Abstract
Recent studies have reported that founder viruses play unique roles in establishing HIV-1 infection. Understanding the biological and immunological features of envelope glycoproteins (Env) from such viruses may facilitate the development of effective vaccines against HIV-1. In this report, we evaluated the immunogenicity of gp120 immunogens from two pairs of clade B and two pairs of clade C mother-to-child transmitted (MTCT) HIV-1 variants that had various levels of sensitivity to broadly neutralizing monoclonal antibodies. Individual gp120 DNA and protein vaccines were produced from each of the eight MTCT Env antigens included in the current study. Rabbits were immunized with these gp120 immunogens by the DNA prime-protein boost approach. High level Env-specific antibody responses were elicited by all MTCT gp120 immunogens. However, their abilities to elicit neutralizing antibody (NAb) responses differed and those from relatively neutralization-resistant variants tended to be more effective in eliciting broader NAb. Results of this pilot study indicated that not all MTCT Env proteins have the same potential to elicit NAb. Understanding the mechanism(s) behind such variation may provide useful information in formulating the next generation of HIV vaccines.
Introduction
HIV-1 mother-to-child HIV-1 transmission (MTCT) is the primary mode of pediatric infection. Women in their childbearing years represent over 50% of HIV-1 infected individuals around the world.Citation1,Citation2 MTCT accounts for up to 14% of all HIV-1 transmission, with 370,000 infants infected in 2009 alone.Citation1,Citation3 Seventy-five percent of HIV-1 infected children die by the age of three without access to antiretroviral treatment and in developing countries, HIV-1 infection is responsible for one in three children who die before age of five.Citation1,Citation4,Citation5 Therefore, the development of effective preventive measures to control MTCT infection will have a significant impact on the control of the HIV-1 epidemics.
HIV-1 isolated from MTCT patients have unique virological features. Studies in multiple cohorts have demonstrated limited diversity of founder viruses from mucosally-infected individuals, including infants infected through MTCT.Citation6-Citation11 This restricted diversity suggests the dominance of a single donor variant in the majority of recipients.Citation4,Citation12-Citation14 The genetic and biologic determinants of the transmission bottleneck are largely unknown as are the roles of NAb in this process. Such findings support the idea of focusing on founder viruses as the main targets for HIV-1 vaccine development.
The envelope glycoprotein (Env) is the ligand for the HIV-1 receptor and co-receptors, is responsible for virus entry,Citation15 and is the target for NAb. Env is also the most variable HIV-1 protein. Understanding the unique immunogenic features of founder virus Env will have broad implications including the development of vaccines based on such naturally selected Env antigens.
In the current report, a pilot study was conducted to test the immunogenicity of MTCT Env antigens. Based on our recently reported work in characterizing two MTCT cohortsCitation16 (Kishko et al., manuscript in preparation), pairs of MTCT antigens were tested in rabbits for their immunogenicity.
Previously, we demonstrated that priming immunizations with DNA vaccines expressing Env antigens can significantly enhance the quality, especially the neutralizing activities, of antibody responses elicited by subunit Env protein vaccinesCitation17-Citation21 when random HIV-1 Env antigens were selected. We used the same immunization approach to study the immunogenicity of primary MTCT Env antigens in the current report. Our results revealed that not all MTCT Env antigens have the same degree of immunogenicity to elicit broad NAb. While the sample size of Env included in the current study is small, information learned from this pilot study is valuable in designing future studies to further identify those MTCT Env antigens that can be more effective in eliciting broad NAb.
Results
MTCT Env immunogens. MTCT Env proteins used in the current study have been extensively analyzed as recently reported.Citation16 (Kishko et al., manuscript in preparation). DNA sequences for the full-length Envs are available from GenBank (see ‘Materials and Methods’ for accession numbers). Based on the phylogeny data, the representative maternal and infant env clones were selected for analysis of their biological properties. Pseudoviruses (PV) expressing these unique env have been generated.Citation22 The CD4 and co-receptor usage of these env clones were determined by titration on cell lines engineered to express different levels of these molecules.Citation16 In previous studies, MTCT Env demonstrated a wide range of neutralizing sensitivities when tested against broadly neutralizing monoclonal antibodies and patient plasma.Citation16 In the current study, we selected MTCT Env proteins that have demonstrated different neutralizing sensitivity profiles for immunogenicity analysis.
Two pairs each were selected from clade B and clade C MTCT isolates (Table 1). The amino acid alignments of the Envs are provided in and . Monoclonal antibody epitopes for 4E10, 2F5 and 2G12 are well established.Citation23-Citation27 For clade B isolates, one pair was relatively sensitive (M1002 T6 Env clone from mother and P1031 H2 clone from infant) while the other pair was more resistant (M1007 Q6 isolated from mother and P1046 C4 isolated from infant) based on their responses to the broadly neutralizing monoclonal antibodies (mAb) b12, 2G12, and 4E10. Their sensitivity to mAb 2F5 was similar between two pairs of MTCT Env antigens. Clones sensitive to mAb were also sensitive to soluble CD4 (sCD4) (). Both pairs of clade B Env have the signature GPGR sequence at the V3 crown position ().
Figure 1. Amino acid alignments of Env protein sequences from clade B mother-infant pairs (M1002-T6/P1031-H2, M1007-Q6/P1046-C4). Epitopes of the mAbs 4E10 (NWFDITXXLW) and 2F5 (ELDKWA), and the HXB2 positions of the PNGS (Nxxx) that play a role in 2G12 binding are shown below the alignments with essential PNGS residues shown in red.
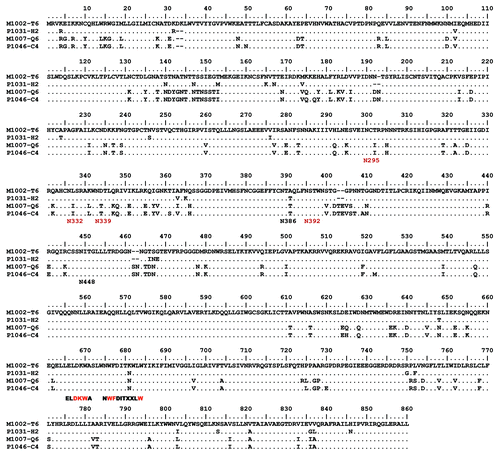
Figure 2. Amino acid alignment of Env protein sequences from clade C mother-infant pairs (CM3-M11/CP3-D2, CM4-N1/CP4-L2). Epitopes of the mAbs 4E10 (NWFDITXXLW) and 2F5 (ELDKWA), and the HXB2 positions of the PNGS (Nxxx) that play a role in 2G12 binding are shown below the alignments with essential PNGS residues shown in red.
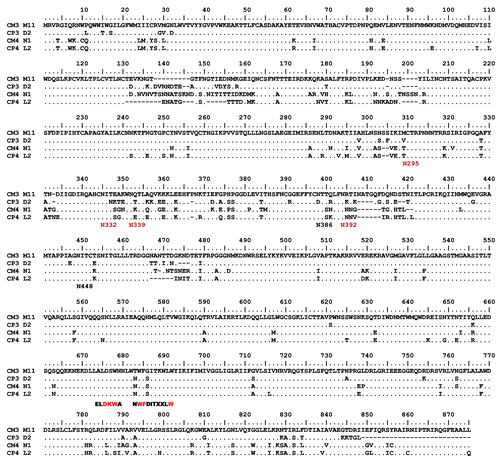
Table 1A. Neutralization and inhibition sensitivity of selected subtype B MTCT env
Similarly, two pairs of clade C Env antigens were selected. However, clade C Env antigens collected from our cohort were overall more resistant to the same panel of broadly neutralizing mAb than the clade B Env antigens () (Kishko et al., manuscript in preparation). However, a unique profile between the two pairs of clade C Env antigens selected for this study was observed. While both pairs were resistant to mAbs 2G12 and 2F5, one pair (CM4-N1 isolated from mother and CP4-L2 isolated from infant) was slightly more sensitive to mAb b12 than the other pair (CM3-M11 isolated from mother and CP3-D2 isolated from infant); both pairs were moderately sensitive to mAb 4E10. When measured against sCD4, the CM4-N1/CP4-L2 pair was slightly more resistant (). As expected, clade C Env antigens have GPGQ as the V3 crown motif (). There was no significant difference between the clade B and clade C pairs on their sensitivity to pooled plasma from HIV-1 positive patients (Table 1).
Table 1B. Neutralization and inhibition sensitivity of selected clade C MTCT env
None of the selected Env antigens exhibited mutations in the critical recognition determinants of 4E10 (). Resistance of subtype C viruses to 2F5 is thought to be due primarily to the substitution K to S in the critical recognition determinant DKW,Citation23,Citation26 which is found in all four selected clade C Env antigens (). Each of the six Env antigens resistant to 2G12 lacks at least one of the four Potential N-Linked Glycosylation sites (PNGS) most important for 2G12 bindingCitation27 (). Differences in sensitivity to b12 are not completely explained by mutations in its contact residues.Citation28 Rather, sensitivity to this mAb correlates with numerous context dependent residues outside the presumed epitope.Citation29 Similarly, sCD4 sensitivity cannot be predicted by sequence analysis.
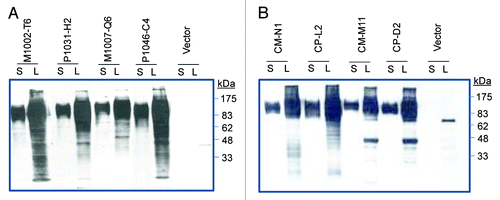
Production of DNA and protein vaccines expressing selected clade B and C MTCT gp120 immunogens. In the current study, the gp120 form of Env was used to test the immunogenicity of MTCT antigens. Our previous studies have identified gp120 as the most immunogenic form of Env antigens.Citation30 Genes coding for gp120 subunits were PCR amplified from the full length Env clones of both clade B and clade C MTCT pairs, and then were sub-cloned into the DNA vaccine vector pJW4303 to produce MTCT gp120 DNA vaccines. The abilities of these DNA vaccine constructs to properly express gp120 proteins were determined by in vitro transfection of 293T cells (). All four pairs of MTCT gp120 DNA vaccines showed good expression of gp120 proteins as shown by western blot analysis using samples expressed in 293T cells. There was a high level of gp120 expression in the secreted form in supernatant of 293T cell cultures. To produce proteins used for the boost immunization, all eight MTCT gp120 proteins secreted from 293T cells were purified using a Lectin column and verified by SDS-PAGE and western blot analysis (data not shown).
Figure 3. Western blot analysis of gp120 protein expressions in supernatant (S) and lysate (L) of 293T cells transfected by either a DNA vaccine expressing MTCT gp120 or empty vector. (A) gp120 DNA vaccines expressing HIV-1 clade B MTCT Env from paired M1002-T6/P1031-H2 and M1007-Q6/P1046-C4. (B) gp120 DNA vaccines expressing HIV-1 clade B MTCT Env from paired CM-N1/CP-L2 and CM-M11/CP-D2. The letters “M” and “P” indicate the Env from mother and her descendent infant in each pair. The rabbit serum specific for HIV-1 gp120 was used as the detecting antibody at 1:500 dilution.
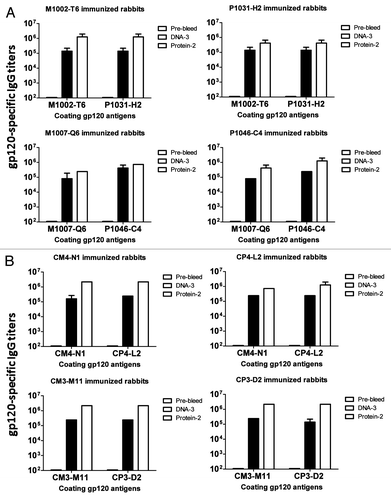
Levels of gp120-specific antibody responses induced by clade B and clade C MTCT gp120 DNA prime-protein boost in rabbits. To investigate the immunogenicity of the selected gp120 clones in this pilot study, eight groups (2 rabbits/group) of NZW rabbits were immunized using a DNA prime-protein boost regimen including three biweekly DNA immunizations followed by two monthly protein boosts with matched MTCT gp120 immunogens between DNA prime and boost vaccinations. The DNA immunizations were performed via gene gun and the protein immunizations were formulated with Incomplete Freund’s Adjuvant (IFA). Each group of rabbits received one particular MTCT gp120 immunogen. The immunogenicity of these MTCT gp120 immunogens was first determined by ELISA as measured by binding titers of serum IgG for each individual rabbit against gp120 antigens (). Overall, these MTCT gp120 immunogens were highly immunogenic as shown by the high titer gp120-specific antibody responses. By the end of the three gene gun-delivered immunizations, titers were around 1:105. Following protein boost immunizations, the final titers increased by another half log to one log for almost all groups. ELISA analyses were done by coating the plates with either the autologous gp120 antigens or a gp120 antigen from the same pair of MTCT viral isolates. There is limited difference in IgG titers when either the autologous or the other gp120 from the same pair of MTCT gp120 immunogens was used as the coating antigen.
Figure 4. HIV-1 gp120-specific antibody responses in rabbit sera induced by MTCT gp120 DNA vaccine (DNA-3), prime-protein boost (Protein-2), or pre-immune rabbit sera (Pre-bleed). (A) gp120-specific antibody titers induced by clade B MTCT gp120 vaccines. The upper panel shows paired gp120 vaccines M1002-T6/P1031-H2 immunized rabbit sera against their autologous gp120 proteins. The lower panel shows the paired gp120 vaccines M1007-Q6/P1046-C4 immunized rabbit sera against their autologous gp120 proteins. (B) gp120-specific antibody titers induced by clade C MTCT gp120 vaccines. The upper panel shows the paired gp120 vaccines CM4-N1/CP4-L2 immunized rabbit sera against their autologous gp120 proteins. The lower panel shows the paired gp120 vaccines CM3-M11/CP3-D2 immunized rabbit sera against their autologous gp120 proteins. The ELISA coating antigens are indicated below each graph. Each column represents the mean antibody titers with standard deviations of each group of 2 rabbits.
NAb responses induced by clade B MTCT gp120 vaccines. To evaluate NAb responses elicited by the clade B gp120 vaccines, we first examined the neutralizing activity against HIV-1 clade B Tier 1 pseudoviruses expressing Env antigens from isolates, SF162 and NL4-3 (). While both viruses are considered relatively sensitive to neutralization; NL4-3 is generally more resistant to neutralization than SF162. All rabbit sera elicited by clade B MTCT gp120 immunogens demonstrated positive NAb activities against SF162 although the NAb titers (mean titer 1:3591) in the rabbits receiving the clade B MTCT pair (M1007-Q6/P1046-C4) that were sensitive to neutralization () were higher than that in rabbits receiving the relatively resistant to neutralization clade B MTCT pair (M1002-T6/P1031-H2) (mean titer 1:378) (p < 0.05).
Table 2. Neutralizing antibody titers induced by subtype B MTCT gp 120 vaccines
However, when neutralization against NL4-3 was measured, all four rabbits that received gp120 immunogens from the resistant to neutralization clade B MTCT pair had positive NAb against NL4-3 with NAb titers ranging between 1:21 to 1:63 but none of the rabbits receiving gp120 immunogens from the sensitive to neutralization clade B MTCT pair neutralized NL4-3.
NAb responses were then evaluated against the autologous clade B MTCT viruses. Overall, these viruses are hard to neutralize with rabbit immune sera and only 8 positive NAb results were obtained following a total of 32 assays (25%). However, rabbit sera generated from gp120 immunogens from the resistant to neutralization MTCT pair had a higher frequency of NAb responses (7 positive out of 16 assays, 44%) against this panel of viruses (NAb titers ranging 1:11 to 1:55). Rabbit sera elicited by gp120 immunogens from the sensitive to neutralization MTCT pair had a much lower frequency of NAb responses (1 positive out of 16 assays (6%) with NAb titer 1:11) (). The difference in the positive NAb frequency between those from the resistant vs. sensitive to neutralization clade B pairs was significant (p < 0.05).
The third panel of neutralization assays was performed against five heterologous clade B primary isolates SS1196, 6535.3, QH0692.42, B33, and LN40 as used in previous studies.Citation31,Citation32 The NAb responses induced by gp120 immunogens from the resistant to neutralization clade B MTCT pair achieved a higher frequency (12 positive out of 20 assays, 60%) and higher titers (ranging 1:15–1:257) than rabbit sera elicited by gp120 immunogens from sensitive to neutralization clade B MTCT pairs, which showed neutralization frequency of 9/20 (45%) and NAb titers ranging 1:10 to 1:74.
NAb responses induced by clade C MTCT gp120 vaccines. The NAb responses in rabbit immune sera induced by the clade C MTCT gp120 immunogens were evaluated by three panels of pseudotyped viruses. Neutralizing antibody responses were first measured against the clade B Tier 1 pseudoviruses SF162 and NL4–3. All rabbit sera, regardless of which clade C gp120 immunogens were received, neutralized SF162 with relatively high NAb titers (except one rabbit R786) (). However, none of the rabbits receiving the clade C MTCT gp120 immunogens could neutralize NL4-3, suggesting NL4-3 is a more typical clade B virus, resistant to rabbit sera elicited by clade C gp120 immunogens.
Table 3. Neutralizing antibody titers induced by subtype C MTCT gp120 vaccines
NAb responses were then evaluated against the autologous clade C MTCT viruses. Interestingly, only one gp120 immunogen from one virus (CM3-M11) collected from these two pairs showed no NAb activity while three other clade C MTCT gp120 immunogens were able to elicit high frequency positive NAb against autologous viruses (23 positive results among 32 assays, 72%). This rate is higher than that observed for the clade B MTCT gp120 immunogens against their autologous Env viruses (25%) (p < 0.01).
The third panel of neutralization assays was performed against 11 heterologous clade C Tier 2 primary isolates.Citation33,Citation34 Similar to the NAb pattern observed against the autologous viruses, three clade C MTCT gp120 immunogens were able to elicit a high frequency of positive NAb (48 out of 66 assays, 73%) while the other clade C MTCT gp120 immunogen (CM3-M11) had only two positive results among 22 assays (9%). The difference was significant (p < 0.001).
Discussion
In the current study, we tested the immunogenicity of Env immunogens from MTCT viral isolates. These viruses and their Env proteins demonstrated a wide range of biological features.Citation16 (Kishko. et al., manuscript in preparation). We focused on the neutralizing sensitivity of these MTCT viral Env proteins to select candidate gp120 immunogens for our study in rabbits due to previous work showing that one Env immunogen, LN40, which is more resistant to neutralization by broadly neutralizing mAb, showed better potential in eliciting broader NAb than another Env immunogen, B33, which is sensitive to mAb neutralization.Citation32 LN40 and B33 have high sequence homology as they were isolated from the same patients. Therefore, we selected one pair of clade B MTCT Env, which is resistant to mAb neutralization and one pair of clade B MTCT Env, which is sensitive to mAb neutralization to understand if the ability of eliciting broadly NAb is related to the sensitivity of Env to mAb. For clade C MTCT pairs, they are overall more resistant to neutralization by mAb. We could only select two pairs of clade C MTCT Env that showed slight differences in their sensitivity to mAb neutralization.
All eight MTCT gp120 immunogens included in our study were able to elicit high levels of binding antibodies against either homologous or heterologous gp120 antigens based on ELISA. However, their abilities to elicit NAb exhibited a very interesting pattern.
For clade B MTCT gp120 immunogens, there is a clear difference between the neutralization sensitive and resistant pairs as reflected in the following three points: (1) against the sensitive virus SF162, the neutralization sensitive pair elicited more potent NAb titers; (2) against the more resistant lab stain and autologous Env, the neutralization resistant pair was more effective in eliciting NAb, and (3) against heterologous clade B isolates, the neutralization resistant pair was more effective in eliciting NAb although the difference was not statistically significant.
For clade C MTCT pairs, since both pairs are relatively resistant to neutralization, no difference was observed between two pairs of clade C gp120 immunogens. However, one gp120 immunogen, CM3-M11, was much less effective in eliciting NAb than the other three gp120 immunogens and CM3-M11 happened to be the most sensitive among the four clade C Env antigens when tested for neutralization by mAb 4E10, HIV-1 positive patient plasma, and sCD4. If judged by individual gp120 immunogens, rather than pairs, it is then clear that gp120 immunogens from all resistant to neutralization isolates were able to elicit broad NAb against both autologous clade C and heterologous clade C viral isolates. Not surprisingly, NAb against SF162 was similar among sensitive and resistant to neutralization clade C isolates as SF162 is a clade B virus that is overly sensitive to neutralization. Clade specificity was further demonstrated as NAb was lacking against this highly clade B-specific NL4-3 isolate despite it being a laboratory adapted virus.
While this is only a pilot study to explore the immunogenicity of MTCT Env proteins, data generated from this study suggested that not all MTCT Env immunogens have the same ability to elicit NAb. These differences are further enhanced when different panels of targeted HIV-1 isolates are tested. However, the finding that the neutralization resistant Env immunogens have a better potential to elicit NAb confirmed the initial observation made with Env antigens collected from adult patientsCitation32 and will have a major impact on the understanding of the structural basis of such differences. More importantly, this finding will guide us in the testing of more Env immunogens to further validate the discovery reported in the current study as part of the effort to develop the next generation HIV-1 vaccine formulation. We need to understand the key epitopes for neutralizing anti-gp120 antibodies that are unique in animal groups that received Env immunogens from more resistant to neutralization viruses.
Materials and Methods
Amplification of MTCT Env genes.
The optimized one-step RT-PCR and nested PCR techniques were adapted from previous studies to amplify env genes from plasma viral RNA with PCR primers.Citation35,Citation36 The detailed steps were recently reported.Citation16 Briefly, env genes from HIV-1 clade B or clade C intrapartum-infected mother-infant pairs (maternal plasma obtained at delivery and infant plasma obtained at the time of HIV-1 diagnosis, within 1–2 mo of birth except for clade B P1046 sample which was drawn on day 66 after birth)Citation37 were amplified in independent limiting dilution PCR reactions. The env genes were cloned into the pcDNA3.1-directional TOPO mammalian expression vector. Each cloned env was tested for functionality and sequenced. Genotypic and phylogenetic analyses of these env clones were performed to show probable transmission of a single variant and infant variants.Citation16 GenBank accession numbers for clade B mother-infant pairs M1002/P1031 and M1007/P1046 are HM368234, HM 368230, HM368249, and HM368245, respectively. The accession numbers for clade C mother-infant pairs CM3/CP3 and CM4/CP4 are JX845593, JX845601, JX845597, and JX845600, respectively.
Vaccines. HIV-1 MTCT gp120 DNA vaccines
Four pairs of MTCT Env antigens (two pairs from clade B and 2 pairs from clade C) were chosen to produce gp120 vaccines used in this study. The gp120 genes were PCR-amplified from the gp160 genes using primers (gp120-P-F1 and gp120-P-B1) as previously describedCitation19 and cloned into DNA vaccine vector pJW4303 at the NheI and BamHI sites, under the tissue plasimogen activator (tPA) leader sequence.Citation38,Citation39 The gp120 DNA vaccine plasmids were prepared in large amounts from E. coli HB101 strain with a Mega purification kit (Qiagen) for both in vitro transfection and in vivo animal immunization studies.
HIV-1 gp120 MTCT protein vaccines
The same MTCT gp120 DNA vaccines described above were also used to produce gp120 proteins in transiently transfected 293T cells.Citation21 Each of the secreted gp120 proteins in the supernatant of transfected 293T cells were harvested and purified by FPLC using Lentil lectin sepharose 4B (GE Healthcare, Waukesha, WI). The amount of purified gp120 proteins was determined before being used for rabbit immunizations and ELISA.
Western blot analysis
The gp120 immunogens produced in transiently transfected 293T cells were confirmed by western blot. Samples were subjected to standard SDS-PAGE and blotted onto PVDF membrane (BioRad). Blocking was done with 0.1% I-Block (Tropix, Bedford, MA). A gp120-specific rabbit serum produced in previous report was used as the detecting antibody at 1:500 dilution for this assay.Citation19 The membranes were washed with blocking buffer and then reacted with AP-conjugated goat anti-rabbit (Tropix) at 1:5000 dilution. After final wash, Western-light substrate was applied to the membranes for 5 min. Once the membranes were dry, Kodak films were exposed to the membrane and developed with an X-Omat processor.
Rabbit immunizations
New Zealand White (NZW) rabbits at 6–8 weeks of age were purchased from Millbrook Breeding Lab (Amherst, MA) and housed in the animal facility at the University of Massachusetts Medical School in accordance of the IACUC approved protocol. Each rabbit received three times of gp120 DNA vaccine prime immunizations by gene gun as previously describedCitation19 at Weeks 0, 2 and 4, and two times gp120 protein boost at Weeks 8 and 12, which was autologous to the DNA vaccine used in the priming immunizations. For gene gun immunization, 36 ∝g of gp120 DNA vaccine coated on 1 ∝m gold beads (1∝g of DNA per 0.5 mg of gold beads) was delivered at shaved abdominal skin. A total of 36 non-overlapping shots were delivered by a helium-driven gene gun (BioRad). Protein immunizations were administered consisting of 50 ∝g recombinant gp120 protein in 500 ∝L PBS and mixed with 500 ∝L Incomplete Freund’s Adjuvant (IFA). The 1 mL adjuvanted protein solution was then injected subcutaneously into the back of rabbits. Sera were collected for antibody studies prior to the first immunization and at two weeks after each animal immunization to evaluate Env-specific binding and NAb responses.
Enzyme linked immunosorbent assay (ELISA)
Con-A (5 ∝g/mL) was pre-coated onto 96 well microtiter plates (Costar #3369). Recombinant gp120 protein was then added at 1 ∝g/mL in 100 ∝L of PBS and incubated for 1 h at room temperature. Plates were then washed 5 times in phosphate buffered saline (PBS) containing 0.1% Triton-X (EWB) and blocked overnight at 4°C in PBS containing 4% whey by weight (whey dilution buffer) and 5% powdered milk. Plates were then washed 5 times in EWB and serially diluted rabbit sera, collected at 2 weeks following the final protein immunization, were added to the wells in a volume of 100 ∝L. Plates were washed 5 times in EWB and 100 ∝L of biotinylanted anti-rabbit secondary antibody (Vector Labs BA-1000) at 1.5 ∝g/mL was incubated on the plate for 1 h at room temperature. Plates were washed 5 times with EWB and incubated with 100 ∝L of a streptavidin horseradish peroxidase construct (Vector Labs SA-5004) at 500ng/mL. Plates were washed 5 times with EWB and developed for 3 min in 100 ∝L of a 3,3'5,5'-tetramethylbenzidine substrate solution (Sigma T3405). The reaction was stopped with addition of 25 ∝L of 2N H2SO4. Endpoint titers as reported are defined as the last dilution of a serially diluted serum sample with greater than double the background optical density of a preimmune serum sample.
HIV-1 neutralization assays
Two types of neutralization assays were conducted: (1) to evaluate the neutralization sensitivity of individual MTCT Env antigens isolated from the mother-infant pairs, and (2) to evaluate the NAb responses induced by MTCT gp120 DNA prime—protein boost immunization regimen.
To evaluate the neutralization sensitivity of MTCT Env antigens
A modified TZMbl infectivity assay was used to allow for a more efficient neutralization analysis. As an alternative to using a luminometer to quantify infectious or neutralization titers, ®-galactosidase activity was used as the readout, and foci forming units (FFU) were directly visualized using an ELISPOT reader. Stained cells were enumerated mechanically on the immunospot reader, and results were expressed as the concentration (monoclonal antibodies) or reciprocal dilution (serum antibodies) at which the FFU were reduced by 50% from the levels found in the uninhibited control.Citation16
To analyze the NAb responses in rabbit immune sera
Neutralization assays were done as previously described.Citation20,Citation40 HIV-1 pseudovirions were produced through co-transfection of the pSG3⊗env backbone (NIH AIDS Research Reference and Reagent Program) and a MTCT gp160 Env bearing plasmid in 293T cells. HIV-1 pseudoviruses were collected from transfected 293T cell supernatants. The infectious titers of pseudoviruses were then determined as the median tissue culture infection dose (TCID50) on the TZM-bl cell line. For a typical neutralization assay, 200 TCID50 of pseudovirus was incubated with rabbit sera for 1 h at 37°C. The virus/sera mix was then added to 105 TZM-bl cells in a final concentration of 20 ∝g/mL DEAE Dextran. Plates were incubated at 37°C for 48 h and developed with luciferase assay reagent according to the manufacturer’s instruction (Promega). Neutralization was calculated as the percent change in luciferase activity in the presence of preimmune sera vs. that of luciferase activity in the presence of immune sera [(Preimmune RLUs - Immune RLUs)/(Preimmune RLUs)]*100.
Statistical analysis
Student’s t-test and Fisher exact test were performed to compare the NAb titers and breadth, respectively, in different MTCT gp120 DNA prime—protein boost vaccinated rabbit sera.
| Abbreviations: | ||
| Env | = | envelope glycoprotein |
| FFU | = | foci forming units |
| IFA | = | Incomplete Freund’s adjuvant |
| mAb | = | monoclonal antibody |
| MTCT | = | mother-to-child transmission |
| NAb | = | neutralizing antibody |
| NZW | = | New Zealand white |
| PBS | = | phosphate buffered saline |
| EWB | = | phosphate buffered saline containing 0.1% Triton-X |
| PV | = | pseudoviruses |
| sCD4 | = | soluble CD4 |
| TCID50 | = | 50% tissue culture infective dose |
| tPA | = | tissue plasimogen activator |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was conducted as part of Elizabeth Glaser Pediatric AIDS Foundation Pediatric HIV Vaccine Program, and funded in part by NIH/NIAID grants P01AI082274, U19AI082676 and R21/R33AI087191, and with the support of Clinical, Molecular Virology and Molecular Biology Core facilities of UMMS CFAR grant (P30 AI42845). The authors thank the SAINT investigators for providing the clinical samples and Boehringer Ingelheim for SAINT trial support, and Dr Jill Serrano for her critical reading and editing of the manuscript.
References
- UNAIDS Report on the Global AIDS Epidemic. 2010. Joint United Nations Programme on HIV/AIDS (UNAIDS). http://wwwunaidsorg/globalreport/defaulthtm.
- Quinn TC, Overbaugh J. HIV/AIDS in women: an expanding epidemic. Science 2005; 308:1582 - 3; http://dx.doi.org/10.1126/science.1112489; PMID: 15947174
- Luzuriaga K, Sullivan JL. Pediatric HIV-1 infection: advances and remaining challenges. AIDS Rev 2002; 4:21 - 6; PMID: 11998780
- Ahmad N, Baroudy BM, Baker RC, Chappey C. Genetic analysis of human immunodeficiency virus type 1 envelope V3 region isolates from mothers and infants after perinatal transmission. J Virol 1995; 69:1001 - 12; PMID: 7815476
- Newell ML, Coovadia H, Cortina-Borja M, Rollins N, Gaillard P, Dabis F, Ghent International AIDS Society (IAS) Working Group on HIV Infection in Women and Children. Mortality of infected and uninfected infants born to HIV-infected mothers in Africa: a pooled analysis. Lancet 2004; 364:1236 - 43; http://dx.doi.org/10.1016/S0140-6736(04)17140-7; PMID: 15464184
- Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, et al, CHAVI Clinical Core B. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med 2009; 206:1253 - 72; http://dx.doi.org/10.1084/jem.20090365; PMID: 19487423
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 2008; 105:7552 - 7; http://dx.doi.org/10.1073/pnas.0802203105; PMID: 18490657
- Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol 2008; 82:3952 - 70; http://dx.doi.org/10.1128/JVI.02660-07; PMID: 18256145
- Zhang LQ, MacKenzie P, Cleland A, Holmes EC, Brown AJ, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol 1993; 67:3345 - 56; PMID: 8497055
- Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 1993; 261:1179 - 81; http://dx.doi.org/10.1126/science.8356453; PMID: 8356453
- Wolinsky SM, Wike CM, Korber BT, Hutto C, Parks WP, Rosenblum LL, et al. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 1992; 255:1134 - 7; http://dx.doi.org/10.1126/science.1546316; PMID: 1546316
- Dickover RE, Garratty EM, Plaeger S, Bryson YJ. Perinatal transmission of major, minor, and multiple maternal human immunodeficiency virus type 1 variants in utero and intrapartum. J Virol 2001; 75:2194 - 203; http://dx.doi.org/10.1128/JVI.75.5.2194-2203.2001; PMID: 11160723
- Zhang H, Hoffmann F, He J, He X, Kankasa C, West JT, et al. Characterization of HIV-1 subtype C envelope glycoproteins from perinatally infected children with different courses of disease. Retrovirology 2006; 3:73; http://dx.doi.org/10.1186/1742-4690-3-73; PMID: 17054795
- Wu X, Parast AB, Richardson BA, Nduati R, John-Stewart G, Mbori-Ngacha D, et al. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J Virol 2006; 80:835 - 44; http://dx.doi.org/10.1128/JVI.80.2.835-844.2006; PMID: 16378985
- Doms RW. Unwelcome guests with master keys: how HIV enters cells and how it can be stopped. Topics in HIV medicine: a publication of the International AIDS Society, USA 2004; 12:100-3.
- Kishko M, Somasundaran M, Brewster F, Sullivan JL, Clapham PR, Luzuriaga K. Genotypic and functional properties of early infant HIV-1 envelopes. Retrovirology 2011; 8:67; http://dx.doi.org/10.1186/1742-4690-8-67; PMID: 21843318
- Wang S, Arthos J, Lawrence JM, Van Ryk D, Mboudjeka I, Shen S, et al. Enhanced immunogenicity of gp120 protein when combined with recombinant DNA priming to generate antibodies that neutralize the JR-FL primary isolate of human immunodeficiency virus type 1. J Virol 2005; 79:7933 - 7; http://dx.doi.org/10.1128/JVI.79.12.7933-7937.2005; PMID: 15919951
- Wang S, Kennedy JS, West K, Montefiori DC, Coley S, Lawrence J, et al. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 2008; 26:3947 - 57; http://dx.doi.org/10.1016/j.vaccine.2007.12.060; PMID: 18724414
- Wang S, Pal R, Mascola JR, Chou TH, Mboudjeka I, Shen S, et al. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology 2006; 350:34 - 47; http://dx.doi.org/10.1016/j.virol.2006.02.032; PMID: 16616287
- Vaine M, Wang S, Crooks ET, Jiang P, Montefiori DC, Binley J, et al. Improved induction of antibodies against key neutralizing epitopes by human immunodeficiency virus type 1 gp120 DNA prime-protein boost vaccination compared to gp120 protein-only vaccination. J Virol 2008; 82:7369 - 78; http://dx.doi.org/10.1128/JVI.00562-08; PMID: 18495775
- Vaine M, Wang S, Hackett A, Arthos J, Lu S. Antibody responses elicited through homologous or heterologous prime-boost DNA and protein vaccinations differ in functional activity and avidity. Vaccine 2010; 28:2999 - 3007; http://dx.doi.org/10.1016/j.vaccine.2010.02.006; PMID: 20170767
- Montefiori DC. (2004) Evaluating neutralizing antibodies against HIV, SIV and SHIV in luciferase reporter gene assays. Current Protocols in Immunology, (Coligan, J.E., A.M. Kruisbeek, D.H. Margulies, E.M. Shevach, W. Strober, and R. Coico, eds.), John Wiley & Sons, 12.11.1-12.11.15.
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol 2004; 78:13232 - 52; http://dx.doi.org/10.1128/JVI.78.23.13232-13252.2004; PMID: 15542675
- Brunel FM, Zwick MB, Cardoso RM, Nelson JD, Wilson IA, Burton DR, et al. Structure-function analysis of the epitope for 4E10, a broadly neutralizing human immunodeficiency virus type 1 antibody. J Virol 2006; 80:1680 - 7; http://dx.doi.org/10.1128/JVI.80.4.1680-1687.2006; PMID: 16439525
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol 1993; 67:6642 - 7; PMID: 7692082
- Gray ES, Meyers T, Gray G, Montefiori DC, Morris L. Insensitivity of paediatric HIV-1 subtype C viruses to broadly neutralising monoclonal antibodies raised against subtype B. PLoS Med 2006; 3:e255; http://dx.doi.org/10.1371/journal.pmed.0030255; PMID: 16834457
- Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, et al. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol 2002; 76:7293 - 305; http://dx.doi.org/10.1128/JVI.76.14.7293-7305.2002; PMID: 12072528
- Bublil EM, Yeger-Azuz S, Gershoni JM. Computational prediction of the cross-reactive neutralizing epitope corresponding to the [corrected] monclonal [corrected] antibody b12 specific for HIV-1 gp120. FASEB J 2006; 20:1762 - 74; http://dx.doi.org/10.1096/fj.05-5509rev; PMID: 16940148
- Peters PJ, Duenas-Decamp MJ, Sullivan WM, Brown R, Ankghuambom C, Luzuriaga K, et al. Variation in HIV-1 R5 macrophage-tropism correlates with sensitivity to reagents that block envelope: CD4 interactions but not with sensitivity to other entry inhibitors. Retrovirology 2008; 5:5; http://dx.doi.org/10.1186/1742-4690-5-5; PMID: 18205925
- Lu S, Wyatt R, Richmond JF, Mustafa F, Wang S, Weng J, et al. Immunogenicity of DNA vaccines expressing human immunodeficiency virus type 1 envelope glycoprotein with and without deletions in the V1/2 and V3 regions. AIDS Res Hum Retroviruses 1998; 14:151 - 5; http://dx.doi.org/10.1089/aid.1998.14.151; PMID: 9462925
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 2005; 79:10108 - 25; http://dx.doi.org/10.1128/JVI.79.16.10108-10125.2005; PMID: 16051804
- Vaine M, Duenas-Decamp M, Peters P, Liu Q, Arthos J, Wang S, et al. Two closely related Env antigens from the same patient elicited different spectra of neutralizing antibodies against heterologous HIV-1 isolates. J Virol 2011; 85:4927 - 36; http://dx.doi.org/10.1128/JVI.00081-11; PMID: 21411542
- Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, et al. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol 2006; 80:11776 - 90; http://dx.doi.org/10.1128/JVI.01730-06; PMID: 16971434
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 2010; 84:1439 - 52; http://dx.doi.org/10.1128/JVI.02108-09; PMID: 19939925
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, et al. Antibody neutralization and escape by HIV-1. Nature 2003; 422:307 - 12; http://dx.doi.org/10.1038/nature01470; PMID: 12646921
- Gao F, Morrison SG, Robertson DL, Thornton CL, Craig S, Karlsson G, et al. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID Networks for HIV Isolation and Characterization. J Virol 1996; 70:1651 - 67; PMID: 8627686
- Moodley D, Moodley J, Coovadia H, Gray G, McIntyre J, Hofmyer J, et al, South African Intrapartum Nevirapine Trial (SAINT) Investigators. A multicenter randomized controlled trial of nevirapine versus a combination of zidovudine and lamivudine to reduce intrapartum and early postpartum mother-to-child transmission of human immunodeficiency virus type 1. J Infect Dis 2003; 187:725 - 35; http://dx.doi.org/10.1086/367898; PMID: 12599045
- Lu S, Manning S, Arthos J. Antigen engineering in DNA immunization. Methods Mol Med 2000; 29:355 - 74; PMID: 21374335
- Wang S, Farfan-Arribas DJ, Shen S, Chou TH, Hirsch A, He F, et al. Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine 2006; 24:4531 - 40; http://dx.doi.org/10.1016/j.vaccine.2005.08.023; PMID: 16140431
- Montefiori DC, Metch B, McElrath MJ, Self S, Weinhold KJ, Corey L, HIV Vaccine Trials Network. Demographic factors that influence the neutralizing antibody response in recipients of recombinant HIV-1 gp120 vaccines. J Infect Dis 2004; 190:1962 - 9; http://dx.doi.org/10.1086/425518; PMID: 15529261