.org/10.1086/515629; PMID: 9697722
- De Lorenzo F, Collot-Teixeira S, Boffito M, Feher M, Gazzard B, McGregor JL. Metabolic-inflammatory changes, and accelerated atherosclerosis in HIV patients: rationale for preventative measures. Curr Med Chem 2008; 15:2991 - 9; http://dx.doi.org/10.2174/092986708786848668; PMID: 19075647
- Madge S, Kinloch-de-Loes S, Mercey D, Johnson MA, Weller IVD. Lipodystrophy in patients naive to HIV protease inhibitors. AIDS 1999; 13:735 - 7; http://dx.doi.org/10.1097/00002030-199904160-00020; PMID: 10397574
- Mutimura E, Stewart A, Rheeder P, Crowther NJ. Metabolic function and the prevalence of lipodystrophy in a population of HIV-infected African subjects receiving highly active antiretroviral therapy. J Acquir Immune Defic Syndr 2007; 46:451 - 5; http://dx.doi.org/10.1097/QAI.0b013e318158c0a6; PMID: 18077834
- Shevitz A, Wanke CA, Falutz J, Kotler DP. Clinical perspectives on HIV-associated lipodystrophy syndrome: an update. AIDS 2001; 15:1917 - 30; http://dx.doi.org/10.1097/00002030-200110190-00003; PMID: 11600819
- Trøseid M, Manner IW, Pedersen KK, Haissman JM, Kvale D, Nielsen SD. Microbial translocation and cardiometabolic risk factors in HIV infection. AIDS Res Hum Retroviruses 2014; 30:514 - 22; http://dx.doi.org/10.1089/aid.2013.0280; PMID: 24521167
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008; 453:620 - 5; http://dx.doi.org/10.1038/nature07008; PMID: 18509436
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A 2010; 107:12204 - 9; http://dx.doi.org/10.1073/pnas.0909122107; PMID: 20566854
- Dillon SM, Manuzak JA, Leone AK, Lee EJ, Rogers LM, McCarter MD, Wilson CC. HIV-1 infection of human intestinal lamina propria CD4+ T cells in vitro is enhanced by exposure to commensal Escherichia coli. J Immunol 2012; 189:885 - 96; http://dx.doi.org/10.4049/jimmunol.1200681; PMID: 22689879
- Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife (Cambridge) 2013; 2:e01202; http://dx.doi.org/10.7554/eLife.01202; PMID: 24192039
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011; 145:745 - 57; http://dx.doi.org/10.1016/j.cell.2011.04.022; PMID: 21565393
- Larsen JM, Steen-Jensen DB, Laursen JM, Søndergaard JN, Musavian HS, Butt TM, Brix S. Divergent pro-inflammatory profile of human dendritic cells in response to commensal and pathogenic bacteria associated with the airway microbiota. PLoS One 2012; 7:e31976; http://dx.doi.org/10.1371/journal.pone.0031976; PMID: 22363778
- Elinav E, Henao-Mejia J, Flavell RA. Integrative inflammasome activity in the regulation of intestinal mucosal immune responses. Mucosal Immunol 2013; 6:4 - 13; http://dx.doi.org/10.1038/mi.2012.115; PMID: 23212196
- Benjdia A, Martens EC, Gordon JI, Berteau O. Sulfatases and a radical S-adenosyl-L-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron. J Biol Chem 2011; 286:25973 - 82; http://dx.doi.org/10.1074/jbc.M111.228841; PMID: 21507958
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 2010; 107:14691 - 6; http://dx.doi.org/10.1073/pnas.1005963107; PMID: 20679230
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature 2012; 486:222 - 7; PMID: 22699611
- Ruengsomwong S, Korenori Y, Sakamoto N, Wannissorn B, Nakayama J, Nitisinprasert S. Senior Thai fecal microbiota comparison between vegetarians and non-vegetarians using PCR-DGGE and real-time PCR. J Microbiol Biotechnol 2014; PMID: 24743571
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011; 334:105 - 8; http://dx.doi.org/10.1126/science.1208344; PMID: 21885731
- Cabrero E, Griffa L, Burgos A, HIV Body Physical Changes Study Group. Prevalence and impact of body physical changes in HIV patients treated with highly active antiretroviral therapy: results from a study on patient and physician perceptions. AIDS Patient Care STDS 2010; 24:5 - 13; http://dx.doi.org/10.1089/apc.2009.0191; PMID: 20095903
- Galescu O, Bhangoo A, Ten S. Insulin resistance, lipodystrophy and cardiometabolic syndrome in HIV/AIDS. Rev Endocr Metab Disord 2013; 14:133 - 40; http://dx.doi.org/10.1007/s11154-013-9247-7; PMID: 23700046
- Leclercq P, Goujard C, Duracinsky M, Allaert F, L’henaff M, Hellet M, Meunier JP, Carret S, Thevenon J, Ngo Van P, et al. High prevalence and impact on the quality of life of facial lipoatrophy and other abnormalities in fat tissue distribution in HIV-infected patients treated with antiretroviral therapy. AIDS Res Hum Retroviruses 2013; 29:761 - 8; http://dx.doi.org/10.1089/aid.2012.0214; PMID: 23268562
- Hadigan C, Jeste S, Anderson EJ, Tsay R, Cyr H, Grinspoon S. Modifiable dietary habits and their relation to metabolic abnormalities in men and women with human immunodeficiency virus infection and fat redistribution. Clin Infect Dis 2001; 33:710 - 7; http://dx.doi.org/10.1086/322680; PMID: 11486294
- Hendricks KM, Dong KR, Tang AM, Ding B, Spiegelman D, Woods MN, Wanke CA. High-fiber diet in HIV-positive men is associated with lower risk of developing fat deposition. Am J Clin Nutr 2003; 78:790 - 5; PMID: 14522738
- Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci U S A 2012; 109:21307 - 12; http://dx.doi.org/10.1073/pnas.1215689109; PMID: 23151509
- Mansfield KG, Carville A, Wachtman L, Goldin BR, Yearley J, Li W, Woods M, Gualtieri L, Shannon R, Wanke C. A diet high in saturated fat and cholesterol accelerates simian immunodeficiency virus disease progression. J Infect Dis 2007; 196:1202 - 10; http://dx.doi.org/10.1086/521680; PMID: 17955439
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335 - 6; http://dx.doi.org/10.1038/nmeth.f.303; PMID: 20383131
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26:2460 - 1; http://dx.doi.org/10.1093/bioinformatics/btq461; PMID: 20709691
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012; 6:610 - 8; http://dx.doi.org/10.1038/ismej.2011.139; PMID: 22134646
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met 1995; 57:289 - 300
