Abstract
Previously, we have shown that DNA prime-protein boost is effective in eliciting neutralizing antibodies (NAb) against randomly selected HIV-1 isolates. Given the genetic diversity of HIV-1 viruses and the unique predominant subtypes in different geographic regions, it is critical to test the DNA prime-protein boost approach against circulating viral isolates in key HIV endemic areas. In the current study, the same DNA prime-protein boost vaccine was used as in previous studies to investigate the induction of NAb responses against HIV-1 clade BC, a major subtype circulating in China. A codon optimized gp120-BC DNA vaccine, based on the consensus envelope (Env) antigen sequence of clade BC, was constructed and a stable CHO cell line expressing the same consensus BC gp120 protein was produced. The immunogenicity of this consensus gp120-BC was examined in New Zealand White rabbits by either DNA prime-protein boost or protein alone vaccination approaches. High levels of Env-specific antibody responses were elicited by both approaches. However, DNA prime-protein boost but not the protein alone immune sera contained significant levels of NAb against pseudotyped viruses expressing HIV-1 BC Env antigens. Furthermore, high frequencies of CD4 binding site-targeted antibodies were found in the DNA prime- protein boost rabbit sera indicating that the positive NAb may be the result of antibodies against conformationally sensitive epitopes on HIV-1 Env. The findings support that DNA prime-protein boost was effective in eliciting NAb against a key HIV-1 virus subtype in China. This result may lead to the development of regional HIV vaccines through this approach.
Introduction
Given the broad diversity of HIV-1 circulating throughout the world, eliciting broadly cross reactive protective antibodies is a major challenge for any HIV candidate vaccine currently under development. Previous studies have shown that by using a consensus Env gene approach, the breadth of antibody and cell mediated responses may be improved;Citation1-Citation4,Citation5 however, the level of improvement is still somewhat limited. One possible reason is that the viruses tested are collected from different geographic areas with highly diverse genetic backgrounds and use of one consensus Env may be unable to cover such broad diversity.
Currently and to our knowledge, there has not been any report on the production of consensus Env immunogens that can target viruses of the same subtype that are circulating in the same geographic region. Study on such “subtype-based” or “region-based” consensus will allow for the establishment of the “consensus” concept in developing a useful immunogen for HIV vaccine development. Information gained from such “proof-of-concept” studies will guide the development of additional consensus Env antigens to expand the coverage of HIV isolates in the world.
Previously, we demonstrated that priming immunizations with DNA vaccines expressing Env antigens can significantly enhance the quality, especially the neutralizing activities, of antibody responses elicited by subunit Env protein vaccines 7891011 when random HIV-1 Env antigens were selected; however, there is no information on whether this prime-boost approach is able to further enhance the neutralizing activities elicited by a consensus Env antigen.
In the current report, an immunogenicity study was conducted in rabbits using the DNA prime-protein boost approach to test the immunogenicity of a consensus gp120 immuogen targeting recombinant subtype BC, the dominant subtype viruses in China.Citation12-Citation18 Results from our study will determine whether a consensus immunogen can elicit antibodies that neutralize a wide range of viruses of the same subtype and whether the DNA priming step is helpful for further improving the quality of antibody responses.
Results
Expression of BC subtype consensus gp120 protein by DNA vaccine and stably transfected CHO cells
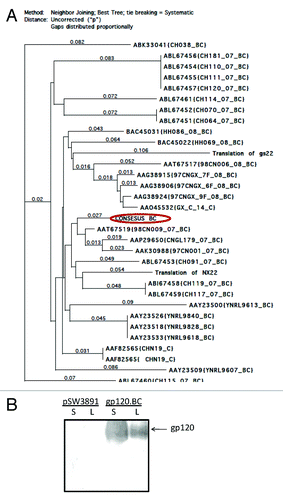
Based on more than 30 HIV-1 subtype BC gp120 protein sequences isolated from China, a consensus gene coding for BC gp120 was designed () and the BC gp120 DNA vaccine was produced by using this synthetic gene insert. The recombinant gp120 protein expressed by this BC gp120 DNA vaccine was examined by transient transfection of 293T cells with this DNA vaccine construct and Western blot analysis was conducted to verify the expression (). The BC gp120 protein produced from CHO cells was purified using a lectin column and verified by SDS-PAGE and Western blot analysis (data not shown).
Figure 1. (A) Phylogenetic tree of HIV-1 subtype BC natural and consensus gp120 amino acid sequences. The arrow indicates the consensus gp120-BC sequence produced in this study. (B) Western-blot analysis of gp120-BC protein expression in supernatant (S) and lysate (L) of 293T cells transfected by either a DNA vaccine expressing codon optimized consensus gp120-BC or the empty vector pSW3891. A rabbit serum specific for HIV-1 gp120 was used as the detecting antibody at 1:500 dilution.
gp120-specific antibody responses induced by BC gp120 protein alone and DNA prime-protein boost in rabbits
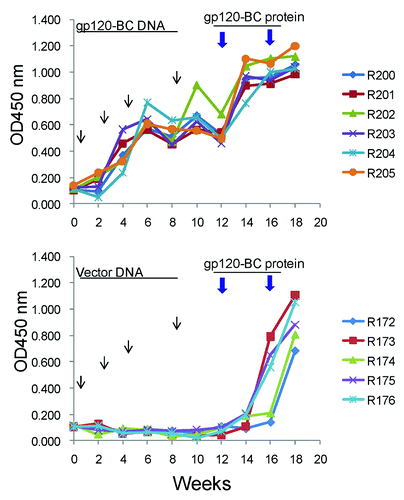
In this study, groups of rabbits (5 or 6 rabbits/group) were immunized using one of two schedules in order to provide a direct comparison of protein alone to prime-boost vaccinations: 1) four empty DNA vector inoculations followed by two gp120-BC boosts, and 2) four BC gp120 DNA vaccine prime immunizations followed by two gp120 protein boosts. The overall immunogenicity for each animal group was first determined by the binding titers of serum IgG for each individual rabbit against BC gp120 antigen by ELISA (). All rabbits, regardless of immunization regimen, generated similar high levels of gp120-specific antibody responses.
Figure 2. HIV-1 gp120-specific temporal antibody responses in rabbit sera induced by either gp120-BC DNA prime-protein boost (A) or protein alone immunizations (B). The arrows indicate the time points of DNA or protein immunizations. The antibody responses were measured by ELISA against gp120-BC as coating antigen at 1:1000 serum dilution. Each curve represents the antibody responses in one rabbit.
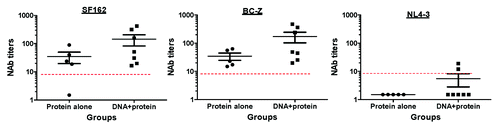
Neutralization of HIV-1 subtype B Tier 1 viruses
To evaluate neutralizing antibody responses elicited by the consensus BC subtype gp120 vaccine, we first examined the neutralizing activity against the HIV-1 clade B Tier 1 pseudoviruses, SF162 and NL4–3 (). SF162 is relatively sensitive to neutralization and NL4–3 is slightly resistant to neutralization. All rabbit sera, regardless of whether they received the BC subtype gp120 protein alone or DNA prime-protein boost, could neutralize SF162. However, the potency at which this was achieved differed between immunization groups. The neutralizing antibody titers against SF162 and chimeric virus BC-Z (autologous gp120-BC in SF162 backbone) in the protein alone group were between 1:20 to 1:100, with geometric mean titer of 1:25 and 1:29, respectively, while the NAb titers against SF162 and BC-Z in the DNA prime-protein boost group were up to 1:418 and 1:470 with geometric mean titer of 1:73 and 1:88, respectively. Although there was no statistical significance, likely due to the large variation of NAb titers, the pattern was clear that the DNA prime – protein boost group induced better NAb responses against SF162 and BC-Z viruses. Two of six rabbits in the DNA prime-protein boost group had NAb activity against NL4–3. In contrast, none of the rabbits in the gp120-BC protein alone group had NAb activity against NL4–3. Because both SF162 and NL4–3 are subtype B viruses and BC subtype is a recombinant virus mainly expressing Env from subtype C viruses, the difference in cross subtype neutralizing activities elicited by the DNA prime-protein boost vs. protein alone approach was quite significant.
Neutralization of HIV-1 subtype BC pseudotyped viruses
Next, rabbit sera were tested against a pseudotyped virus expressing the chimeric Env with consensus BC subtype gp120 fused with the gp41 portion of Env protein from SF162. Although all rabbits had NAb titers against this autologous pseudotyped viruses, the rabbit sera that received gp120DNA prime-protein boost elicited significantly more potent NAb responses (group geometric mean NAb titer 1:173) than those elicited by the protein alone group (geometric mean titer 1:37) (p = 0.0303) ().
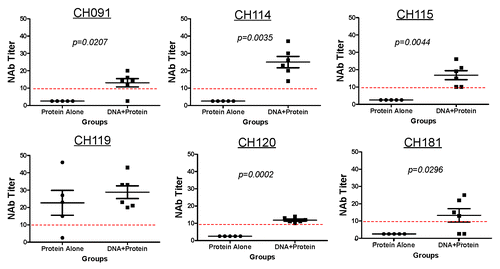
Further NAb analysis was conducted against a panel of eight HIV-1 pseudotyped viruses expressing the full length gp160 from each of the following viruses: CH064, CH091, CH110, CH114, CH115, CH119, CH120, and CH181, These viruses were isolated from patients in China and belonged to the same subtype BC as the consensus subtype BC gp120 antigen used in the current study. Rabbits that received protein alone immunization were completely incapable of neutralizing seven of the eight viruses tested. In contrast, animals receiving the DNA prime-protein boost neutralized six of the eight viruses tested. Four of five animals in the protein alone group neutralized the only virus in this subtype BC panel, CH1119, with NAb titers between 1:14 to 1:46. The group receiving DNA prime-protein boost immunizations neutralized six subtype BC viruses CH091, CH114, CH115, CH119 and CH120, but not CH064 and CH110, with NAb titers 1:10 to 1:42 (). The percent of positive neutralization events against this panel of eight subtype BC viruses were 69% in DNA primed rabbit sera and 12% in protein alone sera, respectively, which was statistically significant between the groups (p < 0.0001). The tier classification of these pseudotyped viruses was not determined. Our data demonstrates that the DNA prime-protein boost approach significantly improved neutralizing antibody responses against subtype BC viruses when compared with the protein alone immunization. However, no NAb activities were detected against subtype B and AE primary viruses as examined (data not shown).
Figure 4. Neutralizing antibody titers in rabbit sera receiving gp120-BC protein alone or DNA prime-protein boost immunizations against a panel of HIV-1 clade BC pseudotyped viruses expressing Env protein from subtype BC primary isolates: CH091, CH114, CH115, CH119, CH120 and CH181. Each dot represents one rabbit in each vaccination group. The statistical differences are shown when p value is significant (p < 0.05).
Analysis of antibody specificity elicited by gp120-BC vaccinations
In order to understand why significantly enhanced neutralization activities with the DNA prime-protein boost approach were observed (as described above) while the gp120-specific binding antibody titers were similar in both protein alone and DNA prime-protein boost groups, two studies were conducted to map the epitope specificity of rabbit sera from these two groups.
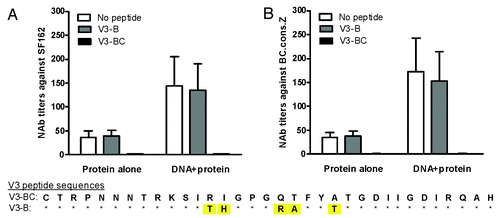
The first study measured neutralization activities with V3 peptide absorption. It is known that HIV-1 SF162 is sensitive to V3-specific antibody- mediated neutralization. Therefore, we employed V3 peptide absorption to dissect the potential contribution of V3-mediated neutralization using two V3 peptides: (1) subtype B V3 (V3-B) peptide matching to the SF162 virus, and (2) subtype BC V3 (V3-BC) peptide matching to the consensus subtype BC gp120 vaccines used in the current report. It is very interesting to note that the V3-B peptide could not inhibit neutralization in rabbits that received the gp120-BC protein alone or DNA prime-protein boost immunization while the V3-BC peptide inhibited the neutralization against SF162 over 90% ().
Figure 5. Neutralization against HIV-1 pseudotyped viruses SF162 (A) or chimeric virus BC-Z (gp120-BC in SF162 backbone) with or without V3 peptide absorption. Subtype B consensus (V3-B) or subtype BC consensus (V3-BC) V3 peptides were used for absorption before testing the neutralizing activities in either the gp120-BC protein alone or DNA prime-protein boost rabbit sera as indicated. The V3 peptide sequences are shown under the graphs.
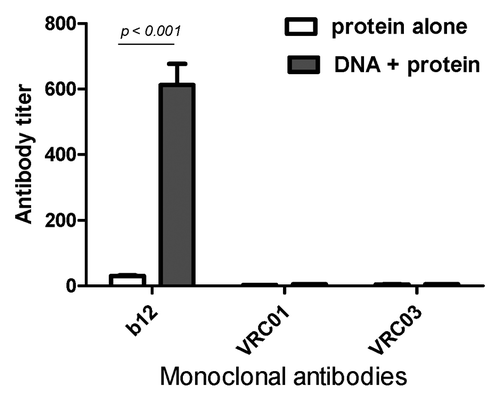
We also chose to use a pseudotyped virus-based competitive binding assay to examine the specificity of antibodies capable of binding to the important gp120 epitopes on the HIV-1 viral envelope spike as previously reported,Citation9,Citation19,Citation20 with the hope of determining the specificity differences between antibodies elicited by protein alone or DNA prime-protein boost immunization approaches. Previously, we reported that DNA prime-protein boost could induce improved antibody responses targeting the CD4bs, as such sera could compete well against the broad neutralizing mAb, b12 (Vaine, Wang et al., 2008; Vaine, Wang et al., 2010). In the current study, the presence of CD4bs-specific antibodies in rabbit immune sera was also evaluated against the broadly neutralizing antibodies b12, VRC01, and VRC03.Citation21-Citation23 When the mAb b12 was used, all animal sera in the DNA prime protein boost group had high titers (mean titers ~1:600) of antibodies competing against b12 and very low titers (mean ~1:30) of antibodies competing against b12 in the protein alone group (p < 0.001). However, none of these rabbit sera could compete against mAbs VRC01 or VRC03 (). In this assay, pseudotyped virus expressing JR-FL gp120 was used. Human mAbs b12, VRC01 and VRC03 could neutralize this virus but not by rabbit immune sera (data not shown).
Figure 6. Antibody titers competing against CD4bs-specific monoclonal antibodies b12, VRC01 and VRC03, respectively, in rabbit sera receiving gp120-BC protein alone or DNA prime-protein boost immunizations. Titers are show as the geometric means of rabbit sera from each immunization group. Statistically significance is indicated (p < 0.001).
Discussion
In the current report, we demonstrated that a consensus gp120 immunogen based on the sequence analysis of subtype BC viruses was able to elicit broad neutralizing antibody responses against a high percentage of viruses in the same subtype while the ability of such immune sera to neutralize viruses belonging to other subtypes was limited. More significantly, the DNA prime-protein boost approach was effective in achieving broadly reactive neutralizing activities while the recombinant gp120 protein immunogen alone, with the same consensus subtype BC gp120 sequences, was not able to achieve the same level of antibody responses.
Antibody specificities involved in improved neutralizing activities may be more than a one component processes. V3-based neutralizing epitopes are clearly present and may be responsible for neutralizing activities against sensitive viruses, including those not from subtype BC, but did not play a major role in cross-neutralizing activities against other more resistant subtype BC viruses.
Antibodies against conformationally sensitive epitopes were also present as shown by the ability of rabbit sera to compete against mAb, b12. This result confirmed our previous data that a CD4 binding site antibody, similar to b12, was elicited in rabbit studies using gp120 immunogens from subtypes other than subtype BC.Citation9,Citation11 At the same time, rabbit sera elicited by subtype BC gp120 were unable to compete against VRC01 or VRC03. This finding is not completely unexpected as VRC01 or VRC03 has much broader neutralizing activities than b12 and b12 can only neutralizing viruses from certain subtypes.
The most interesting finding is that rabbit sera immunized with the subtype BC consensus gp120 immunogen was able to neutralize multiple viral isolates from the same BC subtype but not other primary viral isolates from different subtypes, except those known to be sensitive. Previous studies have not found a similar high preference of neutralizing activities against a particular subtype, probably because previous consensus Env designs were aimed at neutralizing a wide range of diverse subtypes. Therefore, our data presented another option that has not been well explored in the HIV vaccine field, i.e., to use the consensus approach to target a subtype rather than the global spectrum of many subtypes. In a given geographic area, a limited number of subtypes are dominant, creating an opportunity to develop HIV vaccines that focus on local subtypes. While HIV isolates within a subtype still go through a high degree of mutation, the magnitude of such mutations will be less than the mutations occurring at the global level. While not every region or country will have the resources to develop vaccines for their own region, those with significant resources, such as South Africa, China, and India, for example, will be able to use this approach to develop vaccines against the dominant subtype(s) in their respective country. Any success in such a limited scale will offer hope for the development of vaccines against HIV for the entire world.
The current study used a consensus immunogen against one of China’s dominant HIV subtypes and suggested that viruses within certain subtypes may be more sensitive to this approach, although the DNA prime-protein boost method was instrumental in eliciting protective antibody responses. Future studies should determine whether the same consensus Env immunogens will be effective in eliciting neutralizing antibodies against viral isolates from other subtypes. The concept of polyvalent vaccines can then be tested by combining multiple consensus Env immunogens, each covering one subtype circulating in one region/country, to see if such a cocktail of consensus Env immunogens can neutralize viral isolates from all the subtypes circulating in that region/country.
Materials and Methods
Vaccines
HIV-1 gp120 DNA vaccine
Based on sequence and bioinformatics analyses of a collection of HIV-1 subtype BC sequences (> 30) from China that were available in GenBank in 2008, a consensus gp120-BC protein sequence was designed (). A codon optimized gene insert encoding this consensus gp120-BC sequence was then produced based on the codon preference of Homo sapiens using a previously reported approach with the assistance of Accelrys Gene software. Citation24,Citation25 Sequence optimization was also performed by avoiding the following cis-acting sequence motifs: internal TATA-boxes, chi-sites and ribosomal entry sites; AT-rich or GC-rich sequence stretches; ARE, INS, CRS sequence elements; cryptic splice donor and acceptor sites; and branch points. The final codon optimized gp120-BC sequence was chemically synthesized by Geneart (Regensburg, Germany), with added restriction enzyme sites of NheI and BamHI for subcloning purposes immediately upstream of the C1 region of gp120 and downstream of the stop codon, respectively.
The codon optimized gp120-BC gene insert was cloned into DNA vaccine vector pSW3891 at the NheI and BamHI sites, under the tissue plasimogen activator (tPA) leader sequence.Citation24,Citation25 The gp120-BC DNA vaccine plasmid was prepared in large amounts from E. coli HB101 strain with a Mega purification kit (Qiagen, Valencia, CA, USA) for both in vitro transfection and in vivo animal immunization studies.
HIV-1 gp120 protein vaccine
The same codon optimized gp120-BC gene above was cloned into the CHO expression vector under tPA leader sequence. A stable Chinese Hamster Ovary (CHO) cell line was constructed to express the recombinant gp120-BC protein as previously described.Citation8 Secreted gp120-BC protein from a stably transfected CHO cell line was harvested and purified by FPLC using Lentil lectin sepharose 4B column (GE Healthcare, Waukesha, WI).
Antibodies and peptides
The CD4 binding site directed monoclonal antibody, b12, was purchased from Polymun (Donaustrasse, Austria). The VRC01 and VRC03 monoclonal antibodies were obtained from the NIH AIDS Research and Reference Reagent Program. The HIV-1 subtype BC consensus V3 loop sequence (CTRPNNNTRKSIRIGPGQTFYATGDIIGDIRQAHC) and HIV-1 subtype B V3 loop sequence (CTRPNNNTRKSITHGPGRAFYTTGDIIGDIRQAHC) were synthesized by GenScript (Nanjing, China).
Western blot analysis
The gp120 protein produced in transiently transfected 293T cells and stably transfected CHO cells were analyzed by Western blot. Samples were subjected to standard SDS-PAGE and blotted onto PVDF membrane (BioRad). Blocking was done with 0.1% I-Block (Tropix). A gp120-specific rabbit serum produced in a previous report was used as the detecting antibody at 1:500 dilution for this assay.Citation8,Citation9 The membranes were washed with blocking buffer and then reacted with AP-conjugated goat anti-rabbit (Tropix) at 1:5000 dilution. After final wash, Western-light substrate was applied to the membranes for 5 min. Once the membranes were dry, Kodak films were exposed to the membrane and developed with an X-Omat processor.
Rabbit Immunizations
New Zealand White (NZW) rabbits at 6–8 weeks of age were purchased from Shanghai Animal Center, Chinese Academy of Science (Shanghai, China) and housed in the animal facility managed by the Nanjing Medical University (Nanjing, China) in accordance of the protocol approved. As described in the results section, four DNA vaccine immunizations were given to a subset of rabbits at Weeks 0, 2, 4, and 8 by electroporation (EP) using a Caliper Electrodes style electroporator (SCIENTZ-2C) from Scientz Co., Ltd (Ningbo, China). Following intramuscular injection of DNA vaccine at a dose of 100 μg/site at two sites, the injection site areas were electroporated once with the following parameters: 100 V, 60 ms, and 60 Hz. Protein immunizations consisting of 100 µg recombinant gp120-BC protein in 500 µL PBS, mixed with 500 µL Incomplete Freund’s Adjuvant (IFA) were administered. The 1 mL adjuvanted protein solution was then injected subcutaneously into the back of rabbits. Sera were collected for antibody studies at two weeks prior to the first immunization and two weeks after each animal immunization.
Enzyme Linked Immunosorbent Assay (ELISA)
Recombinant gp120-BC protein was coated onto 96 well microtiter plates (Costar #3369) at 1 µg/mL in 100 µL of PBS for 1 h at room temperature. Plates were then washed 5 times in PBS containing 0.1% Triton-X (EWB) and blocked overnight at 4° in PBS containing 4% whey by weight (whey dilution buffer) and 5% powdered milk. The following morning, plates were washed 5 times in EWB and serially diluted rabbit sera, collected at 2 weeks following the final protein immunization, were added to the wells in a volume of 100 µL. Plates were washed 5 times in EWB and 100 µL of biotinylanted anti-rabbit secondary antibody (Vector Labs BA-1000) at 1.5 µg/mL was incubated on the plate for 1 h at room temperature. Plates were washed 5 times with EWB and incubated with 100 µL of a streptavidin horseradish peroxidase construct (Vector Labs SA-5004) at 500 ng/mL. Plates were washed 5 times with EWB and developed for 3 min in 100 µL of a 3,3′5,5′-tetramethylbenzidine substrate solution (Sigma T3405). The reaction was stopped with addition of 25 µL of 2N H2SO4. Endpoint titers as reported are defined as the last dilution of a serially diluted serum sample with greater than double the background optical density of a preimmune serum sample.
HIV-1 Neutralization Assay
Neutralization assays were done as previously described.Citation26 The HIV-1 subtype BC gp160 expression plasmids (CH064, CH091, CH110, CH114, CH115, CH119, CH120, CH181) were provided by Professor Yiming Shao’s lab at the Chinese Center for Diseases Controls and Prevention. In addition, a new chimeric gp160 BC (BC-Z) was constructed by swapping the gp120 region in SF162 gp160 with the consensus gp120-BC gene produced in this study, as previously described.Citation27 HIV-1 pseudovirions were produced through co-transfection of the pSG3Δenv backbone (NIH AIDS Research Reference and Reagent Program) and a gp160 Env-bearing plasmid in 293T cells. HIV-1 pseudoviruses were collected from the transfected 293T cell supernatants and then titered out the median tissue culture infection dose (TCID50) on the TZM-bl cell line before use. For a typical neutralization assay, 200 TCID50 of pseudovirus was incubated with rabbit sera for 1 h at 37°C. The virus/sera mix was then added to 105 TZM-bl cells in a final concentration of 20 µg/mL DEAE Dextran. Plates were incubated at 37°C for 48 h and developed with luciferase assay reagent according to the manufacturer’s instruction (Promega). Neutralization was calculated as the percent change in luciferase activity in the presence of preimmune sera vs. that of luciferase activity in the presence of immune sera [(Preimmune RLUs – Immune RLUs)/(Preimmune RLUs)]*100.
In peptide adsorption experiments the same neutralization protocol was applied as described above except, prior to the exposure of sera to the pseudovirus, the sera were incubated with 25 µg/mL of a consensus HIV-1 subtype BC or B V3 peptide as described above for 30 min at 37°C. In these assays, percent neutralization was calculated with the light signal of HIV-1 pseudovirus SF162 or BC-Z in the presence of sCD4 serving as our baseline light signal.
Competitive binding assays
Competitive binding assays were performed as previously described 2820 with minor modifications. Pseudovirions bearing the JR-FL Envelope and Vesicular Stomatitis Virus (VSV) glycoprotein were produced with the pSG3ΔEnv backbone in 293T cells. Microtiter plates were coated with 50 µL of monoclonal antibody (mAb) at 5 µg/mL for 1 h at room temperature. Plates were then blocked in PBS with 3% BSA overnight at 4°C. Rabbit sera heat inactivated at 56°C for 30 min, serially diluted, and incubated with pseudovirus correlating to 2.5 ng of p24/well for 1 h prior to the addition to the virus/sera mixture to the ELISA wells. Virus/sera mixture was then incubated on the ELISA wells for 3 h at room temperature. Plates were washed 5 times with sterile PBS and overlayed with 10,000 TZM-bl cells per well. Plates were incubated for 48 h at 37°C. Luciferase activity was determined per the manufacturer’s instruction (Promega). Data are reported as the serum dilution at which the luciferase signal is reduced by 50% compared with a serum negative control.
Statistical analysis
All statistical analyses were performed in the Graphpad Prism software program. Reported p values were derived from data subjected to a two-tailed Mann-Whitney test.
| Abbreviations: | ||
| BSA | = | bovine serum albumin |
| EWB | = | PBS containing 0.1% Triton-X |
| ELISA | = | enzyme-linked immunosorbent assay |
| Env | = | envelope glycoprotein |
| HIV-1 | = | human immunodeficiency virus type 1 |
| NAb | = | neutralizing antibody |
| PBS | = | phosphate buffered saline |
| VSV | = | vesicular stomatitis virus |
Acknowledgments
This study was conducted as part of Jiangsu Province Infectious Diseases Key Laboratory program (#2007–158) and was funded in part by China’s 11–5 (2008ZX10001–010 and 2008ZX10001–006) and 12–5 (2012ZX10001–008) National Major Infectious Diseases Programs. This study was also funded in part by NIH grant 5 P01 AI082274–03. The authors thank Dr. Jill Serrano for her critical reading and editing of the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Gao F, Liao HX, Hahn BH, Letvin NL, Korber BT, Haynes BF. Centralized HIV-1 envelope immunogens and neutralizing antibodies. Curr HIV Res 2007; 5:572 - 7; http://dx.doi.org/10.2174/157016207782418498; PMID: 18045113
- Gao F, Weaver EA, Lu Z, Li Y, Liao HX, Ma B, et al. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J Virol 2005; 79:1154 - 63; http://dx.doi.org/10.1128/JVI.79.2.1154-1163.2005; PMID: 15613343
- Kothe DL, Decker JM, Li Y, Weng Z, Bibollet-Ruche F, Zammit KP, et al. Antigenicity and immunogenicity of HIV-1 consensus subtype B envelope glycoproteins. Virology 2007; 360:218 - 34; http://dx.doi.org/10.1016/j.virol.2006.10.017; PMID: 17097711
- Kothe DL, Li Y, Decker JM, Bibollet-Ruche F, Zammit KP, Salazar MG, et al. Ancestral and consensus envelope immunogens for HIV-1 subtype C. Virology 2006; 352:438 - 49; http://dx.doi.org/10.1016/j.virol.2006.05.011; PMID: 16780913
- Weaver EA, Camacho ZT, Gao F. Similar T-cell immune responses induced by group M consensus env immunogens with wild-type or minimum consensus variable regions. AIDS Res Hum Retroviruses 2010; 26:577 - 84; http://dx.doi.org/10.1089/aid.2009.0258; PMID: 20438382
- Weaver EA, Lu Z, Camacho ZT, Moukdar F, Liao HX, Ma BJ, et al. Cross-subtype T-cell immune responses induced by a human immunodeficiency virus type 1 group m consensus env immunogen. J Virol 2006; 80:6745 - 56; http://dx.doi.org/10.1128/JVI.02484-05; PMID: 16809280
- Wang S, Arthos J, Lawrence JM, Van Ryk D, Mboudjeka I, Shen S, et al. Enhanced immunogenicity of gp120 protein when combined with recombinant DNA priming to generate antibodies that neutralize the JR-FL primary isolate of human immunodeficiency virus type 1. J Virol 2005; 79:7933 - 7; http://dx.doi.org/10.1128/JVI.79.12.7933-7937.2005; PMID: 15919951
- Wang S, Pal R, Mascola JR, Chou TH, Mboudjeka I, Shen S, et al. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology 2006; 350:34 - 47; http://dx.doi.org/10.1016/j.virol.2006.02.032; PMID: 16616287
- Vaine M, Wang S, Crooks ET, Jiang P, Montefiori DC, Binley J, et al. Improved induction of antibodies against key neutralizing epitopes by human immunodeficiency virus type 1 gp120 DNA prime-protein boost vaccination compared to gp120 protein-only vaccination. J Virol 2008; 82:7369 - 78; http://dx.doi.org/10.1128/JVI.00562-08; PMID: 18495775
- Wang S, Kennedy JS, West K, Montefiori DC, Coley S, Lawrence J, et al. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 2008; 26:1098 - 110; http://dx.doi.org/10.1016/j.vaccine.2007.12.024; PMID: 18243434
- Vaine M, Wang S, Hackett A, Arthos J, Lu S. Antibody responses elicited through homologous or heterologous prime-boost DNA and protein vaccinations differ in functional activity and avidity. Vaccine 2010; 28:2999 - 3007; http://dx.doi.org/10.1016/j.vaccine.2010.02.006; PMID: 20170767
- Wang Z, Wang SX, Liu SY, Bao ZY, Zhuang DM, Li L, et al. Immunogenicities of Env glycoproteins from circulating HIV-1 isolates in China focusing on the strategy of “DNA prime plus protein boost”. Chin Med J (Engl) 2009; 122:2339 - 45; PMID: 20079137
- Yu XF, Chen J, Shao Y, Beyrer C, Lai S. Two subtypes of HIV-1 among injection-drug users in southern China. Lancet 1998; 351:1250; http://dx.doi.org/10.1016/S0140-6736(05)79316-8; PMID: 9643749
- Yu XF, Chen J, Shao Y, Beyrer C, Liu B, Wang Z, et al. Emerging HIV infections with distinct subtypes of HIV-1 infection among injection drug users from geographically separate locations in Guangxi Province, China. J Acquir Immune Defic Syndr 1999; 22:180 - 8; PMID: 10843533
- Graf M, Shao Y, Zhao Q, Seidl T, Köstler J, Wolf H, et al. Cloning and characterization of a virtually full-length HIV type 1 genome from a subtype B’-Thai strain representing the most prevalent B-clade isolate in China. AIDS Res Hum Retroviruses 1998; 14:285 - 8; http://dx.doi.org/10.1089/aid.1998.14.285; PMID: 9491920
- Li Z, He X, Wang Z, Xing H, Li F, Yang Y, et al. Tracing the origin and history of HIV-1 subtype B’ epidemic by near full-length genome analyses. AIDS 2012; 26:877 - 84; http://dx.doi.org/10.1097/QAD.0b013e328351430d; PMID: 22269972
- Xin R, He X, Xing H, Sun F, Ni M, Zhang Y, et al. Genetic and temporal dynamics of human immunodeficiency virus type 1 CRF07_BC in Xinjiang, China. J Gen Virol 2009; 90:1757 - 61; http://dx.doi.org/10.1099/vir.0.009290-0; PMID: 19321756
- Chong H, Hong K, Zhang C, Nie J, Song A, Kong W, et al. Genetic and neutralization properties of HIV-1 env clones from subtype B/BC/AE infections in China. J Acquir Immune Defic Syndr 2008; 47:535 - 43; http://dx.doi.org/10.1097/QAI.0b013e3181663967; PMID: 18209676
- Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J Virol 2006; 80:2515 - 28; http://dx.doi.org/10.1128/JVI.80.5.2515-2528.2006; PMID: 16474158
- Crooks ET, Moore PL, Franti M, Cayanan CS, Zhu P, Jiang P, et al. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology 2007; 366:245 - 62; http://dx.doi.org/10.1016/j.virol.2007.04.033; PMID: 17580087
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 2010; 329:856 - 61; http://dx.doi.org/10.1126/science.1187659; PMID: 20616233
- Douagi I, Forsell MN, Sundling C, O’Dell S, Feng Y, Dosenovic P, et al. Influence of novel CD4 binding-defective HIV-1 envelope glycoprotein immunogens on neutralizing antibody and T-cell responses in nonhuman primates. J Virol 2010; 84:1683 - 95; http://dx.doi.org/10.1128/JVI.01896-09; PMID: 19955308
- Sundling C, O’Dell S, Douagi I, Forsell MN, Mörner A, Loré K, et al. Immunization with wild-type or CD4-binding-defective HIV-1 Env trimers reduces viremia equivalently following heterologous challenge with simian-human immunodeficiency virus. J Virol 2010; 84:9086 - 95; http://dx.doi.org/10.1128/JVI.01015-10; PMID: 20610729
- Wang S, Farfan-Arribas DJ, Shen S, Chou TH, Hirsch A, He F, et al. Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine 2006; 24:4531 - 40; http://dx.doi.org/10.1016/j.vaccine.2005.08.023; PMID: 16140431
- Wang S, Taaffe J, Parker C, Solórzano A, Cao H, García-Sastre A, et al. Hemagglutinin (HA) proteins from H1 and H3 serotypes of influenza A viruses require different antigen designs for the induction of optimal protective antibody responses as studied by codon-optimized HA DNA vaccines. J Virol 2006; 80:11628 - 37; http://dx.doi.org/10.1128/JVI.01065-06; PMID: 16987975
- Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV and SHIV in luciferase reporter gene assays. Curr Protoc Immunol 2004; In press PMID: 18432938
- Wang Z, Zhang M, Wang Y, Jiao Y, Zhang L, Li L, et al. A versatile vector for the production of pseudotyped viruses expressing gp120 antigens from different clades of primary HIV-1 isolates. J Virol Methods 2011; 171:183 - 9; http://dx.doi.org/10.1016/j.jviromet.2010.10.022; PMID: 21034776
- Derby NR, Kraft Z, Kan E, Crooks ET, Barnett SW, Srivastava IK, et al. Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J Virol 2006; 80:8745 - 62; http://dx.doi.org/10.1128/JVI.00956-06; PMID: 16912322