Abstract
Numerous molecular effects have been attributed to histone deacetylase inhibitors (HDACI’s), including the induction of major histocompatibility (MHC) genes. Here we report that one FDA approved HDACI, Vorinostat, and a second HDACI currently in clinical trials, Entinostat, reduce the ratio of class II associated invariant peptide (CLIP) to the MHC class II molecule, HLA-DR, indicating an increase in the non-CLIP peptides bound to HLA-DR. The HDACI effects are apparent with immortalized B-cells, HLA-DR constitutive melanoma cells and with melanoma cells expressing HLA-DR due to transformation with an expression vector for the HLA-DR gene co-activator, CIITA. Entinostat treatment leads to upregulation of Cathepsin L1, and the HLA-DR peptidome of the Entinostat treated cells is consistent with increased Cathepsin L1 mediated proteolysis. These results indicate that HDACI treatments may alter the HLA-DR peptidome of cells in patients and provide a way to identify novel immunogens for vaccinations and the study of autoantigens.
Introduction
Histone deacetylases remove acetyl groups from histones that constitute the nucleosome, thereby increasing the positive charges on histones and the histone-DNA electrostatic contacts, particularly in the cellular environment where DNA is highly, negatively charged. Removal of histone acetyl groups also removes binding sites for proteins specifically interacting with the acetyl groups. Both functions are important for gene silencing, thus HDACI’s lead to the upregulation of many genes, with greater or lesser specificity depending on the HDACI.Citation1 HDACI’s can also lead to downregulation of transcription, in some cases, most likely due to the induction of negative regulatory molecules.
The FDA approved HDACI, Valproic acid, is used to treat epilepsy,Citation2 and the FDA approved HDACI’s, Vorinostat and Romidepsin, are used to treat cutaneous T-cell lymphoma.Citation3 Also, there are numerous other HDACI’s, such as Entinostat, which have passed all pre-clinical tests and are currently being used in clinical trials. In most cases, the basis for developing the HDACI as a drug is the highly common effect of HDACI mediated apoptosis. However, HDACI’s can potentially have numerous effects owing to their relatively nonspecific effect on gene expression. In particular, HDACI’s have been shown to upregulate MHC molecules,Citation4-Citation6 raising the question of whether these drugs could have an impact on the immune response. We considered the possibility that, in addition to regulation of MHC levels, that HDACI’s could have an impact on MHC function, namely the binding of antigenic peptide, which leads to activation of T-cell receptors and myriad immune response effects, including vaccination.
Thus, we assayed for the ratio of the binding of CLIP (class II associated invariant peptide) to surface HLA-DR, the canonical human MHC class II protein, in the presence or absence of HDACI’s. Stable HLA-DR heterodimers can only be transported to the cell surface via binding of the default, CLIP or by binding canonical peptide, analogous to antigenic peptide.Citation7 In most cases, the non-CLIP peptide is self peptide, which does not lead to T-cell receptor activation.
Results
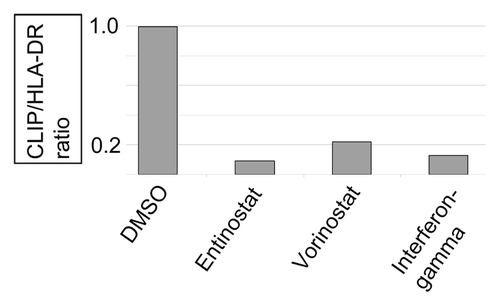
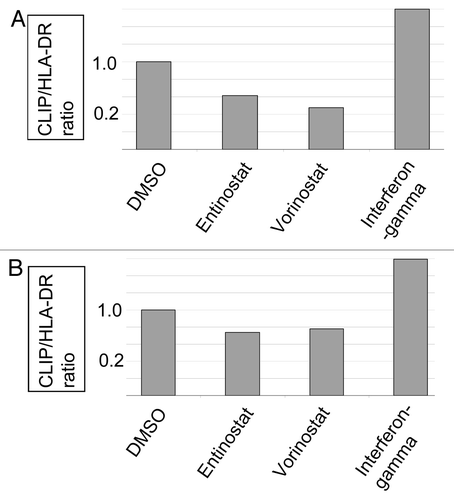
Treatment of immortalized Raji B-cells, representing a Burkitt’s lymphoma, with either 3 μM Vorinostat or 5 μM Entinostat led to a reduction in the CLIP/HLA-DR ratio (), indicating that the HLA-DR of the treated cells had an altered peptidome. HDACI treatment of WM9 melanoma cells also led to a reduced CLIP/HLA-DR ratio (). Interestingly, IFN-γ treatment (400 units/ml) of the B-cells or the WM9 cells did not lead to decreased CLIP/HLA-DR ratios ( and B), despite the fact that IFN-γ has been shown to upregulate gamma interferon inducible lysosomal thiol reductase (GILT), important for HLA-DR peptide availability. This finding raises the question of whether these cells have defects in IFN-γ signaling, a common defect in tumor cells,Citation8-Citation10 that could reduce the availability of tumor-related peptide and thereby reduce the immunogenicity of the tumor from which these cells were derived. Melanoma cells in particular have been noted to have low GILT expression levels.Citation11
Figure 1. CLIP/HLA-DR ratios following HDACI treatments. Raji B-cells (A) and WM9 melanoma cells (B) were cultured in RPMI, antibiotic supplements and 10% fetal calf serum; and treated with either DMSO, 5 μM Entinostat, 3 μM Vorinostat, or 400 units/ml IFN-γ. After 40 h of treatment, cells were recovered and stained with antibodies for cell surface, flow cytometry as detailed in Materials and Methods.
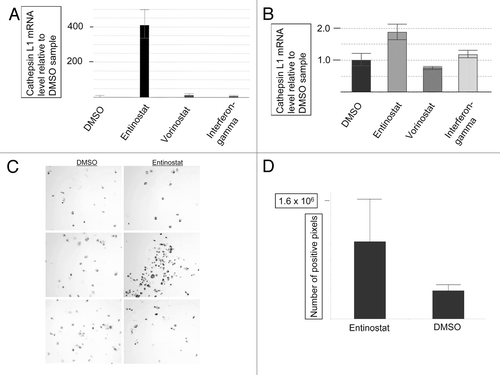
Peptide loading of HLA-DR molecules has been attributed to Cathepsin L1 and S, among other proteases.Citation12,Citation13 Thus, we considered the possibility that HDACI treatment led to an upregulation of Cathepsin L1 or S gene expression. No effect was observed with Vorinostat for either Cathepsin, but Entinostat led to an upregulation of Cathepsin L1 mRNA in both cells types ( and B). IFN-γ led to an upregulation of only Cathepsin S (data not shown; Cathepsin S data not shown for any samples). Taken together, these results suggest that Cathepsin S is not involved in reducing the CLIP/HLA-DR ratios in the above experiments. Entinostat also led to an upregulation of Cathepsin L1 protein in the Raji B-cells ( and D).
Figure 2. HDACI induced expression of Cathepsin L1. RNA was extracted from DMSO, Entinostat, Vorinostat, and IFN-γ treated Raji B-cells (A) and WM9 cells (B) and subject to Real-Time PCR for Cathepsin L1 (ctsl1) mRNA as detailed in “Materials and Methods.” Treatments as indicated in the figure. (C) Immunohistochemical staining: Raji B-cells were treated with DMSO or Entinostat and stained with anti- Cathepsin L1 (ctsl1) as detailed in “Materials and Methods.” (D) Image Analysis: The number of pixels over this threshold per cell was used to determine the differences between anti-Cathepsin L1 staining in the Entinostat and DMSO groups. Each image and analysis was blinded and completed in triplicate, as detailed in “Materials and Methods”. p-value < 0.0004
To verify the effect of Entinostat on the HLA-DR peptidome, and to determine whether the Entinostat-induced, Cathepsin L1 could be involved in altering HLA-DR peptide occupancy, HLA-DR from Entinostat treated Raji B-cells was recovered by immunoprecipitation. HLA-DR bound peptides were eluted and identified by tandem mass spectrometry as previously described.Citation14 Results indicated there were 11 peptides representing 7 proteins, unique to the Entinostat treated cells; and 17 peptides representing 13 proteins, unique to DMSO (control) treated cells (). The Entinostat dependent, HLA-DR bound peptides did not show any significant indication of having an increased dependence on either Cathepsin L1, S, or B cleavages, when compared with peptides from DMSO treated cells.Citation15 However, the peptides from the DMSO treated cells had significantly higher quality Cathepsin L1 sites within the peptides (). Thus, the Entinostat-induced, Cathepsin L1 likely reduced the availability of these peptides, thus explaining the difference in the two peptidomes. It is also possible that Cathepsin L1 has other effects. For example, Cathepsin L1-enhanced CLIP preparation could have altered the frequency of HLA-DR access for certain peptides. In addition, many other HDACI mediated cell processes, not studied here, could have contributed to the alteration of the HLA-DR peptidome, for example, an increase in the amount of the proteins representing the Entinostat unique peptides; or increases or decreases in other proteins regulating the peptidome, besides proteases.
Table 1. Peptides Unique to Entinostat and DMSO (control) treated cells
Table 2. Putative Cathepsin L sites within HLA-DR binding peptides
To determine whether HDACI treatment might also regulate HLA-DR peptide occupancy for cells that do not constitutively express HLA-DR, we examined the effect of Entinostat and Vorinostat on the CLIP/HLA-DR ratio in 1286 melanoma cells transformed with a CIITA expression vectorCitation16 and thereby expressing surface HLA-DR (). Results indicate that the HDACI’s also decreased the CLIP/HLA-DR ratio in these cells. In addition, IFN-γ treatment reduced the CLIP/HLA-DR ratio, in contrast to the result obtained with IFN-γ treatment of the two tumor cell types above, that constitutively express MHC class II. This result is consistent with the idea that peptide loading of tumor cell MHC class II is inconsistent with tumor development. In the absence of MHC class II expression, as was the case for the 1286 melanoma cells prior to the experimental transformation with the CIITA expression vector, there is no selection for peptide processing related, IFN-γ insensitivity, as noted above, an otherwise common tumor cell phenotype. These results also indicate that CIITA transformation can be coupled with small molecule treatment of cells to produce varied HLA-DR peptidomes.
Discussion
The above results demonstrate that HDACI’s can regulate HLA-DR peptide occupancy, with apparent distinct mechanisms for Entinostat and Vorinostat. These two HDACI’s have distinct effects on HDACs, with Entinostat having greater selectivity for the class I HDACs. This different may reflect the differences in the effect of the Entinostat and Vorinostat on Cathepsin L1 induction. Furthermore, other proteins, besides proteases may be regulated differently by the two HDACI’s. While the results of strongly indicate that the induction of Cathepsin L1 plays a role in the Entinostat mediated differences in the HLA-DR peptidome, other bases of differences could include the regulation of HLA-DR itself or the regulation of the amount or availability of proteins that are the source of peptides for HLA-DR binding. As indicated above, there are several, previous reports of HDACI-mediated upregulation of HLA-DR and MHC class II expression,Citation4-Citation6 although this is the first report on the effect of HDACI’s on the MHCII peptidome.
The above results now provide an approach to regulation of the HLA-DR peptidome that is likely applicable to the goals of: (1) identifying new HLA-DR binding epitopes for personalized peptide vaccinesCitation17,Citation18; (2) identifying new HLA-DR binding epitopes in polypeptides that are candidates for vaccines, in HLA-DR-positive cell transformants expressing such polypeptides; (3) vaccine adjuvant discovery; (4) regulating the immune response in chemotherapy; (5) identification of autoantigens, which aid in the understanding of the basis of autoimmune diseases and guide the design of peptidomimetic drugs to block T-cell receptor engagement of autoantigen;Citation19 or (6) increasing the number of known HLA-DR binding peptides, for developing higher quality algorithms for predicting HLA-DR binding peptides. Finally, it is likely that other small molecules, particularly designed to regulate gene expression, will have effects similar to Entinostat and Vorinostat but by regulating distinct components of the MHC class II antigen processing machinery and thereby leading to alternative, regulated MHC class II peptidomes.
Materials and Methods
CLIP/HLA-DR ratios
Fluorescent antibody, cell surface stained samples were analyzed on BD LSR II flow cytometer equipped with 405 nm, 488 nm and 633 nm lasers using BD FacsDiva 6.1.3 software. FITC, PE and DAPI dyes were detected using 530/30, 575/26 and 450/50 bandpass filters, respectively. Ratios are expressed as the ratio of mean fluorescence index (MFI) differentials, with DMSO treatment normalized to one. The MFI values of PE-labeled isotype control were subtracted from PE-labeled anti-HLA-DR. The MFI values of FITC-labeled isotype control were subtracted from FITC-labeled anti-CLIP. (All antibodies were purchased from BD Pharmigen). The MFI differentials for CLIP and HLA-DR were converted to ratios, i.e., the CLIP MFI differential was divided by the HLA-DR MFI differential, as detailed in the supporting online material.
Real-Time PCR assay
RNA was extracted from DMSO, Entinostat, Vorinostat, and IFN-γ treated Raji B-cells and WM9 cells using an RNeasy Mini Kit < Qiagen cat #74106. RNA quality was verified using a bioanalyzer (Agilent). Reverse transcription of total RNA (1 µg) to cDNA was done with a High Capacity RNA-to-cDNA Master Mix (Applied Biosystems part of Life Technologies, Foster City, CA. catalog number 4390777). In a 0.2 ml PCR tube, 5X Complete Master Mix (4.0 µl) and nuclease-free H20 were mixed with 1 µg RNA for a total reaction volume of 20 µL. The sample was incubated at 25°C for 5 min, 42°C for 30 min and then 85°C for 5 min in a DNA Engine Peltier thermal cycler (Bio-Rad). qPCR target related: Catepsin L1, CTSL1, RefSeq- NM_001912.4, amplicon length-85 bp, Assay ID Details-Hs00377632_m1; Cathepsin S, CTSS, RefSeq- NM_004079.4, amplicon length-70 bp, Assay ID Details Hs00175407_m1; GAPDH-glyceraldehyde-3-phosphate dehydrogenase, Gapdh, RefSeq- NM_00246.3, amplicon length-58 bp, Assay ID Details- Hs03929097_g1. All primers and probes are from Applied Biosystems Assay- on- Demand. Probe is FAM-NFQ. cDNA was diluted in nuclease free water (Ambion, part of Life Technologies, catalog number AM9937) as follows: Template for target CTSL1 at 2-fold, template for target CTSS1 at 10-fold; and template for target GAPDH at 10-fold. Amplification for each of the above mRNA targets was done in triplicate for each sample and a no-template control was completed for each assay mix. The GAPDH was used as an endogenous control for relative qPCR quantification. Amplification of cDNA that was diluted with water was done with 2X Gene Expression Master Mix (Applied Biosystems part of Life Technologies, catalog number 4369016). In each well of an Optical 96-well Fast Thermal Cycling Plate (Applied Biosystems catalog number 4346906) is a 10 µl reaction containing 2X Gene Expression Master Mix (5 µL), 20X TaqMan Assay-on-Demand primer and probe mix (0.5 µL), diluted cDNA (2.0 μl) and nuclease-free H20 (2.5 μl). The plate is sealed with MicroAmp Optical Adhesive Film (Applied Biosystems catalog # 4313663) The sample was incubated at 50°C for 20 min, 95° 10 min, than 40 cycles of 95°C for 15 sec and 60°C for 1 min in a 7500FAST Real-time PCR thermocycler (Applied Biosystems). The qPCR analysis program is 7500 Software v2.0.5 from Applied Biosystems. ΔΔCq determination used; all no-template controls were negative.
Immunohistochemistry quantification
Raji B-cells were treated with DMSO or Entinostat and embedded in paraffin blocks after 40 h. Slides were stained using a Ventana Discovery XT automated system (Ventana Medical Systems, Tucson) as per manufacturer's protocol. Briefly, slides were deparaffinized with EZ Prep solution followed by antigen retrieval. The rabbit polyclonal antibody against ctsl1 (#orb37018, Biorbyt, Cambridge) was used at a 1:100 concentration in Dako antibody diluent and incubated for 2hr. The Ventana UltraMap anti-rabbit alkaline phosphatase secondary Antibody was used for 12 min. Ventana ChromoMap Red kit was used for detection of stain and slides were then counterstained with hematoxylin. For image acquisition, samples were viewed with a Leica DMLB upright brightfield microscope and a 20X/0.5NA HC PL fluortar objective (Leica Microsystems GmbH, Wetzlar, Germany). Transmitted bright light was used without contrasting methods. Gain, offset, and other acquisition settings were identical for all samples within each group. Image Analysis: (1) The number of nuclei was determined with auto-threshold segmentation on the cell morphology. The nuclei segmentation was further refined by size and shape of the objects created from the initial segmentation. (2) ImagePro Plus v6.1 (Media Cybernetics) software was used to determine the ctsl1 staining intensity. Positive cells were established as > 20DR over background for each 8bit image (0–255DR). The number of pixels over this threshold per cell was used to determine the differences between the Entinostat and DMSO groups. Each image and analysis was blinded and completed in triplicate.
Additional material
Download Zip (869.8 KB)Acknowledgments
Authors would like to thank Sharon Norton, Herman Hernandez, and Noel Clark for technical assistance; and thank the personnel of the core facilities at the University of Florida (ICBR) and the Moffitt Cancer Center (Tissue procurement, Research histology services). Work was support by funds from the Department of Molecular Medicine at the Morsani College of Medicine and ARUP Institute for Clinical and Experimental Pathology at the University of Utah.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be downloaded here: www.landesbioscience.com/journals/vaccines/article/23085
Reference
- Lemoine M, Younes A. Histone deacetylase inhibitors in the treatment of lymphoma. Discov Med 2010; 10:462 - 70; PMID: 21122478
- Gerstner T, Bell N, König S. Oral valproic acid for epilepsy--long-term experience in therapy and side effects. Expert Opin Pharmacother 2008; 9:285 - 92; http://dx.doi.org/10.1517/14656566.9.2.285; PMID: 18201150
- Akilov OE, Geskin L. Therapeutic advances in cutaneous T-cell lymphoma. Skin Therapy Lett 2011; 16:1 - 5; PMID: 21591544
- Magner WJ, Kazim AL, Stewart C, Romano MA, Catalano G, Grande C, et al. Activation of MHC class I, II, and CD40 gene expression by histone deacetylase inhibitors. J Immunol 2000; 165:7017 - 24; PMID: 11120829
- Osborne A, Zhang H, Yang WM, Seto E, Blanck G. Histone deacetylase activity represses gamma interferon-inducible HLA-DR gene expression following the establishment of a DNase I-hypersensitive chromatin conformation. Mol Cell Biol 2001; 21:6495 - 506; http://dx.doi.org/10.1128/MCB.21.19.6495-6506.2001; PMID: 11533238
- Woan KV, Sahakian E, Sotomayor EM, Seto E, Villagra A. Modulation of antigen-presenting cells by HDAC inhibitors: implications in autoimmunity and cancer. Immunol Cell Biol 2012; 90:55 - 65; http://dx.doi.org/10.1038/icb.2011.96; PMID: 22105512
- Riberdy JM, Newcomb JR, Surman MJ, Barbosa JA, Cresswell P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature 1992; 360:474 - 7; http://dx.doi.org/10.1038/360474a0; PMID: 1448172
- Lu Y, Ussery GD, Muncaster MM, Gallie BL, Blanck G. Evidence for retinoblastoma protein (RB) dependent and independent IFN-gamma responses: RB coordinately rescues IFN-gamma induction of MHC class II gene transcription in noninducible breast carcinoma cells. Oncogene 1994; 9:1015 - 9; PMID: 8134104
- Lu Y, Boss JM, Hu SX, Xu HJ, Blanck G. Apoptosis-independent retinoblastoma protein rescue of HLA class II messenger RNA IFN-gamma inducibility in non-small cell lung carcinoma cells. Lack of surface class II expression associated with a specific defect in HLA-DRA induction. J Immunol 1996; 156:2495 - 502; PMID: 8786310
- Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA 1998; 95:7556 - 61; http://dx.doi.org/10.1073/pnas.95.13.7556; PMID: 9636188
- Haque MA, Li P, Jackson SK, Zarour HM, Hawes JW, Phan UT, et al. Absence of gamma-interferon-inducible lysosomal thiol reductase in melanomas disrupts T cell recognition of select immunodominant epitopes. J Exp Med 2002; 195:1267 - 77; http://dx.doi.org/10.1084/jem.20011853; PMID: 12021307
- Hsieh CS, deRoos P, Honey K, Beers C, Rudensky AY. A role for cathepsin L and cathepsin S in peptide generation for MHC class II presentation. J Immunol 2002; 168:2618 - 25; PMID: 11884425
- Watts C. The exogenous pathway for antigen presentation on major histocompatibility complex class II and CD1 molecules. Nat Immunol 2004; 5:685 - 92; http://dx.doi.org/10.1038/ni1088; PMID: 15224094
- Escobar H, Reyes-Vargas E, Jensen PE, Delgado JC, Crockett DK. Utility of characteristic QTOF MS/MS fragmentation for MHC class I peptides. J Proteome Res 2011; 10:2494 - 507; http://dx.doi.org/10.1021/pr101272k; PMID: 21413816
- Biniossek ML, Nägler DK, Becker-Pauly C, Schilling O. Proteomic identification of protease cleavage sites characterizes prime and non-prime specificity of cysteine cathepsins B, L, and S. J Proteome Res 2011; 10:5363 - 73; http://dx.doi.org/10.1021/pr200621z; PMID: 21967108
- Le E, Zhang H, Blanck G. CIITA transformation rescues the apoptotic function of MHC class II in melanoma cells. Anticancer Res 2005; 25:6B 3889 - 92; PMID: 16312044
- Clive KS, Tyler JA, Clifton GT, Holmes JP, Ponniah S, Peoples GE, et al. The GP2 peptide: a HER2/neu-based breast cancer vaccine. J Surg Oncol 2012; 105:452 - 8; http://dx.doi.org/10.1002/jso.21723; PMID: 22441896
- Itoh K, Yamada A. Personalized peptide vaccines: a new therapeutic modality for cancer. Cancer Sci 2006; 97:970 - 6; http://dx.doi.org/10.1111/j.1349-7006.2006.00272.x; PMID: 16984371
- Rosloniec EF, Brandstetter T, Leyer S, Schwaiger FW, Nagy ZA. Second-generation peptidomimetic inhibitors of antigen presentation effectively treat autoimmune diseases in HLA-DR-transgenic mouse models. J Autoimmun 2006; 27:182 - 95; http://dx.doi.org/10.1016/j.jaut.2006.09.005; PMID: 17081730