Abstract
HIV preferentially infects activated T cells, and activated mucosal CD4+ T cells are the primary sites of viral replication. One potential explanation for increased HIV acquisition rates in the STEP study is that vaccination with adenoviral (Ad) vectors increased CD4+ T cell activation levels at the site of infection, a concept that others and we continue to explore.1,2 Whether vaccination with HIV vaccine platforms increases the activation state of CD4+ T cells within peripheral tissues, such as the gastro-intestinal (GI) mucosa, is exceptionally important to determine as a vaccine safety measure, given the susceptibility of activated CD4+ T cells to HIV infection.
In this study we examined whether vaccination with DNA plasmids and chemokine adjuvants alter the activation state of T cells within the GI mucosa, inguinal LN, and peripheral blood. T cell activation state was measured by expression of CD25, CD69, and HLA-DR over the course of the prime/boost study. DNA plasmid vaccination did not increase expression of any of these markers in the 3 tissues studied. Addition of the gut-homing chemokine TECK during DNA plasmid vaccination did not alter activation levels of CD4+ T cells at any of these sites. These findings indicate that DNA vaccines do not elicit generalized mucosal T cell activation. Thus, DNA platforms may be especially suitable for HIV vaccine development, where bystander activation could promote increased HIV transmission.
Keywords: :
Introduction
Since acquired immunodeficiency syndrome (AIDS) was first identified in the early 1980s, the disease has grown into a global pandemic and resulted in the death of over 25 million people. Despite advances in therapeutic interventions, there remains a need for an effective vaccine to either prevent or control human immunodeficiency virus (HIV) infection, the causative agent of AIDS. Several recent HIV vaccine candidates have been designed to induce cell-mediated immunity since maintenance of CD8+ T cell effector function has been linked to partial control of HIV infection and delayed progression to AIDS.Citation3-Citation6
The STEP Study was a promising HIV clinical trial candidate designed to test the CD8+ T cell vaccine concept. Unfortunately, rather than preventing infection, the STEP vaccine increased HIV acquisition rates, particularly among individuals seropositive for the vaccine’s adenoviral (Ad) vector.Citation7,Citation8 HIV preferentially infects and replicates in activated T cellsCitation9 and it is possible that pre-existing immunity in Ad-seropositive individuals increased transmission rates as a result of cellular activation. Generalized immune activation has been linked to HIV disease progression and nonhuman primate SIV models have identified immune activation as a key determinant of progressive disease.Citation10-Citation12 Many cellular activation markers have been identified and are commonly used in monitoring immune responses in humans and nonhuman primates. This study will focus on CD25 (α-chain IL-2 receptor), CD69 (early activation marker), and HLA-DR (MHC class II receptor).
Studies seeking to explain the results of the STEP Study have not identified cellular activation as a contributing factor for increased transmission, but the majority of these studies have only studied peripheral blood.Citation13-Citation15 However, a recent study by the Patterson laboratory demonstrated that Ad can upregulate α4β7 expression on CD4+ T cells through interaction with DCs and they this could induce expansion of susceptible T cells and trafficking to mucosal sites.Citation16 Additionally, a recent preliminary study from our laboratory, also examining CD25, CD69, and HLD-DR expression, found vaccination with Ad-vectors can increase mucosal CD4+ T cell activation in the rectal mucosa, an effect not mirrored in the peripheral blood, suggesting important compartmental differences in T cell response after vaccination (manuscript in preparation).Citation1
DNA-based HIV vaccine platforms represent another, perhaps safer, strategy for eliciting cellular immunity. Plasmid DNA has been shown to induce cellular responses and, to varying degrees, provide protection from infection in both small animal models and nonhuman primates, particularly when chemokine or cytokine adjuvanted.Citation17-Citation19 While these types of adjuvants can boost the cellular response to DNA-based vaccines, they can also redirect immune cells in vivo.Citation20,Citation21 Tissue-homing chemokines interact with chemokine receptors (CCR) and direct immune cells to specific compartments in response to homeostatic or inflammatory signals. The chemokine receptor CCR9 is important for T cell homing to the GI mucosa and is expressed on almost all small intestine-resident T cells.Citation22-Citation24 CCR9 binds to the chemokine TECK, also known as CCL25. In this study, TECK was used as an adjuvant for a SIV-specific DNA plasmid vaccine with the intention of directing SIV-specific T cells to the gut, where HIV infection and pathogenic damage occurs.
One concern with our adjuvant strategy was that the TECK chemokine might induce an influx of activated CD4+ T cells into the gut mucosa, thereby potentially exacerbating viral infection upon challenge. Therefore, the goal of this preliminary study was to characterize the impact of DNA plasmid vaccination, with or without TECK adjuvant, on CD4+ T cell activation in various anatomical compartments in vivo in vaccinated rhesus macaques. Our results suggest that any affect of TECK or DNA vaccination in general on increasing CD4+ T cell activation in the gut mucosa is minimal.
Results
Female Indian rhesus macaques were divided into 4 groups: baseline, naïve, DNA, and DNA + TECK (). The animals followed a prime/boost immunization schedule with SIV-specific plasmids and periodic jejunum (GI mucosa), inguinal lymph node (LN), and blood collection at biopsy and necropsy time points (). Baseline animals received no treatment prior to whole tissue collection at necropsy. All naïve, DNA, and DNA + TECK animals received immunizations with water or plasmids and in vivo electroporation at day 0. Biopsy samples were collected from 2 animals from each group at day 10. At day 14, the 2 biopsied animals from each group were euthanized and whole tissue collection occurred at necropsy. Biopsy samples were collected from the remaining 2 animals from each group at day 21. Animals received an immunization boost and in vivo electroporation at day 42 and whole tissues were collected at the day 49 necropsy. Sample tissues were processed and immunostained for flow cytometry analysis. Non-naïve CD3+CD4+ T cells were examined for expression of the activation markers CD25, CD69, and HLA-DR ().
Table 1. Experimental groups and DNA vaccines adminitstered
Figure 1. Immunization schedule indicating prime (day 0) and boost (day 42) time points. GI mucosa, inguinal LN, and blood samples were collected at both biopsy (circle) and necropsy (square) time points.
Figure 2. (A) Activation marker gating strategy. Top row, left to right: singlets, viable cells (exclude LIVE/DEAD Fixable Aqua), CD3+ cells, Dump- cells (CD14 and CD20), and lymphocytes. Bottom row, left to right: CD4+CD8- cells, non-naïve cells (excluding CD28+CD95- population), CD25 vs. HLA-DR expression, and CD69 vs. HLA-DR expression. (B) Total activation marker expression in GI mucosa (left), inguinal LN (center), and blood (right). Y-axis indicates percentage of CD3+CD4+ non-naïve T cells expressing CD25, CD69, and/or HLA-DR. Open shapes indicate biopsies and closed shapes indicate necropsies. Solid gray lines separate experimental groups. Note missing data points, due to poor cell recovery, for GI mucosa, days 14 and 21, all experimental groups, and for inguinal LN, day 10, DNA group. *p < 0.01, decreased compared with baseline.
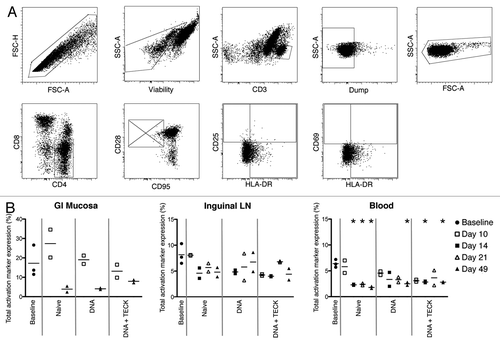
First, we quantified total activation marker expression on non-naïve CD3+CD4+ T cells in these tissues (). Cells were analyzed for expression of CD25, CD69, HLA-DR, or any combination of these 3 markers. Since cells can simultaneously express multiple markers, this type of analysis gave a more complete picture of the potential activation level in each tissue.
Immunization, with either DNA plasmids or water, did not increase activation levels in the GI mucosa (, left graph). Over the course of the study, activation levels in experimental groups were either equivalent to baseline or slightly decreased. The animals immunized with water alone (naïve group) showed a slight increase in activation at day 10, but this did not reach statistical significance due to small sample size. Both DNA and DNA + TECK groups showed lower levels of activation compared with the naïve group. Unfortunately, as a result of limited cell recovery, we did not have data for the GI mucosa for day 14 or day 21.
While inguinal LN and blood showed lower levels of overall activation, when compared with the GI mucosa, the expression patterns were the same (, center and right graphs). Activation levels were either maintained or decreased for all experimental groups for both tissues when compared with baseline. This level of expression suggests vaccination with SIV-specific DNA plasmids did not increase CD4+ T cell activation. Further, this suggests that the TECK mucosal homing chemokine adjuvant plasmid also did not increase CD4+ T cell activation in these sites. Basal activation levels in the GI mucosa were higher than in the LN or blood;, likely due to repetitive exposure to pathogens and pathogen-associated molecular patterns. While nearly all time points in the blood demonstrate statistically significant lower activation levels than baseline expression, the small sample size in this study does not provide conclusive evidence for vaccine-induced reduction of activation.
Next we analyzed individual activation marker expression on non-naïve CD3+CD4+ T cells in GI mucosa, inguinal LN, and blood (, left, center, and right columns, respectively). This type of analysis will identify the largest contributors to overall activation in these tissues as well as detect any modulation of individual marker expression that could be masked by looking at activation as a whole.
Figure 3. Individual activation marker expression in GI mucosa (left), inguinal LN (center), and blood (right). Y-axis indicates percentage of CD3+CD4+ non-naïve T cells expressing CD25 (top row), CD69 (middle row) or HLA-DR (bottom row). Open shapes indicate biopsies and closed shapes indicate necropsies. Solid gray lines separate experimental groups. Note missing data points, due to poor cell recovery, for GI mucosa, days 14 and 21, all experimental groups, and for inguinal LN, day 10, DNA group. * p < 0.01, decreased compared with baseline. ** p < 0.02, increased compared with baseline.
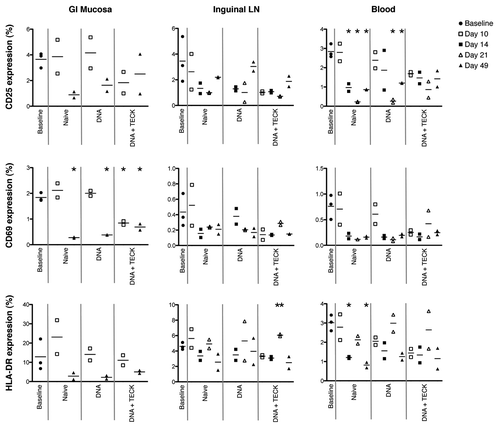
Similar to what was observed for total activation marker expression in , individual expression of CD25, CD69, and HLA-DR (, top, middle, and bottom row, respectively) was not dramatically elevated in any tissue in response to immunization with DNA plasmids, with or without TECK adjuvant. For all groups, and all tissues, individual activation marker expression was at or below that observed in the baseline group and generally declined over time. The only exception to this was a slight increase in CD69 and HLA-DR expression in the GI mucosa and inguinal LN in the naïve group compared with the baseline group at day 10. These increases did not reach statistical significance. Of note, HLA-DR expression in the inguinal LN in the DNA + TECK group was increased compared with the baseline group at day 21, p < 0.02. Low overall expression and small sample size suggest this time point represents a statistical outlier that may not persist in a repeat study with larger animal groups.
Of the 3 activation markers examined in this study, HLA-DR seems to be the largest contributor to overall activation, especially in the GI mucosa. CD25 is also a large contributor to tissue activation, while CD69 was only expressed at low levels. The pattern of expression across all 3 markers is similar across all experimental groups, suggesting neither SIV-specific DNA plasmid nor TECK plasmid immunization increase cellular activation.
Discussion
This study demonstrates that DNA plasmids and mucosal chemokine adjuvants can be administered without eliciting potentially harmful CD4+ T cell activation in the GI mucosa, inguinal LN, or peripheral blood. Both total and individual activation marker expression remained at or below baseline levels over the course of the study. This data are in contrast to recent data from our laboratory that shows vaccination with Ad vectors increases CD4+ T cell activation, as measured by expression of CD25, CD69, HLA-DR, and other markers, in the GI mucosa of rhesus macaquesCitation1 and in the work of Qureshi et al.Citation2 The GI mucosa is a particularly vulnerable compartment during HIV infection and altered basal activation levels in this cell population could increase viral acquisition and replication.
Together, these results suggest DNA plasmids may represent a safer platform for HIV vaccine development compared with viral based vector systems. The preliminary conclusions from this study are promising and highlight a potential advantage of HIV-specific DNA-based vaccines. However, this small pilot study does not contain enough animals to consistently reach statistical significance or to make definitive conclusions about vaccine safety. Due to the inclusion of necropsies in the study design, we do not have data that spans baseline through day 49 for any of the experimental groups. Since basal CD4+ T cell activation varies between animals, similarly designed future studies need to include more animals to confirm these results.
Additionally, our study suggests DNA plasmids adjuvanted with the mucosal chemokine TECK are unlikely to increase CD4+ T cell activation levels. This data suggests TECK may be a safe adjuvant for vaccine candidates. Current studies in our laboratory, and in collaboration with others, are exploring the functional and protective qualities of HIV-specific DNA vaccines with mucosal chemokine vaccine adjuvants.
While we have not found that DNA vaccine strategies increase mucosal CD4+ T cell activation, this remains a significant concern for other HIV vaccine vector systems. Since vaccine-induced T cell responses may vary greatly between tissues, it is important expand vaccine candidate studies into all compartments of relevance. This will give a better idea of the overall vaccination response and may help to identify undesirable side effects. Eliciting strong cellular responses may be important for HIV control, but the benefits may not outweigh the risk if vaccination also increases susceptibility to infection by highly activating target cells.
Materials and Methods
Animals
Fifteen female Indian rhesus macaques (Macaca mulatta) were used for this study. Animals were housed at the AAALAC-accredited Tulane National Primate Research Center and the Institution’s Animal Care and Use Committee approved all animal protocols.
DNA plasmids, immunizations, and tissue collection
The macaques were divided into 4 groups: Baseline, Naïve, DNA, and DNA + TECK, as depicted in . Animals received (or not) SIV-specific antigenic plasmid constructs, with or without adjuvant, at days 0 (prime) and 42 (boost). Antigenic constructs were specific for SIV smGAG1 (pGX1013), SIV smGAG2 (pGX1014), SIV smPOL (pGX1008), and SIVsmENV (pGX1009, SIVmac239). The plasmid adjuvant TECK (pGX6007), also known as CCL25, was utilized for the DNA + TECK treatment group at both prime and boost. All plasmid constructs were formulated in sterile water for injection with 1% wt/wt poly-L-glutamate sodium salt (Inovio Biomedical Corporation). One mg of each plasmid was delivered by injection followed by in vivo electroporation with a constant current CELLECTRA® (Inovio Biomedical Corporation) into the quadriceps muscle at 2 separate sites. Site 1 received GAG1, POL, ENV, +/− TECK and site 2 received GAG2, POL, ENV, +/− TECK.
The immunization and tissue collection schedule is depicted in . All animals from the naïve, DNA, and DNA + TECK groups received either plasmids or water at the day 0 immunization. Two animals from each group were anesthetized at day 10 for biopsy collection and euthanized at day 14 for whole tissue collection. The remaining 2 animals from each group were anesthetized at day 21 for biopsy collection, received an immunization boost at day 42, and were euthanized at day 49 for whole tissue collection. Three baseline animals received no treatment prior to euthanization and whole tissue collection.
Whole blood, inguinal lymph node (LN), and jejunum (GI mucosa) samples were collected at both biopsy and necropsy timepoints. All samples were shipped overnight from the Tulane National Primate Research Center to the University of Pennsylvania for processing.
Tissue processing and cell isolation
Peripheral blood mononuclear cells (PBMC) were isolated from whole blood by Ficoll-hypaque centrifugation and red blood cells were removed with ammonium chloride-potassium (ACK) lysis buffer (Lonza BioWhittaker). LN were macerated, filtered through a 100 μm cell strainer, and treated with ACK lysis buffer. Jejunum intraepithelial lymphocytes and lamina propria lymphocytes were isolated by a combination of mechanical disruption (16–20 gauge blunt needles, Fisher) and collagenase treatment (0.5 mg/ml collagenase type II, Sigma) followed by Percoll (Amersham Biosciences) step-gradient purification.
Isolated cells were suspended in complete medium (RPMI-1640, 2 mM/L L-glutamine, 10% heat-inactivated fetal bovine serum (BSA), 100 IU/ml penicillin, 100 μg/ml streptomycin, and 55 μM/ml β-mercaptoethanol) and allowed to rest in a 37°C, 5% CO2 incubator for 5 h prior to immunostaining for flow cytometry.
Immunostaining and flow cytometry
Following rest, cells were washed with phosphate-buffered saline (PBS) and incubated with LIVE/DEAD Fixable Aqua Dead Cell Stain (Invitrogen) for 10 min. Surface stain antibodies were pooled in FACs buffer (PBS, 1% BSA, and 0.1% sodium azide), added to cells, and incubated for 30 min. Surface stain monoclonal antibodies include: (i) anti-CD14 QD655 [Tuk4], anti-CD20 QD655 [HI47], anti-CD8 QD605 [3B5], and anti-HLA-DR PE-TxRd [TU36] (Invitrogen); (ii) anti-CD4 PerCPCy5.5 [L200], anti-CD95 PE-Cy5 [DX2], and anti-CD25 APC-Cy7 [M-A251] (BD Biosciences); (iii) anti-CD28 PE-Cy7 [CD28.2] (eBioscience); (iv) anti-CD38 PE [OKT10] (NIH Nonhuman Primate Reagent Resource, R24 RR016001, N01 AI040101). Cells were washed with FACs buffer and treated with Cytofix/Cytoperm (BD Biosciences) for 17 min. Cells were washed with Perm/Wash buffer (BD Biosciences). Intracellular stain antibodies were pooled in Perm/Wash buffer, added to cells, and incubated for 1 h. Intracellular stain monoclonal antibodies include: anti-CD3 PacBlue [SP34–2] and anti-CD69 APC [FN50] (BD Biosciences). Cells were washed with Perm/Wash buffer and fixed in 1% paraformaldehyde. All incubations were kept in the dark, at room temperature.
Data collection and analysis
0.5–1x106 events were collected for each sample on a modified LSR-II flow cytometer (BD Immunocytometry Systems) configured for detection of 18 fluorescent parameters. Data was analyzed using FlowJo software (Treestar, Inc., Ashland, OR) and graphed using GraphPad Prism (v. 5.0a for Mac OS X, GraphPad Software, San Diego, CA). Figures were prepared using Adobe® Photoshop® CS5 Extended software (v. 12.0 x64). Two-tailed t-test statistical analyses were performed using GraphPad Prism and compared all time points to baseline. Limited sample size (two or three monkeys per group) resulted in a failure to reach statistical significance, except where noted. Small samples size combined with a necropsy-style study design prevented further statistical analyses.
| Abbreviations: | ||
| AIDS | = | acquired immunodeficiency virus |
| HIV | = | human immunodeficiency virus |
| Ad | = | adenovirus |
| GI | = | gastrointestinal |
| LN | = | lymph node |
| PBMC | = | peripheral blood mononuclear cell |
| CCR | = | chemokine receptor |
Acknowledgments
The authors thank Arielle A. Ginsberg, Albert J. Sylvester, Devin Myles, and Noshin Kathuria for aid in tissue processing and T cell isolation. We also thank Meredith Hunter at the Tulane National Primate Research Center and Amir S. Khan from Inovio for animal handling and immunizations, respectively.
Disclosure of Potential Conflicts of Interest
D.B.W. has commercial relationships, including grant funding, collaborations, stock ownership, and/or consulting, with Pfizer, Inovio, Bristol Myers Squibb, VGXI, VIRxSYS, Ichor, Merck, Althea, Aldevron, Novartis, and possibly others. The remaining authors have no conflicts of interest.
Financial Support
This research was supported by NIH P01 AI071739. This project was supported by the National Center for Research Resources and the Office of Research Infrastructure Programs (ORIP) of the National Institutes of Health through grant number P51 RR00164–50. M.A. Reuter was supported by NIH T32 AI007632.
References
- Bukh I, Calcedo R, Roy S, Carnathan DG, Grant R, Ratcliffe SJ, et al. Increased mucosal CD4+ T cell activation following vaccination with an adenovirus vector in rhesus macaques. 29th Annual Nonhuman Primate Models for AIDS Symposium. Seattle, WA, 2011:Conference Presentation.
- Qureshi H, Ma ZM, Huang Y, Hodge G, Thomas MA, DiPasquale J, et al. Low-dose penile SIVmac251 exposure of rhesus macaques infected with adenovirus type 5 (Ad5) and then immunized with a replication-defective Ad5-based SIV gag/pol/nef vaccine recapitulates the results of the phase IIb step trial of a similar HIV-1 vaccine. J Virol 2012; 86:2239 - 50; http://dx.doi.org/10.1128/JVI.06175-11; PMID: 22156519
- Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol 2002; 3:1061 - 8; http://dx.doi.org/10.1038/ni845; PMID: 12368910
- Hess C, Altfeld M, Thomas SY, Addo MM, Rosenberg ES, Allen TM, et al. HIV-1 specific CD8+ T cells with an effector phenotype and control of viral replication. Lancet 2004; 363:863 - 6; http://dx.doi.org/10.1016/S0140-6736(04)15735-8; PMID: 15031033
- Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 2006; 107:4781 - 9; http://dx.doi.org/10.1182/blood-2005-12-4818; PMID: 16467198
- Hersperger AR, Migueles SA, Betts MR, Connors M. Qualitative features of the HIV-specific CD8+ T-cell response associated with immunologic control. Curr Opin HIV AIDS 2011; 6:169 - 73; http://dx.doi.org/10.1097/COH.0b013e3283454c39; PMID: 21399496
- Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, et al, Step Study Protocol Team. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 2008; 372:1881 - 93; http://dx.doi.org/10.1016/S0140-6736(08)61591-3; PMID: 19012954
- McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, et al, Step Study Protocol Team. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet 2008; 372:1894 - 905; http://dx.doi.org/10.1016/S0140-6736(08)61592-5; PMID: 19012957
- Zhang Z, Schuler T, Zupancic M, Wietgrefe S, Staskus KA, Reimann KA, et al. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 1999; 286:1353 - 7; http://dx.doi.org/10.1126/science.286.5443.1353; PMID: 10558989
- Deeks SG, Kitchen CM, Liu L, Guo H, Gascon R, Narváez AB, et al. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood 2004; 104:942 - 7; http://dx.doi.org/10.1182/blood-2003-09-3333; PMID: 15117761
- Hazenberg MD, Otto SA, van Benthem BH, Roos MT, Coutinho RA, Lange JM, et al. Persistent immune activation in HIV-1 infection is associated with progression to AIDS. AIDS 2003; 17:1881 - 8; http://dx.doi.org/10.1097/00002030-200309050-00006; PMID: 12960820
- Sodora DL, Silvestri G. Immune activation and AIDS pathogenesis. AIDS 2008; 22:439 - 46; http://dx.doi.org/10.1097/QAD.0b013e3282f2dbe7; PMID: 18301056
- Hutnick NA, Carnathan DG, Dubey SA, Cox KS, Kierstead L, Makadonas G, et al. Vaccination with Ad5 vectors expands Ad5-specific CD8 T cells without altering memory phenotype or functionality. PLoS ONE 2010; 5:e14385; http://dx.doi.org/10.1371/journal.pone.0014385; PMID: 21203546
- Hutnick NA, Carnathan DG, Dubey SA, Makedonas G, Cox KS, Kierstead L, et al. Baseline Ad5 serostatus does not predict Ad5 HIV vaccine-induced expansion of adenovirus-specific CD4+ T cells. Nat Med 2009; 15:876 - 8; http://dx.doi.org/10.1038/nm.1989; PMID: 19620962
- O’Brien KL, Liu J, King SL, Sun YH, Schmitz JE, Lifton MA, et al. Adenovirus-specific immunity after immunization with an Ad5 HIV-1 vaccine candidate in humans. Nat Med 2009; 15:873 - 5; http://dx.doi.org/10.1038/nm.1991; PMID: 19620961
- Benlahrech A, Harris J, Meiser A, Papagatsias T, Hornig J, Hayes P, et al. Adenovirus vector vaccination induces expansion of memory CD4 T cells with a mucosal homing phenotype that are readily susceptible to HIV-1. Proc Natl Acad Sci USA 2009; 106:19940 - 5; PMID: 19918060
- Boyer JD, Robinson TM, Kutzler MA, Vansant G, Hokey DA, Kumar S, et al. Protection against simian/human immunodeficiency virus (SHIV) 89.6P in macaques after coimmunization with SHIV antigen and IL-15 plasmid. Proc Natl Acad Sci USA 2007; 104:18648 - 53; http://dx.doi.org/10.1073/pnas.0709198104; PMID: 18000037
- Chattergoon MA, Saulino V, Shames JP, Stein J, Montaner LJ, Weiner DB. Co-immunization with plasmid IL-12 generates a strong T-cell memory response in mice. Vaccine 2004; 22:1744 - 50; http://dx.doi.org/10.1016/j.vaccine.2004.01.036; PMID: 15068858
- Kutzler MA, Robinson TM, Chattergoon MA, Choo DK, Choo AY, Choe PY, et al. Coimmunization with an optimized IL-15 plasmid results in enhanced function and longevity of CD8 T cells that are partially independent of CD4 T cell help. J Immunol 2005; 175:112 - 23; PMID: 15972637
- Kutzler MA, Weiner DB. Developing DNA vaccines that call to dendritic cells. J Clin Invest 2004; 114:1241 - 4; PMID: 15520855
- McKay PF, Barouch DH, Santra S, Sumida SM, Jackson SS, Gorgone DA, et al. Recruitment of different subsets of antigen-presenting cells selectively modulates DNA vaccine-elicited CD4+ and CD8+ T lymphocyte responses. Eur J Immunol 2004; 34:1011 - 20; http://dx.doi.org/10.1002/eji.200324840; PMID: 15048711
- Marsal J, Svensson M, Ericsson A, Iranpour AH, Carramolino L, Márquez G, et al. Involvement of CCL25 (TECK) in the generation of the murine small-intestinal CD8alpha alpha+CD3+ intraepithelial lymphocyte compartment. Eur J Immunol 2002; 32:3488 - 97; http://dx.doi.org/10.1002/1521-4141(200212)32:12<3488::AID-IMMU3488>3.0.CO;2-E; PMID: 12442331
- Stenstad H, Ericsson A, Johansson-Lindbom B, Svensson M, Marsal J, Mack M, et al. Gut-associated lymphoid tissue-primed CD4+ T cells display CCR9-dependent and -independent homing to the small intestine. Blood 2006; 107:3447 - 54; http://dx.doi.org/10.1182/blood-2005-07-2860; PMID: 16391017
- Svensson M, Johansson-Lindbom B, Wurbel MA, Malissen B, Márquez G, Agace W. Selective generation of gut-tropic T cells in gut-associated lymphoid tissues: requirement for GALT dendritic cells and adjuvant. Ann N Y Acad Sci 2004; 1029:405 - 7; http://dx.doi.org/10.1196/annals.1309.025; PMID: 15681793